Abstract
Many insects feed on blood or tissue from mammalian hosts. One potential strategy for the control of these insects is to vaccinate the host with antigens derived from the insect. The larvae of the fly Lucilia cuprina feed on ovine tissue and tissue fluids causing a cutaneous myiasis associated with considerable host morbidity and mortality. A candidate vaccine antigen, peritrophin 95, was purified from the peritrophic membrane, which lines the gut of these larvae. Serum from sheep vaccinated with peritrophin 95 inhibited growth of first-instar L. cuprina larvae that fed on this serum. Growth inhibition was probably caused by antibody-mediated blockage of the normally semipermeable peritrophic membrane and the subsequent development of an impervious layer of undefined composition on the gut lumen side of the peritrophic membrane that restricted access of nutrients to the larvae. The amino acid sequence of peritrophin 95 was determined by cloning the DNA complementary to its mRNA. The deduced amino acid sequence codes for a secreted protein containing a distinct Cys-rich domain of 317 amino acids followed by a mucin-like domain of 139 amino acids. The Cys-rich domain may be involved in binding chitin. This report describes a novel immunological strategy for the potential control of L. cuprina larvae that may have general application to the control of other insect pests.
Keywords: peritrophic membrane, Lucilia cuprina, immunological control
Many insects feed on blood, tissue, or tissue fluids of mammals. The insect feeding process itself is often detrimental to the host and in addition many of these insects are vectors for the transmission of viral, protozoal, and helminth parasites that cause considerable host morbidity and mortality, particularly in humans and livestock animals. Current strategies for control of insect pests in livestock industries are becoming inadequate because of rapid and widespread development of insecticide resistance and concerns relating to the presence of chemical residues derived from these insecticides in livestock products and the immediate environment. Thus, alternative means of controlling these insects are required.
One possible approach is to vaccinate the host against the offending insect life stage by using an insect gut antigen (1, 2). The gut of most insects is lined with a peritrophic membrane (PM or peritrophic matrix) at least at some stage in the life cycle (3). This semipermeable membrane is composed of chitin, proteoglycans, and proteins. The functions of the PM involve facilitating the digestive process and protecting the gut epithelial cells from invasion by microrganisms and parasites (3). The permeability of the PM is such that it either excludes or severely restricts access of ingested host antibodies to the underlying digestive epithelial cells of the gut (3, 4). The PM, however, is bathed in ingested host antibodies and, therefore, PM proteins may be candidate vaccine antigens. Preliminary studies have shown that sheep vaccinated with crude PM protein extracts from the larvae of the fly Lucilia cuprina induce an anti-larval immune response (2, 5). These larvae feed on ovine tissue fluids, dermal tissue, and blood, eventually causing a severe cutaneous myiasis associated with considerable production losses in the sheep industry. In this study, we report the purification of a protein from L. cuprina larval PM that, when injected into sheep, induces an immune response that inhibits the growth of L. cuprina larvae that subsequently feed on serum from these vaccinated animals. The deduced amino acid sequence of this protein and the probable mechanism of action of the anti-larval immune response are also identified.
MATERIALS AND METHODS
Identification and Purification of Peritrophin 95.
The isolation of PM by culture of L. cuprina larvae has been described elsewhere (5). A previous series of experiments demonstrated that a 4 M urea extract of detergent-washed PM solubilized a group of proteins (peritrophins) that, when injected into sheep, induced an anti-larval immune response measured by in vitro feeding and in vivo bioassays (5). The strategy for the isolation of one antigen, peritrophin 95, entailed successive protein fractionations that were each assessed for anti-larval activity in sheep vaccination trials. Peritrophin 95 was isolated and purified from a 6 M urea extract of detergent-washed PM by a two-step chromatographic procedure involving gel permeation chromatography on Superose 12 (Pharmacia) followed by Mono Q anion exchange chromatography (Pharmacia) (6). Both procedures were performed in the presence of 6 M urea. The yield of purified peritrophin 95 was 250 μg/g (dry weight) of PM. Protein concentrations were determined using the Pierce BCA kit with BSA as a standard. The urea contained within buffers was sufficiently diluted to avoid interference with protein determinations. SDS/PAGE and biotinylated lectin blot analysis were carried out as described elsewhere (6).
Vaccination of Sheep with PM Proteins.
Sheep (eight sheep per vaccination group), which had not previously suffered a cutaneous myiasis were initially injected with peritrophin 95 in the muscle of the rear leg and 4 weeks later in the muscle of the neck. The adjuvant for the first injection was Freund’s complete adjuvant; the second injection used Freund’s incomplete adjuvant (Sigma). Each sheep received a total of 63 μg of peritrophin 95. Two weeks after the second injection, the effect of vaccination was assessed by an in vitro larval growth bioassay that consisted of allowing first-instar larvae to feed on an agar-based medium containing serum from the vaccinated animals (7). The number of surviving larvae and their weights were measured after 20 h. There was no significant difference between the mean weights (and mean survival) of larvae feeding on control serum or individual prevaccination serum.
Isolation of Ig from Serum and Feeding of Concentrated Ig to L. cuprina Larvae.
Total Ig was isolated from the serum of one of the strongest responding sheep and also from pooled sera from control sheep that had been injected with adjuvant and PBS (8). An antigen-capture ELISA was performed to determine the relative concentration of total Ig in the original control serum and after isolation (7). The Ig samples were concentrated and added to 4 ml of normal sheep serum (NSS) to give immune Ig concentrations equal to one, two, and four times those in the original immune serum. The total volume was adjusted to 5 ml with PBS and formulated into agar-based diets on which L. cuprina larvae were grown (7).
In Vitro Feeding of L. cuprina Larvae with Colloidal Gold.
Neonate larvae were allowed to feed for 5 h at 34°C on a diet containing NSS with a 4-fold enrichment of total Ig isolated from control serum or for 20 h on a similar diet with a 4-fold enrichment of immune Ig. The shorter feeding duration (5 h) adopted for control larvae ensured that these larvae were at a similar stage of development (first instar) to the larvae fed on immune Ig for 20 h. After being counted and weighed to confirm the enhanced larval growth inhibition caused by the Ig-enriched serum, some of the larvae were then transferred to the same diets but also containing 15% (vol/vol) of an aqueous suspension of colloidal gold (mean diameter, 6 nm) stabilized by conjugation to the B-chain of insulin (9). These larvae were allowed to continue feeding on these preparations for 2 h at 34°C. The larvae were then removed, carefully dissected open, and processed for transmission electron microscopy (9).
Peptide Amino Acid Sequences and Oligonucleotide Synthesis.
The amino-terminal amino acid sequence of peritrophin 95 and internal peptide sequences were determined by published procedures (6). Peptide amino acid sequences were used to design degenerate oligonucleotide primers (Pharmacia LKB Gene Assembler Plus) suitable for PCR (10). The sense primer derived from the amino-terminal sequence and peptide 1 was 5′-C(TCA)AC(TCA)GG(TCA)AC(TCA)AA(AG)TT(TC)CC(TCA)AG-3′ and the antisense primer derived from peptide 3 was 5′-C(AG)AA(AG)TA(AGT)AT(AGT)GA(AG)AA(AGT)AC(AGT)CC-3′. Bracketed nucleotides show alternatives at a specific position.
Cloning of cDNA Coding for Peritrophin 95.
A peritrophin 95-specific double-stranded DNA probe was prepared by using PCR in conjunction with the oligonucleotide primers described above and L. cuprina first-instar larval cDNA. The reaction mixture contained 5 ng of cDNA, 500 ng of each oligonucleotide primer, all four dNTPs (each at 2 mM), 4 mM MgCl2, 2.5 units of Taq DNA polymerase (Perkin–Elmer Cetus) in 100 μl of 10 mM Tris⋅HCl (pH 8.3) and 50 mM KCl. Amplification was performed with the following conditions: 1 cycle of 5 min at 95°C, 1 min at 50°C, and 5 min at 72°C; 33 cycles of 1 min at 95°C, 1 min at 50°C, and 5 min at 72°C; and 1 cycle of 1 min at 95°C, 1 min at 50°C, and 10 min at 72°C. A single DNA product (595 bp) was amplified and sequenced on both strands (11). The DNA product was labeled and used to screen a L. cuprina first-instar cDNA library constructed in λgt-11 (6). Seven positive plaques were purified. A 1,616-bp DNA insert from one clone was excised and sequenced on both strands (11, 12). DNA inserts from three additional positive clones were sequenced and contained fragments of the sequence derived from the first clone.
RESULTS
Purification of Peritrophin 95.
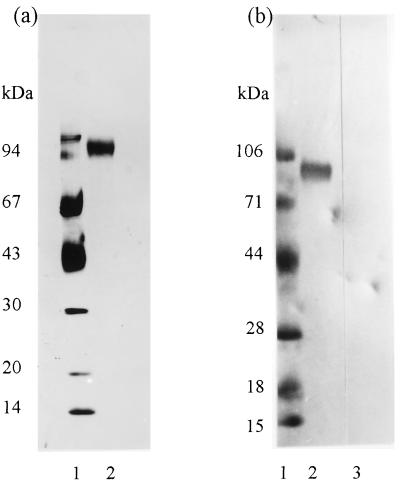
Preliminary experiments using L. cuprina larval PM demonstrated that it induced an immune response in vaccinated sheep that inhibited the growth of larvae that subsequently fed on serum from those sheep. A series of protein fractionations using vaccination trials in sheep as a bioassay were undertaken to purify individual PM proteins responsible for inducing the anti-larval effects. With this bioassay, a 95-kDa integral PM protein (peritrophin 95) was purified from a 6 M urea extract of detergent-washed PM by using a combination of gel-permeation and anion-exchange chromatography (Fig. 1a). The apparent size of peritrophin 95 measured by SDS/PAGE varied according to the percentage of polyacrylamide (Mr = 80,000 ± 8,000 after extrapolation to 0% polyacrylamide). Fig. 1b shows that peritrophin 95 binds biotinylated wheat germ lectin. This interaction was inhibited by incubation of the lectin blot with 0.3 M GlcNAc. Thus, peritrophin 95 contained oligosaccharides with terminal GlcNAc (or NeuAc) sugars. Weak binding was observed with biotinylated lentil lectin; however, Phaseolos vulgaris E lectin, peanut agglutinin, Sophora japonica lectin, and Pisum sativum lectin did not bind (results not shown).
Figure 1.
SDS/PAGE of purified peritrophin 95. (a) Silver-stained SDS/PAGE of peritrophin 95 (reduced). Lanes: 1, standards; 2, peritrophin 95 (2 μg). (b) Biotinylated lectin blot of purified peritrophin 95 (5 μg, lanes 2 and 3). Lanes: 1, standards; 2, biotinylated wheat germ lectin; 3, biotinylated wheat germ lectin after incubation of the filter with 0.3 M GlcNAc.
Anti-Larval Effects of Serum from Sheep Vaccinated with Peritrophin 95.
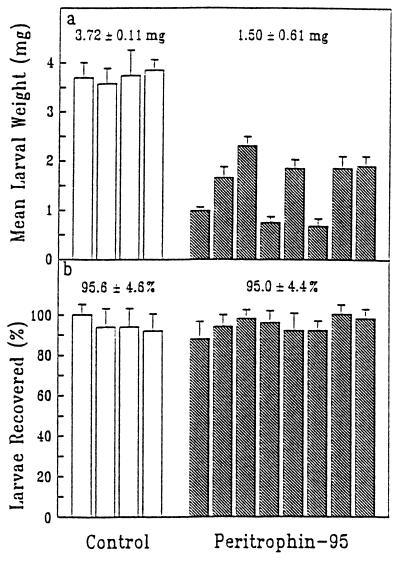
Peritrophin 95 induced an anti-larval immune response in sheep as assessed by the ability of serum from vaccinated sheep to inhibit growth of L. cuprina larvae in an in vitro feeding bioassay (Fig. 2). There was a mean larval weight reduction of 60 ± 16% (P < 0.001) compared with the mean weight of larvae feeding on control serum. There was no significant effect on the survival of larvae. The effect on larval weights is a specific immune response to peritrophin 95 because a number of purified proteins from several diverse species, including L. cuprina, as well as several crude protein extracts from L. cuprina larvae, Boophilus microplus adults and larvae, Escherichia coli, and cultured insect cells did not induce anti-larval effects in serum from appropriately vaccinated sheep (results not shown).
Figure 2.
Anti-larval effects of serum from sheep vaccinated with peritrophin 95. The effects of anti-peritrophin 95 serum from eight sheep on the mean weight (a) and mean survival (b) of L. cuprina larvae were measured (shaded histograms) by using an in vitro feeding bioassay. The unshaded histograms refer to corresponding results for a control group of four sheep. Vertical bars on histograms indicate 1 SD. Numbers above the control and peritrophin 95 groups represent group means ± 1 SD. The mean weight of larvae feeding on the serum from sheep vaccinated with peritrophin 95 was significantly different from the control group (P < 0.001).
Enhanced Anti-Larval Effects with Serum Enriched with Anti-Peritrophin 95 Ig.
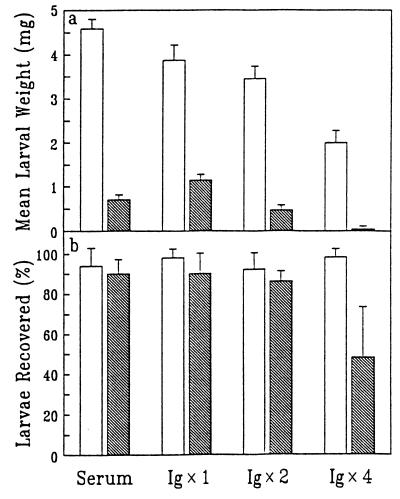
Total Ig was isolated from the serum of a strongly responding sheep vaccinated with peritrophin 95 and from pooled sera from control sheep that received adjuvant alone. The isolated Ig was reconstituted into NSS at 1-fold, 2-fold, and 4-fold enrichments as measured by ELISA. Results of the in vitro larval feeding bioassay undertaken with these modified sera are shown in Fig. 3. The original anti-larval activity in the immune serum resulted in 85 ± 2% mean larval weight reduction. Transfer of total Ig from this serum into NSS at the original Ig concentration (1× Ig) caused 70 ± 7% mean larval weight reduction compared with the corresponding control. Thus, the anti-larval activity was mediated by Ig. There was enhanced inhibition of larval growth when the level of anti-peritrophin 95 Ig in NSS was increased. The mean larval weight reduction was 70 ± 7% at 1× Ig, 86 ± 9% at 2× Ig, and 98 ± 3% at 4× Ig compared with corresponding controls. At the latter Ig concentration, a mean of 52 ± 25% of the larvae were not recovered compared with the corresponding value of 2 ± 4% for the appropriate control. Reduced larval recovery was observed only when the mean larval weight reduction of the surviving larvae was at least 80% (ref. 13 and unpublished results). Heating of the immune serum for 30 min at 55°C did not alter the anti-larval activity, indicating that complement was not involved (results not shown). The high Ig concentration introduced into NSS caused some inhibition of larval growth in control experiments (but had no effect on larval survival). However, this effect was relatively small compared with effects observed with serum enriched with immune Ig. For example, the control mean larval weight at 4× Ig was 48% of the value for the control at 1× Ig; however, the corresponding level of anti-peritrophin 95 Ig reduced the mean larval weight by 95%.
Figure 3.
Enhanced larval growth inhibition using serum enriched with Ig isolated from a sheep vaccinated with peritrophin 95. Larvae were allowed to feed on the Ig-enriched serum for 20 h in an in vitro feeding bioassay after which mean weight (a) and mean survival (b) were measured. Controls, Ig from control sheep supplemented into NSS at 1-, 2-, and 4-fold enrichments (unshaded); Ig, (anti-peritrophin 95) immune Ig supplemented into NSS at 1- (1× Ig), 2- (2× Ig), and 4- (4× Ig) fold enrichments (shaded); serum, original unmodified anti-peritrophin 95 serum. Error bars denote 1 SD.
Effect of Immune Serum on Larval PM.
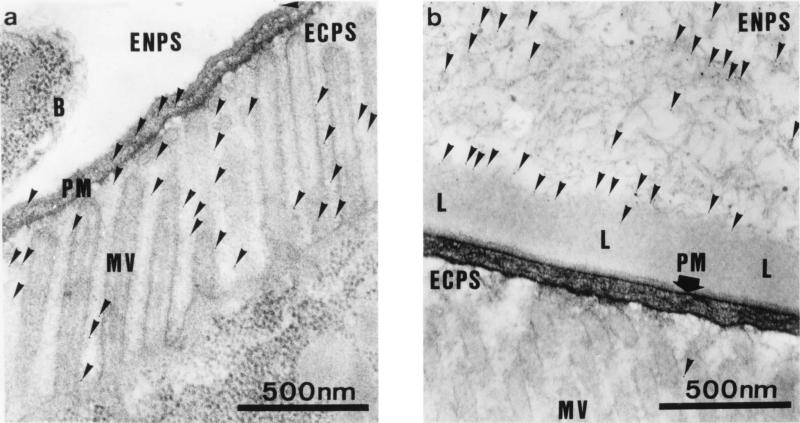
Visual and microscopic examination of the small larvae feeding on immune serum did not reveal any external signs of damage. One possibility was that the ingested antibodies were binding to peritrophin 95 on the PM and interfering with the movement of nutrients through this semipermeable membrane, thereby limiting the availability of nutrients to the digestive gut epithelia. To test this possibility, neonate larvae were fed on NSS enriched 4-fold with immune Ig. This enriched serum reduced larval weights by greater than 90% in an in vitro feeding bioassay. Also included in the Ig-enriched diet for the last 2 h of growth were 6-nm colloidal gold particles. The electron micrograph shown in Fig. 4a demonstrates that the colloidal gold particles ingested by a larva feeding on NSS enriched 4-fold with control Ig were present within the PM and freely distributed among the microvilli of the digestive epithelial cells lining the gut. A similar pattern was seen in larvae allowed to grow on this preparation until second instar. These results indicate that particles of this size can freely traverse the PM and are consistent with other studies that indicate that the permeability limit for the PM is ∼10 nm (unpublished data). In contrast, in a larva fed on immune Ig-enriched serum, the gold particles were almost completely restricted to the endoperitrophic space (Fig. 4b). Very few gold particles (<0.5%) were found within the PM or associated with the digestive epithelial cells. Further, there was now a new layer lining the gut lumen side of the PM that almost totally excluded the ingested gold particles from access to the PM. This layer, of undefined composition, extended from the anterior midgut through the midgut to the hindgut and was in places 1 μm thick compared with a PM thickness of 70–100 nm.
Figure 4.
Reduced permeability of the PM. Larvae were fed on NSS enriched 4-fold with Ig from control serum (a) or anti-peritrophin 95 serum (b). Both Ig-enriched sera contained 6-nm colloidal gold particles. The larval PM was then examined by transmission electron microscopy. PM, peritrophic membrane; MV, microvilli of gut digestive epithelial cells; ENPS, endoperitrophic space; ECPS, ectoperitrophic space; B, bacterium; L, antibody-induced layer on gut lumen side of PM. Colloidal gold particles are indicated by arrows.
cDNA and Deduced Amino Acid Sequences of Peritrophin 95.
Fig. 5 shows the nucleotide and deduced amino acid sequences of cDNA coding for peritrophin 95. The cDNA sequence (1,616 bp) contained an ORF of 1,443 bp that coded for a polypeptide of 480 amino acids containing all peptides derived from peritrophin 95 (Fig. 5). The deduced amino acid sequence of peritrophin 95 contains an amino-terminal signal sequence of 24 amino acids and a mature polypeptide of 456 amino acids. The mature protein has a pIcalc of 4.5 (4.7 ± 0.2 by isoelectric focusing) and a calculated Mr of 50,117, which is significantly different from that measured by SDS/PAGE (Mr = 80,000 ± 8,000). The sequence contains 30 Cys residues that are clustered in the amino-terminal two-thirds of the protein (amino acids 1–341) followed by a carboxyl-terminal domain of 139 amino acids containing several small repeated sequences but no Cys residues.
Figure 5.
Nucleotide and deduced amino acid sequences of peritrophin 95 determined from cDNA. Single underlining denotes peptides determined by amino acid sequencing. Cys residues are circled. A potential polyadenylylation signal sequence is underlined twice. The position of the amino terminus of the mature protein is indicated by an arrow and the amino-terminal signal sequence is underlined with a broken line. Potential N-linked glycosylation sites are boxed. The 5′ and 3′ noncoding regions are represented by lowercase type.
The Cys-rich domain contains five nonidentical but related subdomains each ∼65 amino acids long and characterized by a specific register of six Cys residues (Fig. 6a). The number of Cys residues, their close spacing, and the presence of abundant potential β-turns in the secondary structure of the protein (14) strongly suggest that the Cys residues are involved in intrasubdomain disulfide bonds (15). In addition to the Cys residues, there was also strong conservation of aromatic amino acids at three positions in each of the subdomains. There are four potential N-linked glycosylation sites (16) present within the Cys-rich domain and adjacent to this is a hydrophilic Pro-rich carboxyl-terminal domain of 139 amino acids containing several small imperfectly repeated structures consisting of two general types (Fig. 6b). The first, which is repeated 18 times, is a pentameric structure involving primarily proline, glutamate, and threonine (type I repeat) and the second is a tetrameric structure repeated four times adjacent to carboxyl terminus of the protein and characterized by the presence of lysine, proline, and threonine (type II repeat). The prevalence of prolines and threonines in the type I repeats suggests that this region may contain a number of O-linked glycosylation sites (17). Extensive glycosylation and an abundance of acidic amino acids in this domain could also account for the difference in molecular weight between the native form of peritrophin 95 measured by SDS/PAGE (Mr = 80,000 ± 8,000) and that calculated for the mature protein (Mr = 50,117). The genomic sequence of peritrophin 95 was identical (except for the presence of a 77-bp intron within the Cys-rich domain) to the cDNA sequence (unpublished data), indicating that the protein size disparity is not due to an error introduced into the cDNA during construction of the cDNA library.
Figure 6.
Domain structure of peritrophin 95 and similarities with other proteins. (a) Alignment of five related six Cys subdomains in peritrophin 95 with similar domains from other proteins. PM95 I–V, the five consecutive peritrophin 95 six Cys subdomains; PM44 I–V, the five consecutive six Cys subdomains from the L. cuprina PM protein peritrophin 44; Ms, Ch, Av, Ov, and Bm are single six Cys carboxyl-terminal domains of chitinases from Manduca sexta, Chelonus sp., Acanthocheilonema viteae, Onchocera volvulus, and Brugia malayi, respectively. Cysteines are boxed, potential N-linked glycosylation sites are underlined, and the positions of semiconserved aromatic amino acids are indicated by an asterisk. The symbol > represents the carboxyl-terminal end of a protein. (b) Type I and type II repeated sequences in the carboxyl-terminal 139 amino acid of peritrophin 95. Prolines in the type I repeats are boxed. (c) Sequence similarity between T. brucei insect-stage procyclin (parpAa) and a region in peritrophin 95 (PM95).
Similarities with Other Proteins.
The GenPep protein sequence database was searched (18) for proteins that showed significant sequence similarity to the Cys-rich domain (amino acids 1–341) or the Pro-rich carboxyl-terminal domain (amino acids 342–480) of peritrophin 95. The former domain has a highly significant homology with another L. cuprina PM protein, peritrophin 44 (ref. 6 and Fig. 6a). The seqdp computer program (19) calculated the significance of this homology at 10.8 SD from 20 randomized sequences of the same composition. Peritrophin 44 also has five nonidentical but related subdomains of ∼65 amino acids, each characterized by a six Cys register that is very similar to that present in peritrophin 95. One significant difference between these two proteins is the absence of the Pro-rich carboxyl-terminal domain in peritrophin 44. The characteristic six Cys register in each of the Cys-rich subdomains (C-X12–14-C-X5-C-X8–14-C-X10–14-C-X4–16-C or small variations thereof, where X is not C) from both peritrophin 44 and peritrophin 95 was used to search (20) for related domains in other proteins. This search revealed matches with the single six Cys domain adjacent to the carboxyl terminus in each of the chitinases from Manduca sexta (tobacco hornworm; ref. 21), Chelonus sp. (an endoparasitic wasp; ref. 22), Brugia malayi (a parasitic nematode; ref. 23), Acanthocheilonema viteae (a parasitic nematode; GenBank accession LA1653) and Onchocerca volvulus (a parasitic nematode; GenBank accession LA2021) (Fig. 6a). This region in each of the chitinases has been postulated to be a chitin-binding domain on the basis of analogies with plant chitinases and the demonstration that peritrophin 44 binds reacetylated chitosan in vitro (6). The consensus sequence described above or a slight variation of it also identified several Caenorhabditis elegans hypothetical proteins (24, 25). The C. elegans proteins are not chitinases because they do not contain chitinase catalytic domains. Like peritrophin 95 and peritrophin 44, these C. elegans proteins contain multiple copies of the six Cys subdomain, but these are often interspersed between small repetitive elements. The functions of these C. elegans proteins are unknown although their domain similarities to peritrophin 44 suggest that they could be involved in linkages to chitin. C. elegans does not contain a highly structured PM (3) like that present in L. cuprina larvae but there is evidence for the presence of an undefined matrix lining the gut lumen (26).
The presence in peritrophin 95 of a Cys-rich domain adjacent to a Pro-rich domain potentially containing several O-linked glycosylation sites is similar to the architectural design of mammalian mucins (27). In addition to these structural parallels, both proteins are present in analogous locations (lining digestive epithelial cells) and may, therefore, have similar functions. A search of the Genpep database with the carboxyl-terminal Pro-rich domain of peritrophin 95 revealed similar structures in a large number of proteins. However, apart from the involvement of repeated prolines, there was very little amino acid sequence similarity between these proteins and peritrophin 95. One interesting exception is a 46-amino acid sequence from procyclin (parpAa), a major coat protein of the insect gut stage Trypanosoma brucei parasite (28) that has 65% identity with a region from the type I repeated sequences of peritrophin 95 (Fig. 6c). The significance of the sequence similarity is not known although the presence of these proteins in the gut regions of two flies is suggestive of a functional relationship.
DISCUSSION
The results described in this study demonstrate that the induction of an immune response in the serum of sheep vaccinated with the larval PM glycoprotein peritrophin 95 results in inhibition of normal growth of L. cuprina larvae feeding on that serum. Further, severe larval growth inhibition and reduced larval survival can be induced by increasing the concentration of antibodies to peritrophin 95 in serum. Previous studies demonstrated that anti-larval effects observed by using the in vitro feeding bioassay were also observed by using an in vivo bioassay directly on the backs of sheep albeit to a lesser degree (5). Consequently, this vaccination strategy potentially could be used for immunological control of these larvae. L. cuprina larvae are very resilient to major changes in their growth rate. It is not until the growth rate is reduced by >80% that significant mortality is expressed (refs. 9 and 13 and unpublished results). This observation is consistent with the hypothesis that the larvae are being starved when feeding on the immune serum. The surviving small larvae take longer to develop into pupae although subsequent development is normal (results not shown). The delay in reaching pupation is disadvantageous to the larvae because there is significant natural larval mortality during this period. Thus, immunological control will require strong Ig responses in vaccinated animals to cause severe retardation of the growth of first-instar larvae and consequent increased larval mortality at this stage. Combinations of PM antigens could be used to further enhance the anti-larval activity in vaccinated animals.
The nature of the antibody-induced layer that lines the gut lumen side of the PM of larvae that feed on immune serum is unclear. One possibility is that the layer is composed of partially digested protein that has gelled on the PM surface in response to the clogging of the normally semipermeable PM caused by the binding of ingested anti-peritrophin 95 antibodies. A similar physical effect is observed in concentrating cells using semipermeable membranes that become blocked by partial solute precipitation or solute polarization (29). Analogous effects on larval growth and the same type of induced layer on the PM occur in larvae fed on wheat germ lectin or lentil lectin, which also bind to PM proteins (9). The data indicate a similar mechanism of action and reinforces the proposal that the anti-larval effects are mediated by a biophysical effect involving the formation of an impervious layer on the gut lumen side of the PM triggered by the binding of proteins to the PM that block pores in the PM.
The structure of peritrophin 95 suggests that it has the capacity to bind chitin within the PM and has a mucin-like structure in the carboxyl-terminal one-third of the protein that may be extensively glycosylated and probably highly exposed in the gut lumen. The ability of peritrophin 44 (6) and potentially also peritrophin 95 to bind chitin, a linear polymer of GlcNAc, raises another interesting possibility that these proteins could form a coordinated network in the PM mediated by interactions between the chitin-binding domains of these proteins and oligosaccharides containing GlcNAc present on peritrophin 95. These interactions could occur in addition to the direct binding of these proteins to chitin fibrils present in the PM. It is possible that these combined interactions dictate pore size. The reduction in PM permeability after the binding of antibodies to peritrophin 95 is consistent with this model.
The extension of this potential immunocontrol strategy to the control of other insects that cause cutaneous myiasis or insects feeding directly on mammalian blood will be dictated by the specific physiological characteristics of these insects, particularly the intimacy of their relationship with their mammalian hosts. The key aspect of this potential control strategy is the delivery of large quantities of antibody directed against the PM of the insect. The ability to produce relatively large quantities of functional recombinant antibodies in transgenic plants (30) raises the intriguing possibility that this potential immunocontrol strategy could also be used to control insect pests of plants.
Acknowledgments
We are grateful to Susan Briscoe for technical assistance and Peter Willadsen and Chris Elvin for valuable discussions. We also thank the L. W. Bett Trust and Australian wool growers through the International Wool Secretariat for financial support.
ABBREVIATIONS
- PM
peritrophic membrane
- NSS
normal sheep serum
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. U23828).
References
- 1.Tellam R L, Smith D, Kemp D H, Willadsen P. In: Animal Parasite Control Using Biotechnology. Wong W K, editor. London: CRC; 1992. pp. 303–331. [Google Scholar]
- 2.Willadsen P, Eisemann C H, Tellam R L. Parasitol Today. 1993;9:132–135. doi: 10.1016/0169-4758(93)90177-h. [DOI] [PubMed] [Google Scholar]
- 3.Tellam R L. In: The Biology of the Insect Midgut. Lehane M J, Billingsley P F, editors. London: Chapman & Hall; 1996. pp. 87–113. [Google Scholar]
- 4.Eisemann C H, Pearson R D, Donaldson R A, Cadogan L C, Vuocolo T. Med Vet Entomol. 1993;7:177–185. doi: 10.1111/j.1365-2915.1993.tb00672.x. [DOI] [PubMed] [Google Scholar]
- 5.East I J, Fitzgerald C J, Pearson R D, Donaldson R A, Vuocolo T, Cadogan L C, Eisemann C H, Tellam R L. Int J Parasitol. 1993;23:221–229. doi: 10.1016/0020-7519(93)90144-n. [DOI] [PubMed] [Google Scholar]
- 6.Elvin C M, Vuocolo T, Pearson R D, East I J, Riding G A, Eisemann C H, Tellam R L. J Biol Chem. 1996;271:8925–8935. doi: 10.1074/jbc.271.15.8925. [DOI] [PubMed] [Google Scholar]
- 7.Eisemann C H, Johnston L A Y, Broadmeadow M, O’Sullivan B M, Donaldson R A, Pearson R D, Vuocolo T, Kerr J D. Int J Parasitol. 1990;20:299–305. doi: 10.1016/0020-7519(90)90143-b. [DOI] [PubMed] [Google Scholar]
- 8.Mostratos A, Beswick T S L. J Pathol. 1969;98:17–24. doi: 10.1002/path.1710980103. [DOI] [PubMed] [Google Scholar]
- 9.Eisemann C H, Donaldson R A, Pearson R D, Cadogan L C, Vuocolo T, Tellam R L. Entomol Exp Appl. 1994;72:1–10. [Google Scholar]
- 10.Saiki R K, Gelfand D H, Stoffel S, Scharf S J, Higuchi R, Horn G T, Mullis K B, Erlich H A. Science. 1988;239:487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- 11.Sanger F, Nicklen S, Coulson A R. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 13.East I, Eisemann C H. Immunol Cell Biol. 1993;71:453–462. doi: 10.1038/icb.1993.51. [DOI] [PubMed] [Google Scholar]
- 14.Chou P Y, Fasman G D. Adv Enzymol. 1978;47:45–148. doi: 10.1002/9780470122921.ch2. [DOI] [PubMed] [Google Scholar]
- 15.Thornton J M. J Mol Biol. 1981;151:261–287. doi: 10.1016/0022-2836(81)90515-5. [DOI] [PubMed] [Google Scholar]
- 16.Gavel Y, Von Heijne G. Protein Eng. 1990;3:433–442. doi: 10.1093/protein/3.5.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Williams K, Redmond J, Pisano A, Gooley A. Today’s Life Sci. 1992;4:50–60. [Google Scholar]
- 18.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 19.Needleman S B, Wunsch C D. J Mol Biol. 1970;48:443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- 20.Sibbald P R, Argos P. Comput Appl Biosci. 1990;6:279–288. doi: 10.1093/bioinformatics/6.3.279. [DOI] [PubMed] [Google Scholar]
- 21.Kramer J J, Corpuz L, Choi H K, Muthukrishnan S. Insect Biochem Mol Biol. 1993;23:691–701. doi: 10.1016/0965-1748(93)90043-r. [DOI] [PubMed] [Google Scholar]
- 22.Krishnan A, Nair P N, Jones D. J Biol Chem. 1994;269:20971–20976. [PubMed] [Google Scholar]
- 23.Fuhrman J A, Lane W S, Smith R F, Piessens W F. Proc Natl Acad Sci USA. 1992;89:1548–1552. doi: 10.1073/pnas.89.5.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tellam R L. Parasitol Today. 1996;12:291–292. doi: 10.1016/0169-4758(96)80819-2. [DOI] [PubMed] [Google Scholar]
- 25.Blaxter M. Parasitol Today. 1996;12:42. doi: 10.1016/0169-4758(96)80647-8. [DOI] [PubMed] [Google Scholar]
- 26.Borgonie G, Claeys M, Vanfleteren J, De Waele D, Coomans A. Fundam Appl Nematol. 1995;18:227–233. [Google Scholar]
- 27.Carlstedt I, Sheehan J K, Corfield A P, Gallagher J T. Essays Biochem. 1985;20:40–76. [PubMed] [Google Scholar]
- 28.Mowatt M R, Wisdom G S, Clayton C E. Mol Cell Biol. 1989;9:1332–1335. doi: 10.1128/mcb.9.3.1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bellucci F, Drioli E, Scardi V. J Appl Polymer Sci. 1975;19:1639–1647. [Google Scholar]
- 30.Tavladoraki P, Benvenuto E, Trinca S, De Martinis D, Cattaneo A, Galeffi P. Nature (London) 1993;366:469–472. doi: 10.1038/366469a0. [DOI] [PubMed] [Google Scholar]