Abstract
Despite the importance of mitogen-activated protein kinase (MAPK) signaling in eukaryotic biology, the mechanisms by which signaling yields phenotypic changes are poorly understood. We have combined transcriptional profiling with genetics to determine how the Kss1 MAPK signaling pathway controls dimorphic development in Saccharomyces cerevisiae. This analysis identified dozens of transcripts that are regulated by the pathway, whereas previous work had identified only a single downstream target, FLO11. One of the MAPK-regulated genes is PGU1, which encodes a secreted enzyme that hydrolyzes polygalacturonic acid, a structural barrier to microbial invasion present in the natural plant substrate of S. cerevisiae. A third key transcriptional target is the G1 cyclin gene CLN1, a morphogenetic regulator that we show to be essential for pseudohyphal growth. In contrast, the homologous CLN2 cyclin gene is dispensable for development. Thus, the Kss1 MAPK cascade programs development by coordinately modulating a cell adhesion factor, a secreted host-destroying activity, and a specialized subunit of the Cdc28 cyclin-dependent kinase.
Mitogen-activated protein kinase (MAPK) signal transduction cascades play crucial roles in both normal and abnormal eukaryotic development. Inappropriate activation of one such pathway, the extracellular signal-regulated kinase cascade, promotes a significant fraction of human cancers. Despite their recognized function in driving development in diverse organisms, little is known about how signaling leads to observed changes in cellular behavior. As the ultimate target of a MAPK cascade is often a transcription factor, it is reasonable to suppose that changes in gene expression play an important role. However, until recently, it has been difficult to assess the entire ensemble of target genes activated by MAPK signaling.
In the budding yeast, Saccharomyces cerevisiae, the existence of gene knockout mutants as well as methods for the global profiling of transcript levels presents a unique opportunity to identify the targets of a developmental MAPK signaling pathway. The ability of solitary diploid yeast cells to develop into a multicellular, pseudohyphal form (invasive filaments of elongated cells) under conditions of nitrogen starvation requires a MAPK signaling cascade (1). This cascade culminates in the activation of the heteromeric transcription factor, Tec1-Ste12 (2, 3). The same signaling components control a related filamentation process in the haploid cell type, termed haploid-invasive growth, that occurs on rich medium (4). By examining the transcript profiles of mutants in the signaling pathway under haploid invasive growth and diploid pseudohyphal development conditions, we sought to identify the effectors of MAPK signaling in two developmental contexts.
Materials and Methods
Strain Growth, RNA Isolation, and Microarray Hybridization.
Haploid strains were grown at 30°C in synthetic complete medium lacking uracil to an optical density at 600 nm of 1.0. Diploid strains were grown in minimal medium to the same density, washed twice, and resuspended in yeast nitrogen base (Difco) containing 50 μM ammonium sulfate (synthetic low ammonium dextrose medium) and grown for 4 h. RNA was extracted and the polyadenylated fraction was purified by using polyATract (Promega). Biotinylated cRNA was prepared and hybridized as described (5). Hybridization intensities (which are proportional to RNA levels) were determined by using a trimmed average difference between matched and mismatched probe pairs.
Analysis of Hybridization Data.
The initial steps in data analysis were performed by using a set of custom perl scripts run in a unix operating environment (T.G., A. Saldanha, and G.R.F., unpublished work). Chip scans were scaled to each other by using a bulk scaling method. The derived scaling factors for haploid samples varied between 0.4 and 1.7. Scaling factors for diploid samples varied between 0.3 and 2.5. After scaling, average difference values that measured less than a value of 1 were set to a floor of 1. Transposable elements and unannotated ORFs were removed from the data set. As described previously by Wodicka et al. (5), low average differences yield less reliable measurements. Thus, data sets were passed through a quantitative variation filter that demanded, for a given gene, that at least one of the samples showed an average difference above a given threshold. For our analysis, the threshold was set at 100, because it was shown previously that, for duplicate yeast GeneChip (Affymetrix, Santa Clara, CA) experiments, only two genes above this expression threshold showed a variation in hybridization intensities that exceeded 3-fold (5). We emphasize that the relative sensitivity and specificity of subsequent queries depends highly on this user-set threshold. Based on the fact that Tec1, Ste12, and Ste7 are positively acting components of the MAPK signaling pathway, a subset of genes was chosen for display by sorting genes based on the ratio of gene expression in TEC1-HC versus tec1Δ strains, identifying the subset that showed a ratio of greater than 4 and identifying those genes that also showed reduced expression in ste7Δ and ste12Δ strains. This analysis can be represented as a Boolean query of gene-expression ratios: TEC1-HC/tec1Δ>4, ste7Δ/WT<1, and ste12Δ/WT<1 (WT, wild type). From the haploid data, 11 genes were obtained that satisfied this query, whereas 33 genes from the diploid data were identified. To rule out that these results were caused solely by random noise in the data, we performed Boolean queries of equal quantitative and logical stringency that did not conform to biological expectations. For instance, we looked for genes that were repressed by TEC1 greater than 4-fold (tec1Δ/TEC1-HC>4) and that showed increased or decreased expression in ste12Δ and ste7Δ. In Boolean terms, these were (i) TEC1-HC/tec1Δ<4, ste7Δ/WT>1, and ste12Δ/WT>1; (ii) TEC1-HC/tec1Δ<4, ste7Δ/WT>1, and ste12Δ<1; (iii) TEC1-HC/tec1Δ<4, ste7Δ/WT<1, and ste12Δ>1; as well as (iv) TEC1-HC/tec1Δ<4, ste7Δ/WT<1, and ste12Δ<1. If the gene list derived from the original Boolean query for MAPK targets was derived solely from noise, we would expect these queries to yield as many genes as the original query. However, greatly fewer genes were obtained. For the haploid data set, the queries yielded an average of 2 ± 1 genes, and, for the diploid data set, an average of 10.0 ± 4 genes were obtained. To the extent that the alternative query patterns may have yielded genes that make unexpected biological responses to the signaling mutants, these values may actually overestimate the false-positive frequencies. It is notable that the number of genes that behave as if they are repressed by the pathway (query 1 above) is only 2 and 10 under haploid invasion and pseudohyphal conditions, respectively. Thus, the MAPK pathway serves primarily to activate rather than repress gene expression.
Significance of Overlap Between Diploid MAPK Targets and Cell-Cycle-Regulated Genes.
To determine the significance of the overlap between the diploid MAPK target genes and cell-cycle-regulated genes, we performed a cumulative binomial-distribution calculation. The probability that at least 11 of 33 of the diploid MAPK targets overlap with the 800 reported cell-cycle-regulated genes by chance alone was found to be 5.4 × 10−4. This calculation assumes that the genes previously identified to be cell-cycle-regulated represent all or nearly all of the periodic transcripts encoded by the ≈6,200 yeast genes (6).
Pectinase Assays.
Plate assays were a modification of published procedures (7, 8). Haploid cells (n = 4 × 105) for each of the indicated genotypes were spotted onto a plate containing 1% (vol/vol) gellan gum, 1.5% (vol/vol) fruit pectin (Sigma), 6.7 g/liter yeast nitrogen base (Difco), 2 g/liter ammonium sulfate, and 2% (vol/vol) glucose. Plates were also supplemented with amino acids to cover auxotrophic requirements. After incubation, unhydrolyzed pectin was precipitated overnight with cetyltrimethylammonium bromide (1% vol/vol) and counterstained with the pectin-binding dye ruthenium red (0.1%). The expression of PGU1 in diploids falls below the threshold described above for detection (and thus is not on the gene list shown in Fig. 2). However, pectinase activity is MAPK-dependent in diploid cells, as it is in haploid cells (H.D.M., unpublished observations).
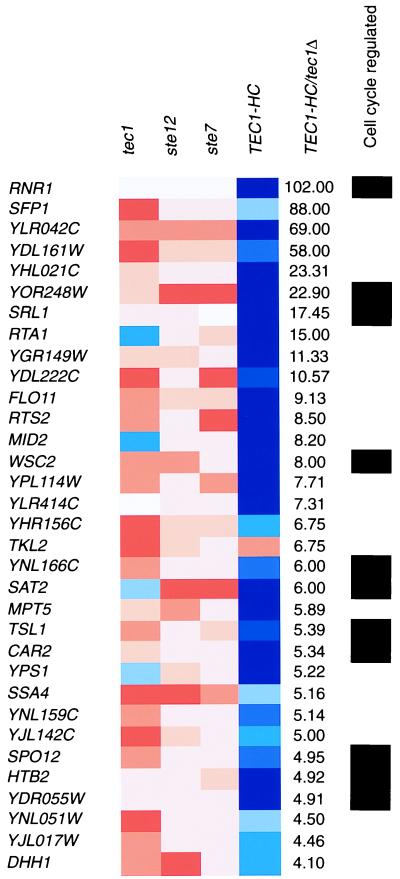
Figure 2.
Display of gene-expression measurements under pseudohyphal-development conditions. Criteria described in Fig. 1 were applied to diploid strains of the indicated genotypes. Genes previously shown to be cell-cycle-regulated are indicated with black rectangles.
PGU1–lacZ Construction and β-Galactosidase Assays.
The intergenic upstream region of PGU1 was amplified and placed upstream of the Escherichia coli lacZ gene on the URA3-marked high-copy yeast shuttle plasmid YEp356R (9) by using the BamHI site, yielding a PGU1 promoter–lacZ fusion. β-galactosidase assays were performed as described (10).
Northern Hybridization.
The CLN1 and ACT1 ORFs were amplified by using the GenePairs (Research Genetics) primers and labeled with [32P]dCTP by using PrimeIt (Stratagene). High-sensitivity Northern analysis was performed with the NorthernMax Product (Ambion, Austin, TX).
Results and Discussion
Identification of MAPK-Regulated Genes by Microarray Hybridization and Boolean Queries.
We used high-density oligonucleotide arrays (Affymetrix GeneChips) to determine transcript levels (5) in strains in which MAPK signaling was altered genetically. In particular, we profiled WT strains, strains containing knockouts of the transcription factor genes TEC1 and STE12, strains deleted for the MAPK kinase gene STE7, and strains that harbor a high-copy plasmid containing the WT TEC1 gene. Before RNA isolation, strains were grown under conditions that induce the relevant developmental switch: rich-medium conditions for haploid cells and nitrogen-starvation conditions for the analogous homozygous diploid cells. After target preparation and hybridization, the data were collected, and transcript levels were determined (5).
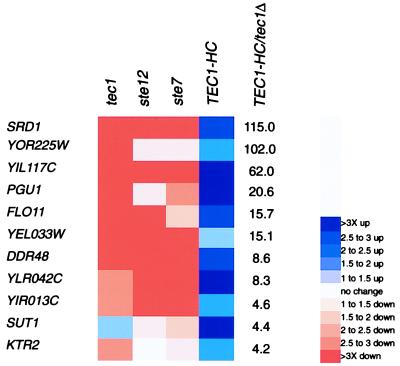
To identify genes regulated by the MAPK pathway, we used a method of Boolean-query analysis (see Materials and Methods). First, we sorted the genome based on the ratio of gene expression in strains overexpressing TEC1 (TEC1-HC) versus strains lacking the gene (tec1Δ). This method was chosen, because, although TEC1 functions specifically in the filamentation pathway, the other components tested are shared with the yeast pheromone-response MAPK pathway (reviewed in ref. 11). We then looked for genes whose expression was also reduced in ste12 and ste7 mutants. Figs. 1 and 2 display those genes that meet the criteria listed above and that show at least a 4-fold expression ratio in TEC1-HC versus tec1Δ cells under haploid invasive and diploid-pseudohyphal conditions, respectively. The data are available at http://staffa.wi.mit.edu/fink_public/mapk.
Figure 1.
Display of gene-expression measurements under haploid-invasive growth conditions. Shown are the fold changes relative to the WT sample. The ratio of gene expression in strains overexpressing TEC1 (TEC1-HC) versus strains deleted for the TEC1 gene (tec1Δ) is shown. Shown is the subset of genes that show a greater than 4-fold TEC1-HC/tec1Δ expression ratio and that are reduced in expression relative to WT in ste12 and ste7 null-mutant strains.
Gene Disruption Analysis of Haploid-Invasive-Growth MAPK Targets.
We first examined the data set for haploid cells grown on rich medium (Fig. 1). A gene strongly regulated by the pathway is FLO11/MUC1, which previously was shown to be essential for both haploid-invasion and diploid-pseudohyphal growth. Flo11 resides on the cell surface and is necessary for cell–cell adhesion, a presumed requirement for the formation of multicellular filaments (12–14). FLO11 was an anticipated target, because previous work had shown that its expression requires the MAPK pathway (14, 15). To determine whether other MAPK target genes were required for invasive growth, we deleted them from the genome. Surprisingly, of nine gene knockouts tested (all genes shown in Fig. 1, except for SRD1 and SUT1), only one (corresponding to YEL033W) had an obvious defect in haploid invasion and pseudohyphal development. The lack of an invasion defect in the other eight knockout strains could result from (i) a function in a coregulated process not measured in the laboratory invasion assay or (ii) a function in invasive growth that is redundant with that of another gene. The former hypothesis presumes that the developmental switch to haploid-invasive growth involves more than the morphological events that produce filamentous differentiation.
A Host-Degrading Enzyme Is Coregulated with Genes Required for Invasive Growth.
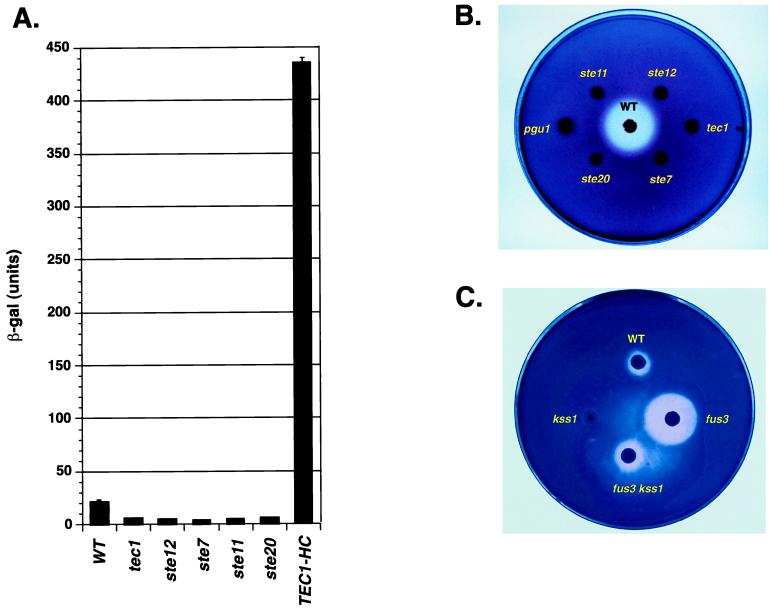
Support for the first hypothesis comes from analysis of the amino acid sequences of the candidate target genes. The sequence of Pgu1 suggested that it functions in invasive growth in the wild, not by promoting filament formation per se, but rather through enzymatic attack on the plant host. PGU1 encodes a secreted endopolygalacturonase that degrades the plant-specific polysaccharide pectin (7, 8). Consistent with the PGU1 transcript profile, a PGU1–lacZ promoter fusion gene depends on the MAPK pathway for its activity (Fig. 3A). Moreover, the secreted pectinase activity of S. cerevisiae absolutely requires components of the MAPK pathway as well as PGU1 (Fig. 3B). Pectinase activity was reduced in cells lacking Kss1, the MAPK that positively and negatively regulates filamentation (Fig. 3C). In contrast, mutation of the mating-specific MAPK Fus3 in haploid cells results in greatly increased pectinolysis compared with WT, and this phenotype requires Kss1 (Fig. 3C). These data are consistent with our previous observations showing that the deletion of FUS3 results in hyperactivation of Tec1-Ste12-dependent transcription and invasion because of inappropriate substitution of Kss1 for Fus3 (16). Thus, Pgu1 activity is regulated by the Kss1 filamentation-specific MAPK. Moreover, as with haploid-invasive growth (16), the positive activity of Kss1 significantly outweighs its inhibitory function.
Figure 3.
The filamentation MAPK pathway controls the expression of the host-degrading enzyme. (A) The filamentation MAPK pathway controls the PGU1 promoter. Shown are the β-galactosidase activities (β-gal) of haploid strains of the indicated genotypes that harbor a PGU1–lacZ fusion. Units correspond to millimoles of product per minute per milligram of protein encoded by PGU1. (B) This pectinase-plate assay shows the requirement for MAPK pathway and PGU1. The plate was incubated for 4 days before staining. (C) This pectinase plate assay shows phenotypes of MAPK gene knockouts. The plate was incubated for 2 days before staining to display the hyperpectinolytic activity of fus3Δ.
The regulation of PGU1 by the Kss1 MAPK pathway is highly significant in the context of the normal host-saprophyte ecology of S. cerevisiae. Pectin is found in all plants and is the primary component of the middle lamella, the layer of extracellular matrix that binds plant cells together (17). Pectin also forms a major structural component of fruits, such as the peel, a layer that S. cerevisiae presumably must penetrate in the wild to access sugars found in rotting fruit, its natural substrate. A number of fungal and bacterial pathogens of plants also secrete pectin-degrading enzymes; these have long been proposed to augment the ability of these microbes to invade their respective hosts. Indeed, in the bacterial soft-rot pathogen Erwinia chrysanthemi, pectinases are proven virulence factors (18). Moreover, the recent disruption of an endopolygalacturonase gene of the eukaryotic phytopathogen Botrytis cinerea established its essential role in the spread of infection from the primary inoculation site (19). In this study, we have shown that secretion of the homologous host-degrading activity by S. cerevisiae is coordinated with expression of genes required for filamentous growth via MAPK signaling. This result is important in light of the proposed relationship between filamentous growth and virulence in pathogenic fungi (see ref. 20). Specifically, because mutants in homologous MAPK cascades are both filamentation-defective and avirulent, it has been proposed that the ability to switch morphologies is required for pathogenesis (e.g., ref. 21). Our data suggest that other relationships between filamentation and virulence are possible. For instance, the avirulence of MAPK signaling mutants may result instead from a defect in the expression of coordinately regulated host-inactivating genes akin to PGU1 or from a combination of defects in filamentation and coregulated genes.
MAPK Regulation of CLN1 Links Signaling and Morphogenesis.
In addition to screening target genes for filamentation phenotypes and informative amino acid sequence relationships, we asked whether they had common modes of regulation, independent of their control by the MAPK pathway. Interestingly, one-third (11 of 33) of the genes regulated under diploid-pseudohyphal conditions (Fig. 2) encode genes previously found to be regulated during the cell cycle (6). This enrichment is highly significant, because only one of eight yeast transcripts are regulated by the cell cycle (see Materials and Methods). Most of the overlapping genes (9 of 11) are induced during the G1 and S phases of the cell cycle (6), suggesting a possible connection between the MAPK pathway and the machinery involved in G1/S-specific gene expression, such as the G1 cyclins. Notably, pseudohyphal growth is associated with an altered cell cycle in which a G2 delay is a prominent feature (22).
Because the escape from G2 is associated with down-regulation of G1 cyclin (Cln) activity and up-regulation of G2 cyclin (Clb) activity (23), the activation of Cln activity and/or repression of Clb activity by the MAPK pathway could account for our results. Because the balance of Cln versus Clb activity also controls the switch between polarized versus isotropic growth of emerging daughter buds (24), regulation of cyclin activity could also profoundly impact cell morphology during pseudohyphal development. In support of this idea, recently, it has been reported that the homozygous double knockout of the G1 cyclin genes CLN1 and CLN2 blocks pseudohyphal growth (25). Thus, the activation of Cln expression by the MAPK pathway would explain the observed induction of G1/S-specific gene expression, the altered cell cycle of pseudohyphal cells, and their elongated morphology. We note that CLN gene expression in nitrogen-starved diploid cells falls below the reliable detection threshold of our microarray system, which would have precluded us from identifying CLN regulation in the experiments described above.
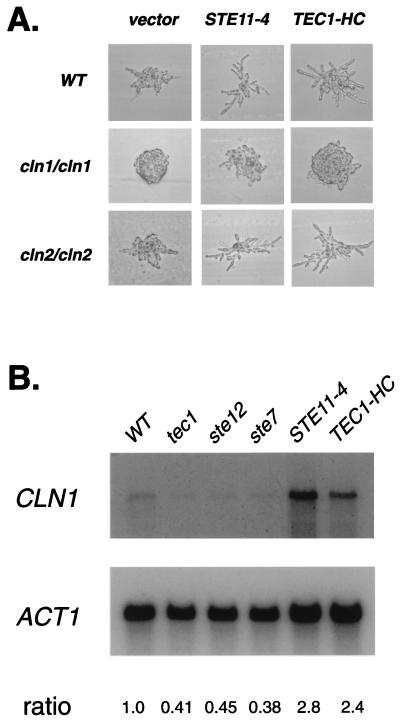
We examined cln1/cln1 and cln2/cln2 strains for effects on the ability of the MAPK pathway to stimulate pseudohyphae formation (26). As shown in Fig. 4A, the cln1/cln1 strain, but not the cln2/cln2 strain, is blocked in its ability to form filaments of elongated cells in response to STE11-4 (an activated allele of the MEKK gene) or high-copy TEC1. This observation is consistent with the model that CLN1 acts downstream of the MAPK pathway to induce filamentation. To test whether CLN1 depends on the MAPK pathway for its expression, we performed quantitative Northern hybridization analysis (Fig. 4B). CLN1 expression was decreased in tec1/tec1, ste7/ste7, and ste12/ste12 mutant diploid strains grown under conditions of nitrogen starvation but increased in strains overexpressing TEC1 or harboring STE11-4. These data show that CLN1 functions as a developmentally specialized cyclin that is regulated by the MAPK pathway and that is necessary for the induction of pseudohyphal filaments. This function of CLN1 is likely to be specific to the diploid cell type, because single knockouts of CLN1 and CLN2 do not block haploid-invasive growth (25).
Figure 4.
The G1 cyclin Cln1 acts downstream of the MAPK pathway. (A) Stimulation of filamentation by the MAPK pathway requires CLN1 but not CLN2. Shown are micrographs of colonies after 18 h of incubation at 30°C on synthetic low ammonium dextrose medium (26). WT, cln1/cln1, and cln2/cln2 strains were transformed with a vector control, STE11-4 or TEC1-HC. (B) The filamentation MAPK pathway controls expression of CLN1. Polyadenylated RNA (10 μg) from WT, tec1/tec1, ste12/ste12, ste7/ste7, STE11-4, and TEC1-HC strains grown under conditions of nitrogen starvation was analyzed by Northern hybridization by using a radioactive CLN1 probe. The blot was reprobed by using a probe derived from the yeast actin gene (ACT1) as a control. Radioactivity was quantitated by using a phosphorimager. Relative ratios of CLN1/ACT1 expression are indicated.
Developmental Context Strongly Influences Target Genes Controlled by the MAPK Pathway.
To determine the extent to which developmental context influences the output of the MAPK pathway, we compared the genes most strongly activated under haploid invasive growth vs. pseudohyphal conditions (Fig. 1 vs. Fig. 2). Only two genes, FLO11 and YLR042C, were determined to be induced strongly in both situations. For FLO11, this makes sense, because it is required for both haploid invasive growth and pseudohyphae formation (disruption of YLR042C did not affect filamentous growth). Some of the differences in transcriptional patterns are likely caused by the fact that genes that are expressed poorly may have been filtered out of one of the two data sets (see Materials and Methods). Nonetheless, the remarkably small amount of overlap observed suggests that context strongly influences the transcriptional response to signal transduction.
Conclusion: Mapping Signaling Targets to Their Functions.
Dissecting the output of a developmental MAPK pathway requires the combination of genomics and genetics. The use of mutants is important to this analysis in two ways. First, by profiling cells that differ in a single component of a signaling pathway under identical environmental conditions, it is possible to examine the contribution of an individual signaling pathway to a developmental process. This study design is the opposite of one in which transcript profiles of cells of the same genotype are determined under different environmental conditions (e.g., ref. 27). In such cases, one is likely to be observing the complex contributions of multiple signaling pathways that are activated by an environmental change. Second, the analysis of strains with null mutations in the regulated target genes permits an assessment of their contributions to the process that the signaling pathway is known to control (e.g., filamentous growth). This observation led us to conclude that most genes strongly regulated by the Kss1 MAPK pathway in haploid cells are not required for invasive growth. A possible explanation for this observation is that many target genes are redundant with other genes. Alternatively, they could represent coregulated genes that function in a process not measured in the laboratory assay.
In this regard, sequence annotation offers a valuable guide for the identification of environmental conditions that reveal the unanticipated functions of target genes. Indeed, our discovery of Pgu1 and pectinolysis as targets of MAPK regulation indicates that the developmental switch to filamentous growth involves more than simply a change in morphology. Because loss of PGU1 function does not affect invasion or filamentation in the standard laboratory assays, pgu1 mutants would not have been identified in previous genetic screens for pseudohyphal mutants (28). However, the demonstrable host-degrading activity of Pgu1 makes biological sense in the context of the starvation-induced switch from growth as solitary yeast cells to pseudohyphae. The coregulation of filamentation genes with Pgu1 by the MAPK cascade is consistent with the proposal that pseudohyphal growth is a coordinated invasive foraging behavior in the wild (26). The switch to pseudohyphal growth can now be thought of more broadly, involving not only the production of multicellular filaments but also a change in the interaction with the plant host. It is possible that other target genes that we have identified are also involved in heretofore unsuspected aspects of this program.
The emerging archive of gene transcript signatures provides an important resource for connecting gene-expression information to gene function; indeed, it is a distinct form of sequence annotation. By comparing our microarray expression data to the cell-cycle expression profiles of Spellman et al. (6), we were able to deduce a connection between signal transduction, the Cdc28 cyclin-dependent kinase, and a morphogenetic event. Although a link between the cell cycle and pseudohyphal growth had been inferred (22), the genome-wide analysis led us to identify a specific cyclin-dependent kinase component (Cln1) that is both a transcriptional target of a MAPK cascade and a genetic requirement for the morphogenetic switch. The success of this approach suggests that, as the gene-expression database grows, it will be possible to resolve complex patterns of coregulation and use these relationships to infer connections between cellular processes.
Acknowledgments
We thank Suzanne Noble and Brian Cali for comments on the manuscript, Alok Saldanha for assistance with informatics, Adrain Halme for strains, and members of the Whitehead/Massachusetts Institute of Technology Genome Center Expression Group for stimulating discussions and technical support. This work was supported by Bristol-Myers Squibb, Affymetrix, and Millenium Pharmaceuticals, as well as by National Institutes of Health Grant GM35010. H.D.M. is supported by a Career Award from the Burroughs–Wellcome Fund. T.G. is a postdoctoral fellow of the Helen Hay Whitney Foundation. G.R.F. is an American Cancer Society Research Professor of Molecular Genetics.
Abbreviations
- MAPK
mitogen-activated protein kinase
- WT
wild type
References
- 1.Liu H, Styles C A, Fink G R. Science. 1993;262:1741–1744. doi: 10.1126/science.8259520. [DOI] [PubMed] [Google Scholar]
- 2.Gavrias V, Andrianopoulos A, Gimeno C J, Timberlake W E. Mol Microbol. 1996;19:1255–1263. doi: 10.1111/j.1365-2958.1996.tb02470.x. [DOI] [PubMed] [Google Scholar]
- 3.Madhani H D, Fink G R. Science. 1997;275:1314–1317. doi: 10.1126/science.275.5304.1314. [DOI] [PubMed] [Google Scholar]
- 4.Roberts R L, Fink G R. Genes Dev. 1994;8:2974–2985. doi: 10.1101/gad.8.24.2974. [DOI] [PubMed] [Google Scholar]
- 5.Wodicka L, Dong H, Mittmann M, Ho M H, Lockhart D J. Nat Biotechnol. 1997;15:1359–1367. doi: 10.1038/nbt1297-1359. [DOI] [PubMed] [Google Scholar]
- 6.Spellman P T, Sherlock G, Zhang M Q, Iyer V R, Anders K, Eisen M B, Brown P O, Botstein D, Futcher B. Mol Biol Cell. 1998;9:3273–3297. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blanco P, Sieiro C, Reboredo N M, Villa T G. FEMS Microbiol Lett. 1998;164:249–255. doi: 10.1111/j.1574-6968.1998.tb13094.x. [DOI] [PubMed] [Google Scholar]
- 8.Gognies S, Ganvoirs A, Aigle M, Belarbi A. Yeast. 1999;15:11–22. doi: 10.1002/(SICI)1097-0061(19990115)15:1<11::AID-YEA336>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 9.Myers A M, Tzagoloff A, Kinney D M, Lusty C J. Gene. 1986;45:299–310. doi: 10.1016/0378-1119(86)90028-4. [DOI] [PubMed] [Google Scholar]
- 10.Adams A, Gottschling D E, Kaiser C. Methods in Yeast Genetics 1997: A Cold Spring Harbor Laboratory Course Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1997. [Google Scholar]
- 11.Madhani H D, Fink G R. Trends Genet. 1998;14:151–155. doi: 10.1016/s0168-9525(98)01425-5. [DOI] [PubMed] [Google Scholar]
- 12.Lambrechts M G, Bauer F F, Marmur J, Pretorius I S. Proc Natl Acad Sci USA. 1996;93:8419–8424. doi: 10.1073/pnas.93.16.8419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lo W S, Dranginis A M. Mol Biol Cell. 1998;9:161–171. doi: 10.1091/mbc.9.1.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lo W S, Dranginis A M. J Bacteriol. 1996;178:7144–7151. doi: 10.1128/jb.178.24.7144-7151.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rupp S, Summers E, Lo H-J, Madhani H D, Fink G R. EMBO J. 1999;18:1257–1269. doi: 10.1093/emboj/18.5.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Madhani H D, Styles C A, Fink G R. Cell. 1997;91:673–674. doi: 10.1016/s0092-8674(00)80454-7. [DOI] [PubMed] [Google Scholar]
- 17.Raven P H, Evert R F, Eichhorn S E. Biology of Plants. New York: Worth; 1986. [Google Scholar]
- 18.Hugouvieux-Cotte-Pattat N, Condemine G, Nasser W, Reverchon S. Annu Rev Microbiol. 1996;50:213–257. doi: 10.1146/annurev.micro.50.1.213. [DOI] [PubMed] [Google Scholar]
- 19.ten Have A, Mulder W, Visser J, van Kan J A. Mol Plant–Microbe Interact. 1998;11:1009–1016. doi: 10.1094/MPMI.1998.11.10.1009. [DOI] [PubMed] [Google Scholar]
- 20.Madhani H D, Fink G R. Trends Cell Biol. 1998;8:348–353. doi: 10.1016/s0962-8924(98)01298-7. [DOI] [PubMed] [Google Scholar]
- 21.Lo H J, Kohler J R, Di Domenico B, Loebenberg D, Cacciapuoti A, Fink G R. Cell. 1997;90:939–949. doi: 10.1016/s0092-8674(00)80358-x. [DOI] [PubMed] [Google Scholar]
- 22.Kron S J, Styles C A, Fink G R. Mol Biol Cell. 1994;5:1003–1022. doi: 10.1091/mbc.5.9.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Amon A, Tyers M, Futcher B, Nasmyth K. Cell. 1993;74:993–1007. doi: 10.1016/0092-8674(93)90722-3. [DOI] [PubMed] [Google Scholar]
- 24.Lew D J, Reed S I. J Cell Biol. 1993;120:1305–1320. doi: 10.1083/jcb.120.6.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oehlen L J, Cross F R. J Biol Chem. 1998;273:25089–25097. doi: 10.1074/jbc.273.39.25089. [DOI] [PubMed] [Google Scholar]
- 26.Gimeno C J, Ljungdahl P O, Styles C A, Fink G R. Cell. 1992;68:1077–1090. doi: 10.1016/0092-8674(92)90079-r. [DOI] [PubMed] [Google Scholar]
- 27.DeRisi J L, Iyer V R, Brown P O. Science. 1997;24:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- 28.Mosch H U, Fink G R. Genetics. 1997;145:671–684. doi: 10.1093/genetics/145.3.671. [DOI] [PMC free article] [PubMed] [Google Scholar]