Abstract
The recent ability to sequence whole genomes allows ready access to all genetic material. The approaches outlined here allow automated analysis of sequence for the synthesis of optimal primers in an automated multiplex oligonucleotide synthesizer (AMOS). The efficiency is such that all ORFs for an organism can be amplified by PCR. The resulting amplicons can be used directly in the construction of DNA arrays or can be cloned for a large variety of functional analyses. These tools allow a replacement of single-gene analysis with a highly efficient whole-genome analysis.
The genome sequencing projects have generated and will continue to generate enormous amounts of sequence data. The genomes of Saccharomyces cerevisiae, Escherichia coli, Haemophilus influenzae (1), Mycoplasma genitalium (2), and Methanococcus jannaschii (3) have been completely sequenced. Other model organisms have had substantial portions of their genomes sequenced as well, including the nematode Caenorhabditis elegans (4) and the small flowering plant Arabidopsis thaliana (5). This massive and increasing amount of sequence information allows the development of novel experimental approaches to identify gene function.
One standard use of genome sequence data is to attempt to identify the functions of predicted open reading frames (ORFs) within the genome by comparison to genes of known function. Such a comparative analysis of all ORFs to existing sequence data is fast, simple, and requires no experimentation and is therefore a reasonable first step. While finding sequence homologies/motifs is not a substitute for experimentation, noting the presence of sequence homology and/or sequence motifs can be a useful first step in finding interesting genes, in designing experiments and, in some cases, predicting function. However, this type of analysis is frequently uninformative. For example, over one-half of new ORFs in S. cerevisiae have no known function (6). If this is the case in a well studied organism such as yeast, the problem will be even worse in organisms that are less well studied or less manipulable. A large, experimentally determined gene function database would make homology/motif searches much more useful.
Experimental analysis must be performed to thoroughly understand the biological function of a gene product. Scaling up from classical “cottage industry” one-gene-oriented approaches to whole-genome analysis would be very expensive and laborious. It is clear that novel strategies are necessary to efficiently pursue the next phase of the genome projects—whole-genome experimental analysis to explore gene expression, gene product function, and other genome functions. Model organisms, such as S. cerevisiae, will be extremely important in the development of novel whole-genome analysis techniques and, subsequently, in improving our understanding of other more complex and less manipulable organisms.
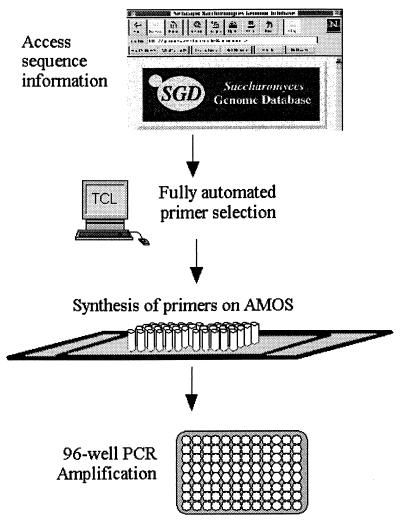
The genome sequence can be systematically used as a tool to understand ORFs, gene product function, and other genome regions. Toward this end, a directed strategy has been developed for exploiting sequence information as a means of providing information about biological function (Fig. 1). Efforts have been directed toward the amplification of each predicted ORF or any other region of the genome ranging from a few base pairs to several kilobase pairs. There are many uses for these amplicons—they can be cloned into standard vectors or specialized expression vectors, or can be cloned into other specialized vectors such as those used for two-hybrid analysis. The amplicons can also be used directly by, for example, arraying onto glass for expression analysis, for DNA binding assays, or for any direct DNA assay (7). As a pilot study, synthetic primers were made on the 96-well automated multiplex oligonucleotide synthesizer (AMOS) instrument (8) (Fig. 2). These oligonucleotides were used to amplify each ORF on yeast chromosome V. The current version of this instrument can synthesize three plates of 96 oligonucleotides each (25 bases) in an 8-hr day. The amplification of the entire set of PCR products was then analyzed by gel electrophoresis (Fig. 3). Successful amplification of the proper length product on the first attempt was 95%. This project demonstrates that one can go directly from sequence information to biological analysis in a truly automated, totally directed manner.
Figure 1.
Overview of systematic method for isolating individual genes. Sequence information is obtained automatically from sequence databases. The data are input into primer selection software specifically designed to target ORFs as designated by database annotations. The output file containing the primer information is directly read by a high-throughput oligonucleotide synthesizer, which makes the oligonucleotides in 96-well plates (AMOS, automated multiplex oligonucleotide synthesizer). The forward and reverse primers are synthesized in the same location on separate plates to facilitate the downstream handling of primers. The amplicons are generated by PCR in 96-well plates as well.
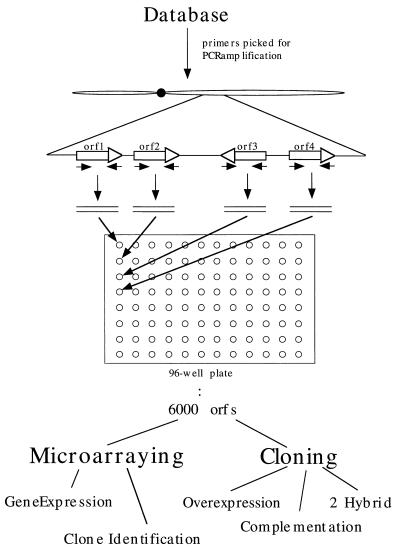
Figure 2.
Overall approach for using database of a genome to direct biological analysis. The synthesis of the 6,000 ORFs (orfs) for each gene of S. cerevisiae can be used in many applications utilizing both cloning and microarraying technology.
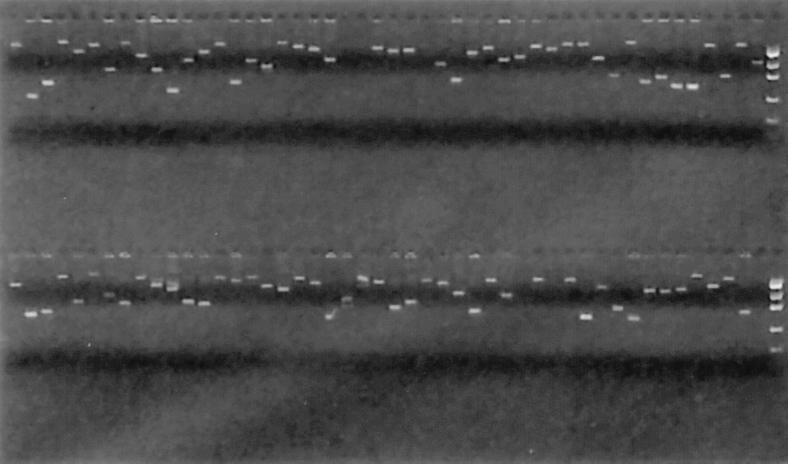
Figure 3.
Gel image of amplifications. Using the method described in Fig. 1, amplicons were generated for ORFs of S. cerevisiae chromosome V. One plate of 96 amplification reactions is shown.
These amplicons can be incorporated directly in arrays or the amplicons can be cloned. If the amplicons are to be cloned, novel sequences can be incorporated at the 5′ end of the oligonucleotide to facilitate cloning. One potential problem with cloning PCR products is that the cloned amplicons may contain sequence alterations that diminish their utility. One option would be to resequence each individual amplicon. However, this is expensive, inefficient, and time consuming. A faster, more cost-effective, and more accurate approach is to apply comparative sequencing by denaturing HPLC (9). This method is capable of detecting a single base change in a 2-kb heteroduplex. Longer amplicons can be analyzed by use of appropriate restriction fragments. If any change is detected in a clone, an alternate clone of the same region can be analyzed. Modifying the system to allow high throughput analysis by denaturing HPLC is also relatively simple and straightforward.
If amplicons are used directly on arrays without cloning, it is important to note that, even if single PCR product bands are observed on gels, the PCR products will be contaminated with various amounts of other sequences. This contamination has the potential to affect the results in, for example, expression analysis. On the other hand, direct use of the amplicons is much less labor intensive and greatly decreases the occurrence of mistakes in clone identification, a ubiquitous problem associated with large clone set archiving and retrieving.
Any large-scale effort to capture each ORF within a genome must rely on automation if cost is to be minimized while efficiency is maximized. Toward that end, primers targeting ORFs were designed automatically using simple new scripts and existing primer selection software. These script-selected primer sequences were directly read by the high-throughput synthesizer and the forward and reverse primers were synthesized in separate plates in corresponding wells to facilitate automated pipetting and PCR amplifications. Each of the resulting PCR products, generated with minimum labor, contains a known, unique ORF.
Large-scale genome analysis projects are dependent on newly emerging technologies to make the studies practical and economically feasible. For example, the cost of the primers, a significant issue in the past, has been reduced dramatically to make feasible this and other projects that require tens of thousands of oligonucleotides. Other methods of high-throughput analysis are also vital to the success of functional analysis projects, such as microarraying and oligonucleotide chip methods (10–14).
Changes in attitude are also required. One of the major costs of commercial oligonucleotides is extensive quality control such that virtually 100% of the supplied oligonucleotides are successfully synthesized and work for their intended purpose. Considerable cost reduction can be obtained by simply decreasing the expected successful synthesis rate to 95–97%. One can then achieve faster and cheaper whole genome coverage by simply adding a single quality control at the end of the experiment and batching the failures for resynthesis.
The directed nature of the amplicon approach is of clear advantage. The sequence of each ORF is analyzed automatically, and unique specific primers are made to target each ORF. Thus, there is relatively little time or labor involved—for example, no random cloning and subsequent screening is required because each product is known. In the test system, primers for 240 ORFs from chromosome V were systematically synthesized, beginning from the left arm and continuing through to the right arm. At no point was there any manual analysis of sequence information to generate the collection. In many ways, now that the sequence is known, there is no need for the researcher to examine it.
These amplicons can be arrayed and expression analysis can be done on all arrayed ORFs with a single hybridization (10). Those ORFs that display significant differential expression patterns under a given selection are easily identified without the laborious task of searching for and then sequencing a clone. Once scaled up, the procedure provides even greater returns on effort, because a single hybridization will ultimately provide a “snapshot” of the expression of all genes in the yeast genome. Thus, the limiting factor in whole genome analysis will not be the analysis process itself, but will instead be the ability of researchers to design and carry out experimental selections.
Current expression and genetic analysis technologies are geared toward the analysis of single genes and are ill suited to analyze numerous genes under many conditions. Additional difficulties with current technologies include: the effort and expense required to analyze expression and make mutants, the potential duplication of effort if done by different laboratories, and the possibility of conflicting results obtained from different laboratories. In contrast, whole genome analysis not only is more efficient, it also provides data of much higher quality; all genes are assayed and compared in parallel under exactly the same conditions. In addition, amplicons have many applications beyond gene expression. For example, one recent approach is to incorporate a unique DNA sequence tag, synthesized as part of each gene specific primer, during amplification. The tags or molecular bar codes, when reintroduced into the organism as a gene deletion or as a gene clone, can be used much more efficiently than individual mutations or clones because pools of tagged mutants or transformants can be analyzed in parallel. This parallel analysis is possible because the tags are readily and quantitatively amplified even in complex mixtures of tags (13).
These ORF genome arrays and oligonucleotide tagged libraries can be used for many applications. Any conventional selection applied to a library that gives discrete or multiple products can use these technologies for a simple direct readout. These include screens and selections for mutant complementation, overexpression suppression (15, 16), second-site suppressors, synthetic lethality, drug target overexpression (17), two-hybrid screens (18), genome mismatch scanning (19), or recombination mapping.
The genome projects have provided researchers with a vast amount of information. These data must be used efficiently and systematically to gain a truly comprehensive understanding of gene function and, more broadly, of the entire genome which can then be applied to other organisms. Such global approaches are essential if we are to gain an understanding of the living cell. This understanding should come from the viewpoint of the integration of complex regulatory networks, the individual roles and interactions of thousands of functional gene products, and the effect of environmental changes on both gene regulatory networks and the roles of all gene products. The time has come to switch from the analysis of a single gene to the analysis of the whole genome.
Acknowledgments
Support was provided by National Institutes of Health Grants R37H60198 and P01H600205.
References
- 1.Fleischmann R D, Adams M D, White O, Clayton R A, Kirkness E F, et al. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 2.Fraser C M, Gocayne J D, White O, Adams M D, Clayton R A, et al. Science. 1995;270:397–403. doi: 10.1126/science.270.5235.397. [DOI] [PubMed] [Google Scholar]
- 3.Bult C J, White O, Olsen G J, Zhou L, Fleischmann R D, et al. Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- 4.Sulston J, Du Z, Thomas K, Wilson R, Hillier L, Staden R, Halloran N, Green P, Thierry-Mieg J, Qiu L, Dear S, Coulson A, Craxton M, Durbin R, Berks M, Metzstein M, Hawkins T, Ainscough R, Waterston R. Nature (London) 1992;356:37–41. doi: 10.1038/356037a0. [DOI] [PubMed] [Google Scholar]
- 5.Newman T, de Bruijn F J, Green P, Keegstra K, Kende H, et al. Plant Physiol. 1994;106:1241–1255. doi: 10.1104/pp.106.4.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oliver S. Nature (London) 1996;379:597–600. doi: 10.1038/379597a0. [DOI] [PubMed] [Google Scholar]
- 7.Lashkari, D. A. (1996) Ph.D. dissertation (Stanford Univ., Stanford, CA).
- 8.Lashkari D A, Hunicke-Smith S P, Norgren R M, Davis R W, Brennan T. Proc Natl Acad Sci USA. 1995;92:7912–7915. doi: 10.1073/pnas.92.17.7912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oefner P J, Underhill P A. Am J Hum Genet. 1995;57:A266. [Google Scholar]
- 10.Schena M, Shalon D, Davis R W, Brown P O. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- 11.Fodor S P, Read J L, Pirrung M C, Stryer L, Lu A T, Solas D. Science. 1991;251:767–773. doi: 10.1126/science.1990438. [DOI] [PubMed] [Google Scholar]
- 12.Chee M, Yang R, Hubbell E, Berno A, Huang X C, Stern D, Winkler J, Lockhart D J, Morris M S, Fodor S P. Science. 1996;274:610–614. doi: 10.1126/science.274.5287.610. [DOI] [PubMed] [Google Scholar]
- 13.Shoemaker D D, Lashkari D A, Morris D, Mittmann M, Davis R W. Nat Genet. 1996;14:450–456. doi: 10.1038/ng1296-450. [DOI] [PubMed] [Google Scholar]
- 14.Smith V, Chou K, Lashkari D, Botstein D, Brown P O. Science. 1996;274:2069–2074. doi: 10.1126/science.274.5295.2069. [DOI] [PubMed] [Google Scholar]
- 15.Magdolen V, Drubin D G, Mages G, Bandlow W. FEBS Lett. 1993;316:41–47. doi: 10.1016/0014-5793(93)81733-g. [DOI] [PubMed] [Google Scholar]
- 16.Ramer S W, Elledge S J, Davis R W. Proc Natl Acad Sci USA. 1992;89:11589–11593. doi: 10.1073/pnas.89.23.11589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rine J, Hansen W, Hardeman E, Davis R W. Proc Natl Acad Sci USA. 1983;80:6750–6754. doi: 10.1073/pnas.80.22.6750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fields S, Song O. Nature (London) 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 19.Nelson S F, McCusker J H, Sander M A, Kee Y, Modrich P, Brown P O. Nat Genet. 1994;4:11–18. doi: 10.1038/ng0593-11. [DOI] [PubMed] [Google Scholar]