Abstract
Human cytomegalovirus (CMV), a herpesvirus that causes congenital disease and opportunistic infections in immunocompromised individuals, encodes functions that facilitate efficient viral propagation by altering host cell behavior. Here we show that CMV blocks apoptosis mediated by death receptors and encodes a mitochondria-localized inhibitor of apoptosis, denoted vMIA, capable of suppressing apoptosis induced by diverse stimuli. vMIA, a product of the viral UL37 gene, inhibits Fas-mediated apoptosis at a point downstream of caspase-8 activation and Bid cleavage but upstream of cytochrome c release, while residing in mitochondria and associating with adenine nucleotide translocator. These functional properties resemble those ascribed to Bcl-2; however, the absence of sequence similarity to Bcl-2 or any other known cell death suppressors suggests that vMIA defines a previously undescribed class of anti-apoptotic proteins.
Apoptosis is an important innate antiviral defense that can result in aborted infection and the elimination of infected cells (1, 2). In response, many viruses encode proteins that suppress host cell apoptosis (1, 2). Suppression of apoptosis is believed to be critical for virus replication and in vivo pathogenesis. Some herpesviruses encode homologs of cellular regulators of apoptosis such as Bcl-2 and FLIP (1). Little is known about the role of apoptosis in infections by human cytomegalovirus (CMV), a herpesvirus widely spread in human populations and pathogenic in immunocompromised individuals. Infection by CMV of human fibroblasts protects these cells from apoptosis induced by an E1B19K-deficient adenovirus, providing evidence that CMV can suppress apoptosis (3). However, none of the proteins predicted to be encoded from the 230-kb CMV genome (4, 5) are homologous to known cellular or viral suppressors of cell death (e.g., the Bcl-2 family). Two CMV-encoded proteins, IE1 and IE2, have been reported to partially suppress apoptosis induced in HeLa cells by tumor necrosis factor α (TNF-α) (3), however the mechanisms by which these nuclear proteins may exert their effects are uncertain.
To better understand how CMV avoids inducing apoptosis, we constructed a genomic CMV DNA library in an expression vector and screened plasmid clones for the ability to rescue HeLa cells from Fas-mediated apoptosis. Here we report the identification of UL37 as a potent anti-apoptotic gene of CMV, a function for a viral gene previously implicated to have an important role in CMV replication (6).
Experimental Procedures
Cells and Viruses.
Human MRC-5 fibroblasts (used between passages 20 and 27), HeLa cells, and CMV(AD169) were purchased from the American Type Culture Collection. 293T cells were a gift from G. Nolan (Stanford University). Cells were cultured in DMEM supplemented with 10% fetal bovine serum. CMV(Towne-RIT), a subclone of CMV(Towne), was obtained from S. Plotkin as vaccine lot 131 from RIT (7). The adenovirus mutant Ad2dl250 (8) was a gift from G. Chinnadurai (St. Louis University). HeLa/Bcl-xL cells were generated by amphotropic retroviral transduction (9) of FLAG-tagged Bcl-xL into HeLa cells with subsequent cloning.
Plasmids.
Mammalian expression vectors pcDNA3, pcR3.1-Uni, pZeoSV2(+), pcDNA3.1GS/human ANT-1, and pcDNA3.1/LacZV5His6 were purchased from Invitrogen. E1B19K/pRC/CMV was a gift from G. Chinnadurai. myc-tagged baculovirus p35 (a gift from M. D. Jacobson, University College, London) was subcloned in pcDNA3. Expression plasmids encoding immediate early CMV proteins IE1491aa and IE2579aa, pON2205 and pON2206, were described previously (10). Expression of IE1 and IE2 in transfected HeLa cells was confirmed by Western blot analysis. pcDNA3myc, a derivative of pcDNA3, contains in its polylinker section a DNA sequence encoding three tandem copies of the human c-myc epitope for fusion at the carboxyl terminus of proteins. The coding region for pUL37x1 was generated by PCR from genomic CMV(AD169) DNA and cloned into pCR3.1-Uni and pcDNA3myc. cDNA clones for gpUL37M and gpUL37 were generated by PCR from cDNA prepared from CMV(AD169)-infected cells (27 h after infection) and cloned into pCR3.1-Uni. The pUL37x1Δ2–23 mutant was constructed in pcDNA3myc. The fidelity of all clones was confirmed by DNA sequencing.
Cell Death/Apoptosis Assays.
TNFR-1-mediated apoptosis was induced by exposure of cells to recombinant human TNF-α (Sigma, 10–60 ng/ml) + cycloheximide (CHX; Sigma, 10–30 μg/ml). Fas-mediated apoptosis was induced by exposure of cells to the 7C11 anti-Fas antibody (Coulter, 0.2–4 μg/ml) + CHX (10–30 μg/ml). Dying cells detached from the substratum and then disintegrated into small fragments, while surviving cells remained flat and attached to the substratum. The degree of cell death was determined by counting the number of surviving cells in representative fields under a phase-contrast microscope 12–24 h after the start of the treatment.
Construction and Screening of a CMV Genomic DNA Library.
Cosmids containing segments of the CMV (AD169) genome, pCM1007, pCM1015, pCM1017, pCM1029, pCM1035, pCM1039, pCM1040, pCM1052, pCM1058, pCM1072 (11), and pON2601, a cosmid containing sequences unique to CMV(Toledo) (12) were mixed in equal amounts and partially digested with Sau3AI, and fragments of 2–5 kb were ligated into the BamHI site of pZeoSV2(+). Library complexity was 3 × 105 colonies with an average insert size of 2.1 kb; 212 primary pools of approximately 500 colonies per pool were generated from the library. Plasmid DNA from each pool (0.7 μg per sample) was mixed with pCMVβ (CLONTECH, 0.3 μg), and then 5 × 104 HeLa cells per well in 12-well plates were transfected with the mixtures by the SuperFect protocol (Qiagen). One day after transfection, cells were exposed to anti-Fas (0.4 μg/ml) + CHX (10 μg/ml) for an additional 24 h. Cell-associated β-galactosidase (β-gal) was detected by an ELISA (Boehringer Mannheim). Individual plasmids with anti-apoptotic activity were isolated from pools by repeated subdivision and β-gal transfection assays.
Immunofluorescence and Electron Microscopy.
For immunofluorescence, cells were fixed with 2% paraformaldehyde in PBS, permeabilized by adding cold methanol, rehydrated in PBS, and then processed in succession with a primary antibody [either 9E10 antibody or rabbit antiserum raised against a synthetic peptide corresponding to the carboxyl-terminal 22 amino acids of pUL37x1 (generated at Bio-Synthesis, Lewisville, TX)] and a secondary antibody plus Hoechst 33258 (Sigma). For MitoTracker staining, cells were incubated in MitoTracker Red (Molecular Probes) prior to fixation. For counterstaining with human anti-mitochondrial autoantiserum (ImmunoVision, Springdale, AR), the antiserum was added simultaneously with 9E10 antibody and was detected with Texas-Red-conjugated goat anti-human IgG mixed with the FITC-conjugated goat anti-mouse IgG used to detect the myc tag. For immunoelectron microscopy, cryosections of HeLa/pUL37x1#3 were stained first with 9E10 antibody and then with a secondary antibody–gold conjugate as described previously (13).
Protein and RNA Blots.
Cytochrome c was detected in S-100 fractions of cells (14). Cell extracts for Western blot analysis were prepared by standard procedures using either 150 mM NaCl/5 mM EDTA/50 mM Tris⋅HCl, pH 8.0/1% Triton X-100 [for detection of pUL37x1, procaspase-8, Bid, and poly(ADP-ribose) polymerase (PARP)] or 150 mM NaCl/50 mM Tris⋅HCl, pH 7.5/1% Triton X-100/1% sodium deoxycholate/0.1% SDS (for detection of procaspase-9), in the presence of protease inhibitors. Protein samples were separated under reducing conditions by SDS/PAGE after being loaded at equal protein amounts per well and were analyzed by a standard Western blot protocol using the ECL (enhanced chemiluminescence) detection system (Amersham). The following antibodies were used: anti-human Bid C-20 (Santa Cruz Biotechnology), 5F7 anti-human caspase-8 (Upstate Biotechnology), C210 anti-PARP (Biomol), 7H8.2C12 anti-cytochrome c and B40 anti-human caspase-9 (both from PharMingen), FITC-anti-mouse IgG (Sigma), horseradish peroxidase (HRP)-anti-mouse IgG Fc (Pierce), HRP-anti-mouse IgG2b (Boehringer Mannheim), and HRP-anti-rabbit IgG and HRP-anti-mouse Ig (Amersham). Hybridization was carried out on blots of total cell RNA with riboprobes specific for UL37x1/UL38 (51805–52631) or exon 3 (50417–50837).
Protein Microsequencing.
Cells were lysed in 150 mM NaCl/5 mM EDTA/50 mM Tris⋅HCl, pH 8.0/1% Triton X-100, in the presence of protease inhibitors, and centrifuged at 10,000 × g at 4°C for 10 min. The supernatants were first cleared with ethanolamine-treated Affi-Prep 10 beads (Bio-Rad), then incubated with 9E10 antibody covalently linked to Affi-Prep 10 beads, and washed with the lysis buffer. Proteins were eluted from beads in nonreducing Laemmli sample buffer, and then separated by SDS/PAGE under reducing conditions. Bands unique for immunoprecipitates from cells expressing viral mitochondria-localized inhibitor of apoptosis (vMIA) were isolated, and sequence analysis was performed at the Harvard Microchemistry Facility (William S. Lane) by microcapillary reverse-phase HPLC nano-electrospray tandem mass spectrometry on a Finnigan LCQ quadrupole ion trap mass spectrometer.
Results
To develop a rapid assay for CMV genes that inhibit cell death, we first tested whether human fibroblasts, which are rapidly killed by either anti-Fas or TNF-α in the presence of CHX, change in their sensitivity to apoptosis induced by these agents during productive infection with CMV(Towne-RIT). Immediately after virus adsorption (day 0) or at day 1 after infection, cells remained sensitive to apoptosis induced by either stimulus, but they became markedly resistant when treatment was initiated on day 2 or 3 of infection (Fig. 1a). These results extend previous reports showing that CMV-infected cells acquire resistance to apoptosis induced by serum withdrawal (15) or by infection with an E1B19K-deficient adenovirus (3).
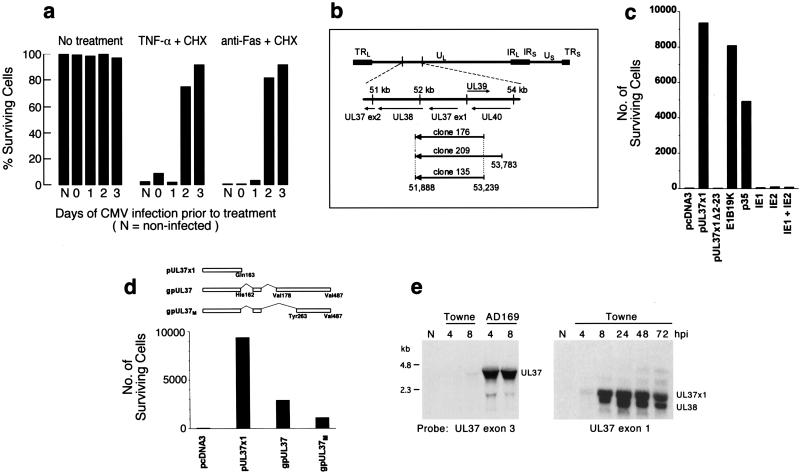
Figure 1.
Identification of a UL37 gene product as an inhibitor of apoptosis. (a) CMV-infected human fibroblasts acquire resistance to Fas- and TNF-α-mediated apoptosis. Cells were infected with CMV(Towne-RIT) (3 plaque-forming units per cell), or left uninfected (N). At the indicated times after infection, cells were exposed for an additional 12 h to medium alone (no treatment), TNF-α + CHX, or anti-Fas antibody + CHX. Viable cells remaining after these treatments were quantified by counting representative fields under a phase-contrast microscope. (b) CMV genomic sequences contained in plasmids with anti-apoptotic activity. (Upper) The structure of the CMV(AD169) genome (5) and UL37 region and associated ORFs are shown schematically. (Lower) CMV DNA inserts in plasmids isolated from three independent library pools (designated 176, 209, and 135). Nucleotide numbering is as described in ref. 4. Arrows indicate the orientation of inserts relative to the promoter in the expression vector. (c) pUL37x1 suppresses Fas-mediated apoptosis in transfected HeLa cells. Cells were transiently transfected with expression plasmids (1 μg) coding for the following proteins: pUL37x1myc, pUL37x1Δ2–23myc, E1B19K, baculovirus p35, CMV IE1491aa, CMV IE2579aa, or a 1:1 mixture of the plasmids containing IE1 and IE2, or they were transfected with an empty vector (pcDNA3). Twenty-four hours after transfection, the cells were exposed to anti-Fas + CHX for an additional 24 h, and surviving cells were scored under a microscope. (d) (Upper) Structures of pUL37x1, gpUL37M, and gpUL37 produced by alternative splicing. The amino acid numbers used in gpUL37M correspond to those in gpUL37. (Lower) Protection against Fas-mediated apoptosis by pUL37x1myc, gpUL37M, and gpUL37. HeLa cells were transfected with 1 μg of plasmid DNA expressing the indicated proteins and assayed as in c. (e) RNA blot analysis of UL37 exon 3 region transcripts expressed by CMV(AD169) and CMV(Towne-RIT) at 4 and 8 h after infection (Left) or UL37 exon 1 region transcripts expressed by CMV(Towne-RIT) at 4, 8, 24, 48, and 72 h after infection (Right).
To identify CMV genes that suppress apoptosis induced by Fas ligation, we established a cell survival assay in which cell viability after treatment with anti-Fas plus CHX correlated with the level of expression of a transfected β-gal indicator. An expression library prepared from fragmented CMV(AD169) DNA was divided into pools and screened for the ability to block apoptosis induced by anti-Fas antibody plus CHX in transfected HeLa cells in this survival assay. Three active pools were identified, and one plasmid with strong anti-apoptotic activity was isolated from each pool (plasmids 135, 176, and 209). The DNA sequence of the CMV DNA inserts in these three plasmids, two of which (nos. 135 and 176) were identical, spanned a region of the CMV genome that includes the entire exon 1 of the UL37 gene (Fig. 1b). UL37 exon 1 encodes a 163-amino acid protein, pUL37x1, previously characterized as an immediate early transactivator (6, 16, 17).
To definitively assign the anti-apoptotic activity detected in these isolated clones to pUL37x1, a DNA segment encoding just the pUL37x1 ORF was introduced into a mammalian expression vector. pUL37x1 protected HeLa cells against Fas-mediated apoptosis in transient transfection assays when expressed either as an unmodified protein (not shown) or as a carboxyl-terminal c-myc-epitope-tagged protein (Fig. 1c). However, deletion of amino acids 2–23 of pUL37x1myc abrogated all protective function (Fig. 1c), indicating that these amino-terminal residues are essential for activity. pUL37x1 inhibited TNF-α-induced apoptosis as well (not shown). Protection was found to be at least as great as that provided by anti-apoptotic proteins from adenovirus (E1B19K) or baculovirus (p35) (Fig. 1c). IE1491aa and IE2579aa, two CMV gene products previously reported to suppress apoptosis in some settings (3), did not suppress anti-Fas or TNF-α-induced apoptosis in our assays.
In addition to pUL37x1, which is expressed from a 1.7-kb unspliced mRNA (16), alternative splicing and polyadenylation of CMV(AD169) UL37 RNA produces a 3.4-kb transcript encoding a 487-amino acid glycoprotein, gpUL37 (18, 19). To test whether this larger UL37 protein can inhibit apoptosis, a cDNA corresponding to the gpUL37 ORF was isolated from CMV(AD169)-infected fibroblasts and inserted into a mammalian expression vector (Fig. 1d). In the process of cloning gpUL37, we isolated a previously undescribed UL37 splice variant, encoding a smaller protein that we designated gpUL37M (medium) (Fig. 1d). Both gpUL37 and gpUL37M exhibited anti-apoptotic activity when tested for their ability to protect transiently transfected HeLa cells against Fas-mediated apoptosis, although neither was as potent as pUL37x1 (Fig. 1d). The first 162 amino-terminal amino acids of pUL37x1 are identical in all UL37 proteins, suggesting that all inhibit apoptosis through this common region.
To determine which UL37 product may play a role in suppression of apoptosis during CMV infection, expression of transcripts from the UL37 region was examined in fibroblasts infected with two virus strains, CMV(Towne-RIT) and CMV(AD169). RNA blot hybridization with exon 1- and exon 3-specific probes demonstrated that both strains expressed the unspliced 1.7-kb UL37 exon 1 mRNA, from 4 h after infection onward (Fig. 1e). Interestingly, CMV(Towne-RIT) did not express the large multiply spliced gpUL37M or gpUL37 mRNA (Fig. 1e), the latter previously characterized as an immediate early transcript in CMV(AD169)-infected cells (18, 20). Thus, pUL37x1 is the only UL37 gene product that appears in CMV(Towne-RIT)-infected cells, suggesting that it is sufficient to block apoptosis during infection with this strain (Fig. 1a).
To further characterize the anti-apoptotic function of pUL37x1, three HeLa cell clones were isolated. Data obtained using two of these clones designated HeLa/pUL37x1myc #3 and #8 are shown in Fig. 2. Western blot analysis with a polyclonal anti-pUL37x1 antibody demonstrated that the level of pUL37x1 expression was significantly higher in CMV(Towne-RIT)-infected fibroblasts (48 h) than in these transfected HeLa cell lines (not shown). In all three cell lines, a majority of pUL37x1myc-expressing cells survived after incubation with anti-Fas or TNF-α in the presence of CHX (Fig. 2b and data not shown), whereas very few cells in control clones isolated after transfection of the empty vector survived either treatment. Treatment of cells with CHX alone did not induce significant levels of apoptosis (Fig. 2b), and the cell surface expression of Fas in pUL37x1-expressing cells was similar to that in control cells (Fig. 2a). HeLa cells expressing pUL37x1 were resistant to apoptosis triggered by infection with the E1B19K-deficient adenovirus mutant Ad2dl250 (Fig. 2c), an apoptosis stimulus previously reported to be inhibited at late times during CMV infection (3), and were resistant to apoptosis induced by the anti-cancer drug doxorubicin (Fig. 2d). Thus, pUL37x1 is a broadly acting inhibitor of apoptosis that can suppress cell death induced by a variety of cytotoxic agents.
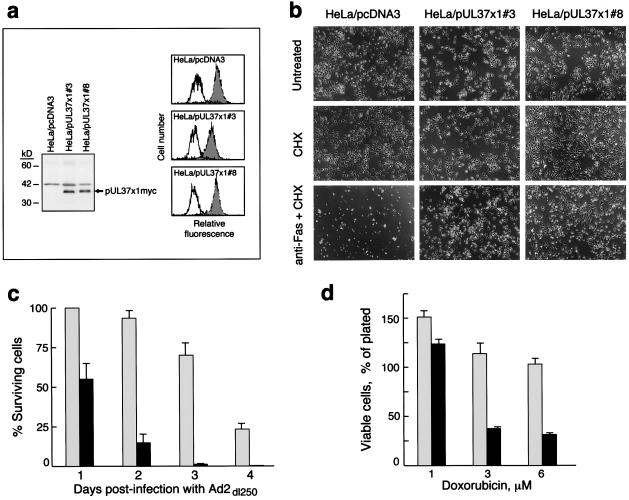
Figure 2.
HeLa cells constitutively expressing pUL37x1myc are resistant to apoptosis. (a) (Left) Western blot analysis of pUL37x1myc expression in stably transfected HeLa cells with the 9E10 anti-myc antibody. (Right) Expression of cell surface Fas on HeLa clones. Fas levels were examined on cells by flow cytometry, after staining with anti-Fas antibody and a secondary FITC-conjugated antiserum (filled histograms). In control samples, the primary antibody was omitted (open histograms). (b) Resistance of pUL37x1myc HeLa clones to Fas-mediated apoptosis. Cells were exposed to anti-Fas antibody plus CHX, or to CHX only, or to medium alone (untreated), and photographed under a phase-contrast microscope 24 h later. (c) Resistance of pUL37x1myc HeLa clones to apoptosis induced by E1B19K-deficient adenovirus. Cells were infected with Ad2dl250 (3 plaque-forming units per cell), and surviving cells were counted under a microscope at the indicated times. Gray bars, HeLa/pUL37x1myc#3; black bars, HeLa/pcDNA3-A control cells (±SEM, n = 2). (d) Inhibition of doxorubicin-induced apoptosis in pUL37x1myc HeLa cells. Cells were exposed to doxorubicin at the indicated concentrations for 24 h, and the viable cells (as determined by trypan blue exclusion) were counted (±SEM, n = 2; legend as for c).
Immunofluorescence analysis of transiently transfected cells (Fig. 3a) and HeLa/pUL37x1myc cells (not shown) demonstrated that pUL37x1myc localized predominantly to mitochondria, as confirmed by its colocalization with a human anti-mitochondrial autoimmune serum (Fig. 3 b and c) and MitoTracker dye (not shown). The pUL37x1Δ2–23myc mutant exhibited a markedly altered staining pattern with some plasma membrane localization (Fig. 3d) and did not colocalize with mitochondrial markers, indicating that the amino-terminal 23-amino acid segment of pUL37x1 is necessary for both anti-apoptotic activity and mitochondrial targeting. Immunoelectron microscopy of HeLa/pUL37x1myc cells further revealed that pUL37x1myc was associated mainly with the outer mitochondrial membrane (two representative fields are shown in Fig. 3 e and f). The broad anti-apoptotic activity of pUL37x1 and its subcellular localization led us to denote this protein as viral mitochondrial inhibitor of apoptosis (vMIA).
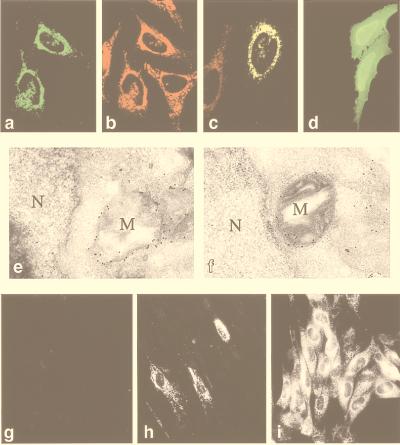
Figure 3.
Mitochondrial localization of pUL37x1 (vMIA). HeLa cells were transiently transfected with plasmids expressing either pUL37x1myc (a–c) or pUL37x1Δ2–23myc (d). The cells were permeabilized and stained with either 9E10 anti-myc antibody (a, c, and d, green fluorescence) or anti-mitochondrial antiserum (b, and c, red fluorescence). The yellow coloring (c) resulted from superimposition of red and green fluorescence. Control cells transiently transfected with pcDNA3 did not stain with the 9E10 antibody (not shown). (e and f) Immunoelectron microscopy of ultrathin cryosections of HeLa/pUL37x1myc #3 cells. Cryosections were stained first with 9E10 antibody and then with a secondary antibody-gold conjugate. Two representative fields are shown (M, mitochondria; N, nuclei). HeLa/pcDNA3-A control cells stained with the 9E10 antibody did not reveal specific mitochondrial labeling (not shown). (g–i) Time course of vMIA expression in MRC-5 fibroblasts infected with CMV(Towne-RIT). Noninfected cells (g), or cells at 1 day (h) or 2 days (i) after CMV infection were permeabilized and stained with a polyclonal anti-vMIA antibody. No immunofluorescent staining was observed with preimmune serum (not shown).
vMIA produced by CMV in the context of productive infection also localized to mitochondria of infected cells, as demonstrated by immunofluorescence analysis with a polyclonal anti-vMIA antibody (Fig. 3 g–i) and costaining with a human anti-mitochondrial serum (not shown). The time course of vMIA expression in CMV(Towne-RIT)-infected fibroblasts was examined by immunofluorescence (Fig. 3 g–i). At 1 day after infection, vMIA was detected in less than 10% of cells (Fig. 3h); however, by 2 days after infection more than 70% of cells expressed vMIA (Fig. 3i). Thus, the kinetics of vMIA protein expression and accumulation in mitochondria of CMV-infected cells coincides with the development of resistance to apoptosis (Fig. 1a), and it further supports a causative role for vMIA in protection of CMV-infected cells from apoptosis.
We examined the impact of vMIA on biochemical events involved in Fas-mediated apoptosis by comparing control HeLa/pcDNA3 cells, which are sensitive to anti-Fas antibody plus CHX, to HeLa clones expressing either vMIA (pUL37x1) or cellular Bcl-xL, both of which are resistant to Fas-mediated apoptosis. Ligation of Fas triggers the recruitment and activation of caspase-8 through the adapter molecule FADD (21). This process was not impaired by vMIA, since procaspase-8 was processed in vMIA-expressing cells similarly to its processing in either Bcl-xL-expressing cells, or control cells within 4 h of the exposure to anti-Fas antibody plus CHX, but not CHX alone (Fig. 4a). Fas signaling is connected to mitochondrial events in apoptosis through Bid, a pro-apoptotic BH3 protein that is proteolyzed by caspase-8 and subsequently translocates to mitochondria, where it triggers cytochrome c release (22, 23). Exposure to anti-Fas antibody plus CHX triggered cleavage of Bid in both control and vMIA-expressing cells, demonstrating that the proteolysis of Bid was not affected by vMIA (Fig. 4b). Li et al. (22) reported that while Bcl-xL has weak affinity to unprocessed Bid, its expression did not prevent the proteolysis of Bid by caspase-8; however, Bid processing appeared to be inhibited in the HeLa/Bcl-xL cells used here as control (a second HeLa/Bcl-xL clone gave similar results). The reasons for this difference are not yet clear, but may reflect a dosage effect—e.g., sufficiently high levels of Bcl-xL may bind unprocessed Bid [to which it has weak affinity (22)], and hinder its cleavage by caspase-8.
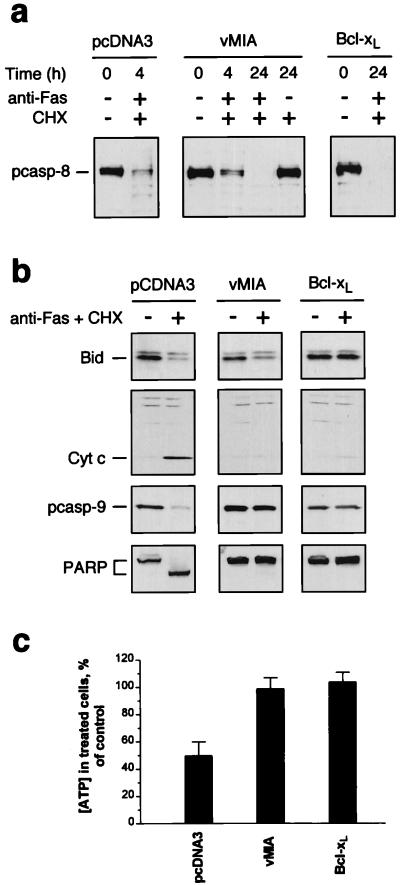
Figure 4.
Effects of vMIA (pUL37x1) expression on biochemical events associated with Fas-mediated apoptosis. (a) HeLa/pcDNA3 cells, HeLa/vMIA cells (pUL37x1myc #3), and HeLa/Bcl-xL cells were treated with CHX, or with anti-Fas antibody plus CHX, or were left untreated. Cell lysates were prepared at the indicated times and procaspase-8 (55 kDa) was detected by Western analysis. (b) Cells were treated with anti-Fas antibody plus CHX for 4 h (+) or were left untreated (−), and the indicated proteins were detected by Western analysis. Cytochrome c was detected in S-100 cell extracts. (c) Cells were treated with anti-Fas antibody plus CHX for 4 h, and the intracellular ATP concentrations were measured by an ATP bioluminescence assay (CLS II, Roche Molecular Biochemicals) and compared with those in control cells (cells exposed to CHX alone). The results are presented as the means ± SEM (n = 4).
In contrast to the inability of vMIA to prevent caspase-8 and Bid processing, several other downstream events in Fas-mediated apoptosis were inhibited in vMIA-expressing HeLa cells. After a 4-h exposure to anti-Fas antibody plus CHX, the control HeLa/pcDNA3 cells underwent efflux of mitochondrial cytochrome c into the cytoplasm, an event almost universally observed during apoptosis (24). Cytochrome c release was not observed, however, in either the vMIA- or Bcl-xL-expressing HeLa cells (Fig. 4b). Processing of procaspase-9, which depends on cytochrome c release, was also inhibited in vMIA- and Bcl-xL-expressing cells, as was cleavage of PARP (Fig. 4b), a substrate of downstream effector caspases. We also examined whether ATP levels in vMIA-expressing and control HeLa cells changed after their exposure to anti-Fas plus CHX (Fig. 4c). Whereas the concentration of ATP in HeLa/pcDNA3 cells dropped to 50% after a 4-h exposure to anti-Fas plus CHX, the ATP levels in vMIA- and Bcl-xL-expressing HeLa cells remained unchanged. Thus, vMIA interferes with Fas-mediated apoptotic signaling in HeLa cells at a point downstream of caspase-8 activation and Bid processing, but upstream of ATP depletion and cytochrome c release.
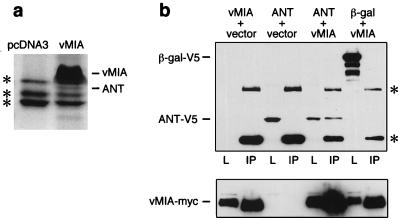
To identify candidate cellular target(s) of vMIA, we examined cellular proteins that coimmunoprecipitated with vMIA expressed in stably transfected cells. Lysates from vMIA-expressing (pUL37x1myc) and control cells were incubated with anti-myc antibody covalently immobilized on beads, and captured proteins were separated by SDS/PAGE. A 30-kDa band present only in immunoprecipitates from vMIA-expressing cells (Fig. 5a) was identified by protein microsequence analysis as mitochondrial adenine nucleotide translocator (ANT). ANT was significantly less abundant than vMIA in Coomassie blue-stained gels of the immunoprecipitates. The interaction between vMIA and ANT was confirmed in transient transfection assays which demonstrated that V5-epitope-tagged ANT-1 specifically co-immunoprecipitated with myc-tagged vMIA (Fig. 5b). Interestingly, ANT is a component of the mitochondrial permeability transition pore complex that interacts with Bax and other Bcl-2 family members (25). Recently it has been reported that Bcl-xL prevents ATP depletion in cells subjected to an apoptotic stimulus (26), suggesting that this protective effect of Bcl-xL may be explained by its interaction with ANT and facilitation of mitochondrial ATP/ADP exchange.
Figure 5.
Association of vMIA with adenine nucleotide translocator (ANT). (a) Lysates from cells constitutively expressing myc-tagged vMIA, or from control cells (pcDNA3 transfected) were immunoprecipitated with the anti-myc antibody 9E10 covalently linked to Affi-Prep-10 beads. Bound proteins were separated by SDS/PAGE and stained with colloidal Coomassie blue (Novex). The bands designated vMIA and ANT were identified by microsequencing. Asterisks indicate nonspecific bands. (b) (Upper) 293T cells were transiently cotransfected as indicated with expression plasmids (0.5 μg each) coding for vMIAmyc, V5-tagged ANT-1, V5-tagged β-gal, or the empty vector. vMIA was then immunoprecipitated (lanes designated IP) with anti-myc antibody and interacting proteins were detected by Western blot analysis with anti-V5 antibody (Invitrogen). Lanes designated L are samples of the transfected cell lysate taken prior to immunoprecipitation. Asterisks indicate bands of antibody heavy and light chains. (Lower) The blot was reprobed with anti-myc to confirm expression of vMIA-myc.
Discussion
The predominant localization of vMIA to mitochondria, physical interaction with ANT, and impact on signaling events that occur in Fas-mediated cell death argue for a mitochondrial mode of action for vMIA in suppressing apoptosis. vMIA suppresses, either directly or indirectly, intracellular ATP depletion and mitochondrial efflux of cytochrome c into the cytoplasm. This point of action at mitochondria, and the ability of vMIA to inhibit apoptosis induced by diverse agents, are functional properties analogous to those of Bcl-2 (25, 27). For both vMIA and Bcl-2, the precise molecular mechanism by which cytochrome c release is blocked has not been elucidated. In particular, it is unclear whether any of these proteins prevent apoptosis by modulating the functional activity of ANT (26), or use ANT as a mitochondrial docking protein. It thus remains possible that vMIA suppresses apoptosis in a manner functionally related to Bcl-2. However, vMIA does not share any notable homology with Bcl-2 and lacks sequences similar to the BH1, BH2, BH3, or BH4 domains characteristic of known Bcl-2 family members. Consistent with the lack of homology to Bcl-2, vMIA does not appear to bind to Bax either in vitro or in cotransfection assays (not shown). This suggests that vMIA may be a biochemically distinct suppressor of apoptosis that may have evolved to efficiently stabilize mitochondrial membrane function. Although searches of databases do not as yet reveal cellular homologs, such homologs may emerge as the human genome sequence nears completion and the functional domains of vMIA are clarified by genetic and biochemical analyses.
vMIA expression during CMV infection closely correlates with the acquired resistance of infected cells to apoptosis. Together with our observations that vMIA functions as a potent inhibitor of apoptosis, these data help to explain the ability of infected cells to support productive CMV replication over prolonged periods in the absence of cell death. Previous studies have suggested an important role for UL37 gene products in both viral replication and transcriptional activation (reviewed in ref. 6), activities that may derive at least in part from the ability of these proteins to inhibit apoptosis. vMIA is likely to have a major role in suppressing apoptotic signals that may arise through the early metabolic changes in CMV-infected cells. Specifically, vMIA may counter the pro-apoptotic effects of CMV infection-induced TNF-α, c-myc, and p53 expression (28–30). Because of its broad anti-apoptotic effects, vMIA is also likely to protect cells from immune surveillance by conferring resistance to Fas-, TNFR-1-, or granzyme B-mediated apoptosis, major components of innate and adaptive immune responses (31). The identification of vMIA establishes a novel target for anti-CMV drug development because agents that interfere with the anti-apoptotic function of vMIA would be predicted to abort infection at an early stage and also facilitate the immune destruction of infected cells that may have escaped innate host defenses.
Our results demonstrate that CMV infection provides resistance to apoptosis induced by anti-Fas or TNF-α, and that expression of vMIA is sufficient to protect cells from these and other apoptotic stimuli. It will now be important to investigate the precise role of vMIA at various stages of CMV infection in vitro and in vivo.
Acknowledgments
We are grateful to our colleagues at Apoptosis Technology, Inc., for support, reagents, and plasmids, and we acknowledge the contribution of Dr. A. Louise McCormick to hybridization analysis of CMV(Towne-RIT)-infected cell RNA samples. We thank Drs. Hartmut Land and Gerard Evan for helpful discussions, Dr. Garry Nolan for his advice and the gift of 293T cells, Dr. Yuhui Xu and Ms. Li Zhang for performing the electron microscopy experiments, Dr. Jerome Ritz for the gift of 7C11 antibody, Dr. G. Chinnadurai for the gift of Ad2dl250 and the plasmid E1B19K/pRC/CMV, Dr. Alan Kingsman for the components of the three-plasmid retrovirus-production system, and Dr. A. Colberg-Poley for critical reading of the manuscript.
Abbreviations
- CMV
human cytomegalovirus
- TNF-α
tumor necrosis factor α
- CHX
cycloheximide
- β-gal
β-galactosidase
- PARP
poly(ADP-ribose) polymerase
- vMIA
viral mitochondria-localized inhibitor of apoptosis
- ANT
adenine nucleotide translocator
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Tschopp J, Thome M, Hofmann K, Meinl E. Curr Opin Genet Dev. 1998;8:82–87. doi: 10.1016/s0959-437x(98)80066-x. [DOI] [PubMed] [Google Scholar]
- 2.O’Brien V. J Gen Virol. 1998;79:1833–1845. doi: 10.1099/0022-1317-79-8-1833. [DOI] [PubMed] [Google Scholar]
- 3.Zhu H, Shen Y, Shenk T. J Virol. 1995;69:7960–7970. doi: 10.1128/jvi.69.12.7960-7970.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chee M S, Bankier A T, Beck S, Bohni R, Brown C M, Cerny R, Horsnell T, Hutchison C A, Kouzarides T, Martignetti J A, et al. Curr Top Microbiol Immunol. 1990;154:125–169. doi: 10.1007/978-3-642-74980-3_6. [DOI] [PubMed] [Google Scholar]
- 5.Mocarski E S. In: Fields Virology. Fields B N, Knipe D M, editors. Philadelphia: Lippincott-Raven; 1996. pp. 2447–2492. [Google Scholar]
- 6.Colberg-Poley A M. Intervirology. 1996;39:350–360. doi: 10.1159/000150506. [DOI] [PubMed] [Google Scholar]
- 7.Plotkin S A, Higgins R, Kurtz J B, Morris P J, Campbell D A, Shope T C, Spector S A, Dankner W M. Transplantation. 1994;58:1176–1178. [PubMed] [Google Scholar]
- 8.Subramanian T, Kuppuswamy M, Gysbers J, Mak S, Chinnadurai G. J Biol Chem. 1984;259:11777–11783. [PubMed] [Google Scholar]
- 9.Soneoka Y, Cannon P M, Ramsdale E E, Griffiths J C, Romano G, Kingsman S M, Kingsman A J. Nucleic Acids Res. 1995;23:628–633. doi: 10.1093/nar/23.4.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jenkins D E, Martens C L, Mocarski E S. J Gen Virol. 1994;75:2337–2348. doi: 10.1099/0022-1317-75-9-2337. [DOI] [PubMed] [Google Scholar]
- 11.Fleckenstein B, Muller I, Collins J. Gene. 1982;18:39–46. doi: 10.1016/0378-1119(82)90054-3. [DOI] [PubMed] [Google Scholar]
- 12.Cha T-A, Tom E, Kemble G W, Duke G M, Mocarski E S, Spaete R R. J Virol. 1996;70:78–83. doi: 10.1128/jvi.70.1.78-83.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu Y, Slayter H S. J Histochem Cytochem. 1994;42:1365–1376. doi: 10.1177/42.10.7930519. [DOI] [PubMed] [Google Scholar]
- 14.Li P, Nijhawan D, Budihardjo I, Srinivasula S M, Ahmad M, Alnemri E S, Wang X. Cell. 1997;91:479–489. doi: 10.1016/s0092-8674(00)80434-1. [DOI] [PubMed] [Google Scholar]
- 15.Kovacs A, Weber M L, Burns L J, Jacob H S, Vercellotti G M. Am J Pathol. 1996;149:1531–1539. [PMC free article] [PubMed] [Google Scholar]
- 16.Colberg-Poley A M, Santomenna L D, Harlow P P, Benfield P A, Tenney D J. J Virol. 1992;66:95–105. doi: 10.1128/jvi.66.1.95-105.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Colberg-Poley A M, Huang L, Soltero V E, Iskenderian A C, Schumacher R F, Anders D G. Virology. 1998;246:400–408. doi: 10.1006/viro.1998.9212. [DOI] [PubMed] [Google Scholar]
- 18.Kouzarides T, Bankier A T, Satchwell S A, Preddy E, Barrell B G. Virology. 1988;165:151–164. doi: 10.1016/0042-6822(88)90668-x. [DOI] [PubMed] [Google Scholar]
- 19.Al-Barazi H O, Colberg-Poley A M. J Virol. 1996;70:7198–7208. doi: 10.1128/jvi.70.10.7198-7208.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tenney D J, Colberg-Poley A M. Virology. 1991;182:199–210. doi: 10.1016/0042-6822(91)90663-v. [DOI] [PubMed] [Google Scholar]
- 21.Ashkenazi A, Dixit V M. Science. 1998;281:1305–1308. doi: 10.1126/science.281.5381.1305. [DOI] [PubMed] [Google Scholar]
- 22.Li H, Zhu H, Xu C-j, Yuan J. Cell. 1998;94:491–501. doi: 10.1016/s0092-8674(00)81590-1. [DOI] [PubMed] [Google Scholar]
- 23.Luo X, Budihardjo I, Zou H, Slaughter C, Wang X. Cell. 1998;94:481–490. doi: 10.1016/s0092-8674(00)81589-5. [DOI] [PubMed] [Google Scholar]
- 24.Green D R, Reed J C. Science. 1998;281:1309–1312. doi: 10.1126/science.281.5381.1309. [DOI] [PubMed] [Google Scholar]
- 25.Marzo I, Brenner C, Zamzami N, Jurgensmeier J M, Susin S A, Vieira H L, Prevost M C, Xie Z, Matsuyama S, Reed J C, Kroemer G. Science. 1998;281:2027–2031. doi: 10.1126/science.281.5385.2027. [DOI] [PubMed] [Google Scholar]
- 26.Heiden M G V, Chandel N S, Schumacker P T, Thompson C B. Mol Cell. 1999;3:159–167. doi: 10.1016/s1097-2765(00)80307-x. [DOI] [PubMed] [Google Scholar]
- 27.Adams J M, Cory S. Science. 1998;281:1322–1326. doi: 10.1126/science.281.5381.1322. [DOI] [PubMed] [Google Scholar]
- 28.Smith P D, Saini S S, Raffeld M, Manischewitz J F, Wahl S M. J Clin Invest. 1992;90:1642–1648. doi: 10.1172/JCI116035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Geist L J, Monick M M, Stinski M F, Hunninghake G W. J Clin Invest. 1994;93:474–478. doi: 10.1172/JCI116995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boldogh I, AbuBakar S, Deng C Z, Albrecht T. J Virol. 1991;65:1568–1571. doi: 10.1128/jvi.65.3.1568-1571.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smyth M J, Trapani J A. J Virol. 1998;72:1–9. doi: 10.1128/jvi.72.1.1-9.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]