Abstract
A cDNA from a novel Ca2+-dependent member of the mitochondrial solute carrier superfamily was isolated from a rabbit small intestinal cDNA library. The full-length cDNA clone was 3,298 nt long and coded for a protein of 475 amino acids, with four elongation factor-hand motifs located in the N-terminal half of the molecule. The 25-kDa N-terminal polypeptide was expressed in Escherichia coli, and it was demonstrated that it bound Ca2+, undergoing a reversible and specific conformational change as a result. The conformation of the polypeptide was sensitive to Ca2+ which was bound with high affinity (Kd ≈ 0.37 μM), the apparent Hill coefficient for Ca2+-induced changes being about 2.0. The deduced amino acid sequence of the C-terminal half of the molecule revealed 78% homology to Grave disease carrier protein and 67% homology to human ADP/ATP translocase; this sequence homology identified the protein as a new member of the mitochondrial transporter superfamily. Northern blot analysis revealed the presence of a single transcript of about 3,500 bases, and low expression of the transporter could be detected in the kidney but none in the liver. The main site of expression was the colon with smaller amounts found in the small intestine proximal to the ileum. Immunoelectron microscopy localized the transporter in the peroxisome, although a minor fraction was found in the mitochondria. The Ca2+ binding N-terminal half of the transporter faces the cytosol.
Peroxisomes and mitochondria are the two organelles present in mammalian eukaryotic cells for which an evolutionary endosymbiotic origin has been proposed (1, 2). Both organelles share a set of similar metabolic pathways, are the site of cellular oxygen consumption, and have similar macromolecular components and membrane phospholipid compositions (3). Mitochondria, in contrast to peroxisomes contain their own DNA and are surrounded by a double membrane rather than a single one. Despite the existence of mitochondrial DNA, most proteins are coded for by nuclear genes, synthesized on free cytosolic polysomes and subsequently posttranslationally sorted to their final mitochondrial location (for a review, see ref. 4). In principle, the same applies for peroxisomal proteins although much less is known about protein import in this organelle (for a review, see ref. 5).
The mitochondrial inner membrane contains three different major classes of proteins: the electron transport chain complex, the ATP synthase complex, and the mitochondrial solute carriers. The first two complexes originated at the prokariotic level whereas the mitochondrial solute carriers must have developed when the ancestor prokaryote became symbiotic in the eukaryotic cell as they fulfill a new demand of the eukaryote for intensive traffic of metabolites between the cytosolic and matrix space (6).
ATP-binding cassette transporters are located in the peroxisomal membrane (7, 8). Although permeability of the peroxisomal membrane for hydrophilic molecules up to 800 Da (9) was originally proposed, more recent work, performed under in vivo conditions, has questioned this (10) and implied the existence of transporters. Two prominent members of the ATP-binding cassette transporter family are the peroxisomal membrane protein of 70 kDa and the adrenoleukodystrophy protein (11, 12). Defects in both are known to be responsible for human metabolic diseases like Zellweger syndrome and X-linked adrenoleukodystrophy (11, 13). Another integral peroxisomal membrane protein in yeast (Candida boidinii), named Pmp47, has been identified as a member of the mitochondrial solute carrier family (14). The function of Pmp47 is related to oleate metabolism, but its loss causes a severe defect in the translocation and folding of a peroxisome matrix protein and finally in peroxisome proliferation (15). The fact that both organelles use solute carriers of the same superfamily underlines the close relationship between mitochondria and peroxisomes. Moreover it raises the possibility that a mutation in a solute carrier located in human peroxisomes could be the cause of a genetic disease with as severe effects as in Zellweger syndrome and adrenoleukodystrophy.
MATERIALS AND METHODS
Isolation of RNA.
RNA was isolated from frozen pulverized tissue of adult rabbit by the acid/guanidinium/thiocyanate method (16). Intestinal RNA was extracted from enterocytes, which were scraped from the luminal side of washed intestine with a microscope slide and frozen directly in liquid nitrogen. The PolyATract mRNA isolation system from Promega was used for the isolation of mRNA.
Construction and Screening of a cDNA Library from Enterocytes.
A directional cDNA library was made with poly(A)-enriched RNA from rabbit small intestinal enterocytes and the Superscript cDNA cloning system of Life Technologies (Paisley, Scotland). The library in the pSPORT vector contained about 1 × 106 independent clones with an average size of 2.6 kb.
The library was screened by colony hybridization using a nick-labeled rat SCP-II cDNA as probe (a generous gift of K. Wirtz, Centre for Biomembranes and Lipid Enzymology, Utrecht, The Netherlands). The final wash was performed with 0.1× NaCl/citrate at 65 C°. A 5′ extension of the cDNA, DNA sequencing, and sequence analysis was performed as described (17).
Expression and Purification of the N-Terminal Fragment.
Two primers and the full-length cDNA were used in a PCR to generate a DNA fragment of the N terminus of the Ca2+-dependent mitochondrial carrier according earlier work (18). After sequencing the fragment was cloned in NdeI/EcoRI-cut pET 28a(+) (Novagen) to produce the final construct with a His-tag at the N terminus. Escherichia coli strain BL21(DE3) was transfected with the final construct. The purification of the polypeptide was essentially done according to the recommendation of the manufacturer of the His-binding Kit (Novagen).
PAGE, 45Ca2+-Overlay Assay.
SDS/PAGE was performed on a 15% gel. After blotting onto a polyvinylidene fluoride membrane Ca2+-binding to the N-terminal fragment was analyzed with 0.1 mCi (1 Ci = 37 GBq) 45Ca2+ in an imidazole-HCl buffer (pH 6.8) according to Maruyama et al. (19).
Circular Dichroism (CD).
CD spectra were measured with a Jasco J-600 spectropolarimeter equipped with a thermostated cell block. Each spectrum was accumulated at least four times, and cells of path length of 0.05 cm for far-UV and 1 cm were used for near-UV measurements, respectively. The titration of mean residue ellipticity intensity of the Ca2+-dependent carrier with increasing concentrations of Ca2+ was performed in the presence of Ca2+/EGTA mixture, 100 mM KCl, and 10 mM Mops (pH 7.2) at 20°C (Calcium Calibration Buffer Kit I, Molecular Probes). Protein solutions were desalted on PD-10 columns (Pharmacia) equilibrated either with solution A (zero free calcium buffer) containing 10 mM EGTA, 100 mM KCl, 10 mM Mops (pH 7.2), or solution B (39.8 μM free calcium buffer) containing 10 mM Ca2+/EGTA, 100 mM KCl, 10 mM Mops (pH 7.2) (20). Identical protein concentrations of 0.13 mg/ml in both solutions were obtained by measuring the absorbance at 280 nm and diluting with the corresponding Ca2+ buffers. The free Ca2+ concentration was calculated as shown in the protocol provided by Molecular Probes. Calcium titration data were fitted with the Hill equation: E = E0 + ΔE(Ka[Ca2+])n/{1 + (Ka[Ca2+])n} using a steepest descent fitting algorithm (20). E0 and ΔE are the initial mean residue ellipticity and maximal change in mean residue ellipticity, respectively; Ka is the apparent binding constant; and n is the Hill coefficient. The mean residue ellipticity intensity for the titration curve at 280 nm wavelength was normalized by taking the fitted values for E0 and ΔE as 0 and 1, respectively.
Immuno-Electron Microscopy.
Polyclonal antibodies against the N-terminal fragment used for the CD measurements were raised in guinea pig. Small intestine was cryoimmobilized by high-pressure freezing immediately after excision (21). For immuno-electron microscopy the frozen samples were freeze-substituted in ethanol containing 0.5% uranylacetate (22) and finally embedded in London resin (LR)-gold at −18°C (Polyscience, Munich). Ultrathin sections were labeled according to the procedure of Schwarz (23). The sections were incubated with the 1/100 diluted serum for 60 min. As gold label 10 nm protein A-gold (dilution 1/100, a generous gift from H. Schwarz, Max-Plank-Institut für Entwicklungsbiologie, Tübingen, Germany) was applied for 60 min. Control sections were incubated in the presence of preimmune serum or only in the presence of gold-labeled protein A. The sections were stained with uranylacetate and lead citrate (24).
Preparation of a Mitochondrial Fraction and Proteolytic Digestion.
Fresh enterocytes from rabbit small intestine were used for the preparation of a mitochondrial fraction. The cells were suspended in 0.225 M mannitol, 0.075 M sucrose, 0.5 mM EDTA, and 0.5 mM Tris (pH 7.4) and homogenized according to Smith (25). The homogenate was centrifuged at 800 × g for 10 min and the supernatant centrifuged at 8,000 × g for 8 min. The pellet was resuspended in the same buffer without EDTA and centrifuged once more at 8,000 × g. The last pellet was resuspended in a small volume. The intactness of the mitochondria was checked using an O2 probe cell. Freshly prepared mitochondrial fractions were subjected to proteolysis with Arg-C endoprotease (Boehringer Mannheim) at 37°C for 1 h. The ratio between total protein and protease was 4:1 (wt/wt). The reaction was stopped by addition of an equal volume of 20% (wt/vol) SDS, 50% (vol/vol) glycerol, 25% 2-mercaptoethanol, and traces of bromophenol blue preheated to 100°C and heated for another 2 min at 100°C. Aliquots were electrophoresed on a 10% PAGE and transferred to a polyvinylidene difluoride membrane. Immunostaining was performed as described (26).
RESULTS
Full-Length Cloning and Sequencing of a Novel Ca2+-Dependent Transporter.
Our initial aim was to isolate a membrane-bound derivative of sterol carrier protein 2 (SCP-2), suggested by earlier work in our group (26). For this purpose, a pSPORT-cDNA library exceeding 2,000 bases was constructed with mRNA from enterocytes, and the library was screened with a SCP-2 cDNA from rat. Based on partial sequence analysis, we confirmed that all eight positive clones contained one SCP-2-like sequence. One of the clones contained an additional cDNA (E) at the 5′ end of the SCP-2 sequence. The coding region of the additional sequence was separated from the SCP-2 cDNA by a poly(A) tail and a long untranslated region. Therefore we concluded that this cDNA was not related to SCP-2 and that the merger of both cDNAs occurred artificially during the preparation of the plasmid library. Partial sequencing of this cDNA revealed a unique motif composition and it was decided to complete the cloning of this cDNA. The full-length cDNA (3,298 nt) has been termed Efinal. The single open reading frame started at nucleotide 14 and encoded a protein of 475 amino acids (53 kDa). At nucleotide 1439, a termination codon was followed by a 1,859-bp 3′ untranslated sequence. A consensus polyadenylylation signal was located 18 bases before a poly(A) tail. The sequence surrounding the initial in-frame ATG is in good agreement with the consensus sequence defined for start codons (27), and the length of the full-length clone corresponded well with the mRNA size of about 3,500 bases detected by Northern blot analysis.
Computer Analysis of the Primary Sequence.
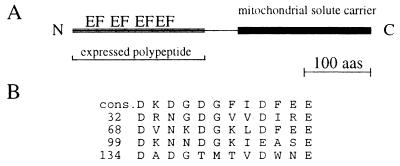
Searching the Swiss protein database with the deduced amino acid sequence of Efinal revealed relationships with two different proteins. Residues 27–150 of the Efinal sequence and troponin C from Japanese horseshoe crab (28) share 28% identical amino acids with an additional 49% of conservative substitutions. In this region, three well-conserved Ca2+-elongation factor (EF)-hand binding loops are located (Fig. 1). A fourth, not so well conserved EF-hand motif spans amino acids 134–146. The C terminus (Fig. 1), starting from amino acid 194, is related to Grave disease carrier protein (30). Here, 39% of the amino acids are identical with another 39% conserved substitutions. When compared with human ATP/ADP carrier protein from liver (31), the identity drops to 27% with additional 40% conserved substitutions. The deduced protein sequence of the C-terminal half was analyzed by Kyte and Doolittle algorithm. The hydropathy profile of the C-terminal half of the protein revealed six stretches rich in hydrophobic amino acids (data not shown). A similar pattern can be found in other mitochondrial solute carriers (32).
Figure 1.
Schematic drawing of the protein and the comparison of the EF-hand motifs. (A) The location of the EF-hand motifs and the mitochondrial solute carrier structure are indicated. A scale for 100 amino acids is given. (B) The EF-hand sequences (EF) are shown in comparison to their consensus sequence drawn from ref. 29.
Tissue Distribution by Northern Blot Studies.
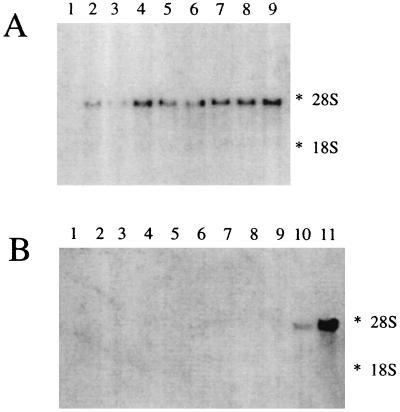
Northern blot analysis was performed on RNA isolated from different rabbit tissues to study the tissue specificity of the Efinal gene transcript. The size of the single transcript was 3,500 bp. The autoradiographs of the same Northern blot probed with the 5′ half, 3′ half, or the full-length Efinal were superimposable (data not shown).
As shown in Fig. 2, no hybridization with RNA from liver, muscle, heart, esophagus, stomach, spleen, testes, and brain was detected. Colon was the tissue with the highest expression rate, followed by small intestine and kidney. In the small intestine the expression rate of the Efinal gene transcript was lowest in duodenum and increased toward the distal part of the intestine.
Figure 2.
Expression of the Ca2+-dependent mitochondrial solute carrier in rabbit tissue. (A) Total RNA samples from nine segments from the small intestine; proximal (lane 1) through distal (lane 9), and (B) RNA samples from esophagus (lane 1), stomach (lane 2), liver (lane 3), spleen (lane 4), skeletal muscle (lane 5), heart (lane 6), lung (lane 7), testes (lane 8), brain (lane 9), kidney (lane 10), and colon (lane 11) were hybridized with the probe for the Ca2+-dependent mitochondrial solute carrier. The amounts of RNA were verified by visualizing the 28S and 18S rRNA bands in the agarose gels after staining with ethidium bromide (data not shown). The position of the 28S and 18S rRNA species are indicated.
Ca2+ Binding.
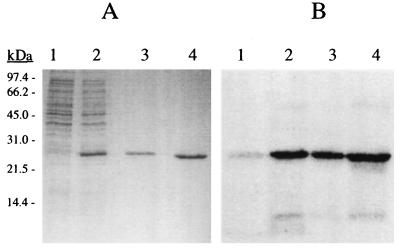
The proposed Ca2+-binding ability of the gene product was tested with a recombinant polypeptide of 25 kDa, which spans all four EF-hand like domains (Fig. 1). The polypeptide was expressed in E. coli and purified to homogeneity (Fig. 3A). The calcium-binding ability of the polypeptide was demonstrated with a Ca2+-overlay method. As shown in Fig. 3, the N-terminal polypeptide of 25 kDa exhibits a strong affinity to calcium even in the SDS denatured state. The sensitivity of its conformation to calcium was investigated spectroscopically. The near-UV CD spectrum of the fragment comprises a broad envelope of intensities in the 250–295 nm spectral range (Fig. 4A). The protein sequence contains nine phenylalanine, three tyrosine, and four tryptophan residues and, without detailed spectroscopic study, it is impossible to assign the intensities to specific residues. In the absence of Ca2+, qualitatively tryptophan and probably tyrosine provide major contributions, whereas the low wavelength region may have components due to phenylalanine. The effect of Ca2+ on the spectrum is marked, even in the low nM range, as shown in Fig. 4A. The changes that occur are complete around 7 μM and are indicative of considerable environmental reorganization of some, or all, of the aromatic residues. The positive intensity completely disappears and is replaced by two significant negative signals at around 283 and 289 nm which are due to one (or more) of the constituent tryptophan side chains.
Figure 3.
Purity of the expressed N-terminal polypeptide and its ability to bind calcium. (A) Coomassie brilliant blue stained SDS/15% PAGE. Shown is E. coli lysate before (lane 1) and after induction (lane 2) with isopropyl β-d-thiogalactopyranoside. In lanes 3 and 4, 2 and 5 μg of purified fragments were loaded. (B) Autoradiograph of the proteins as shown in A transferred on a polyvinylidene difluoride membrane and labeled with 45Ca2+ as described.
Figure 4.
Effect of Ca2+ on the near-UV (A) and far-UV (B) CD spectra of the Ca2+-dependent carrier. (A) The initial spectrum at zero free [Ca2+] was taken in 10 mM K2EGTA, 100 mM KCl, and 10 mM Mops (thick solid line). For the subsequent spectra, several mixtures containing different free calcium concentrations (0.15–7.34 μM) were used. These were prepared as described (20). (B) The same procedure was used to obtain the far-UV spectra at zero free [Ca2+] (thick solid line) and 40 μM free [Ca2+] (thin solid line).
The far-UV spectrum (Fig. 4B) in the absence of Ca2+ is typical of that of a protein containing substantial α-helical and disordered components (33). As the Ca2+ concentration is raised, changes, concomitant with those in the near-UV region, are seen in the spectrum. At saturating Ca2+ concentrations there are changes in the form of the spectrum, qualitatively reminiscent of a diminution of the disordered conformation and suggestive of an increase in, or appearance of, a β-sheet component. This taken together with the far-UV CD spectra suggests that a major conformational change occurs on Ca2+ binding. The effect of Ca2+ on the spectra could be reversed with EGTA, while addition of Mg2+ did not affect the CD spectra. To define the range over which Ca2+ affected the conformation the polypeptide was titrated with Ca2+. The initial measurements were performed in the presence of EGTA, and subsequent titration was performed using Ca2+/EGTA buffers with known free Ca2+ concentrations. The data on changes of mean residue ellipticity intensity of the Ca2+-dependent carrier were plotted as a function of free [Ca2+]. The resulting sigmoid curve was fitted by the Hill equation, as described in Material and Methods, yielding a Kd ≈ 0.37 μM and an apparent Hill coefficient of about 2.0.
Subcellular Location and Topology.
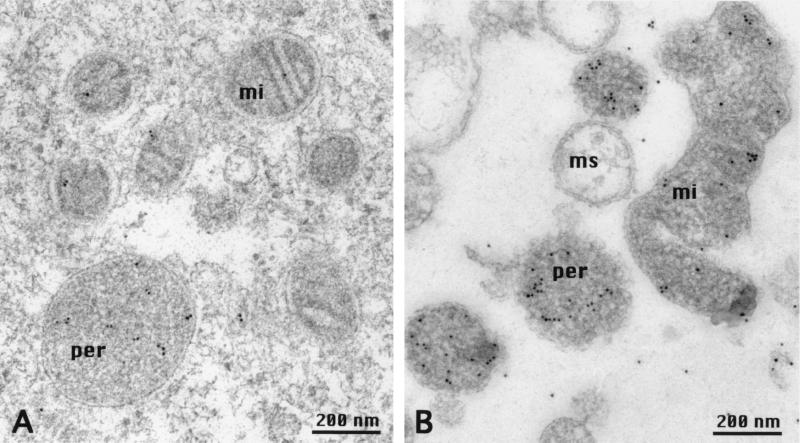
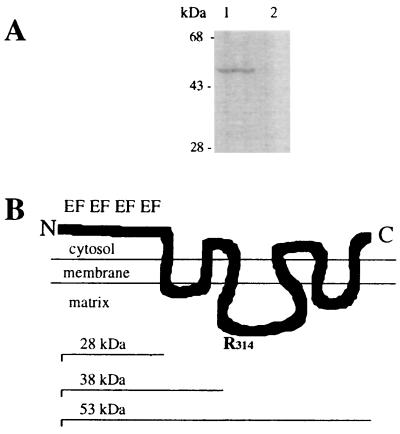
The 25-kDa polypeptide used for the CD measurements was injected into guinea pigs to raise polyclonal antibodies. In immunogold labeled sections of enterocytes (Fig. 5A) the protein was mainly detected in peroxisomes, although some label could be found in mitochondria. Other organelles and structures of the enterocyte were not labeled. This result was confirmed by immunogold-labeled sections of the sediment of a mitochondrial fraction, which contained a substantial amount of peroxisomes (Fig. 5B). Why the gold label was not restricted to the membranes but was also present in the matrix of mitochondria and peroxisomes is not known but, as is shown in Fig. 6, the antiserum specifically labels a protein of 53 kDa in mitochondrial fractions. This size is the same as the calculated molecular weight of the Efinal gene product. To define the location and topology of the protein, mitochondrial fractions were digested with Arg-C protease according to the method used to determine the topology of ADP/ATP translocase (34, 35). Normally, the ADP/ATP transporter is resistant to Arg-C protease cleavage but, if located in inside out mitoplasts, Arg-C cleaves at Arg-140 and/or Arg-152 and creates a 14.5-kDa fragment. Arg-140 is conserved in our 53-kDa transporter at position 314. If the N terminus of our protein faced the inside of peroxisomes, Arg-C should cleave at the conserved Arg-314 and create a proportional amount of an immuno-detectable fragment of 38 kDa. As shown in Fig. 6A, upon digestion with Arg-C, no fragment with a molecular weight around 38 kDa appeared. As there are 7 arginines in the 28-kDa polypeptide that precedes the first membrane-spanning amino acid sequence digestion produces fragments of less than 9 kDa. This result indicates that our protein is localized predominantly in the peroxisomal membrane with the N-terminal Ca2+-binding domain facing the cytosol (Fig. 6B). The rest of the scheme shown in Fig. 6B is based on the topology of the ADP/ATP transporter (34, 35) and the hydropathy profile derived from the Kyte and Doolittle analysis.
Figure 5.
Localization of the Ca2+- dependent solute carrier. Shown is the immunogold labeling of a section of an enterocyte of a villus from rabbit ileum (A) and a section through a sediment of a mitochondrial fraction (B). Immunogold label is found mainly in peroxisomes (per), but to some extent also in mitochondria (mi). Microsomal membranes (ms) carry no gold label (B). Freeze-substitution and LR-gold embedding have been described.
Figure 6.
Topology of the Ca2+- dependent solute carrier. (A) Immunoblotting of 15 μg mitochondrial fraction protein before (lane 1) and after (lane 2) digestion with Arg-C protease is shown. The polyclonal antibodies were generated against a 25-kDa N-terminal polypeptide of the transporter. Note that with the disappearance of the 53-kDa transporter no proportional appearance of a fragment sized between 28 and 53 kDa occurs. Thus, externally administered proteases have access to the N terminus of the transporter. The apparent molecular masses of marker proteins are indicated; the procedure has been described in Materials and Methods. (B) Schematic drawing of a possible transmembrane arrangement of the Ca2+-dependent solute carrier based on our topology studies is shown. The EF-hand motifs of the Ca2+-binding N terminus are indicated. The model is based on the topology of the ADP/ATP transporter (34, 35).The N-terminal and C-terminal regions are exposed to the cytosol and the segment containing Arg-314 (R314) protrudes into the matrix. The size of the entire protein (53 kDa), the N terminus exposed to the cytosol (28 kDa), and of a fragment produced by cleavage at Arg-314 (38 kDa) are indicated.
DISCUSSION
This study describes the complete nucleotide and derived amino acid sequence of a novel Ca2+-dependent member of the mitochondrial transporter superfamily. The possible role of this protein as Ca2+ buffer can be ruled out due to the low abundance of its mRNA in any tissue. Based on the sequence homologies with other cytosolic Ca2+ sensors like calmodulin and troponin C, a role for the N terminus as a Ca2+ sensor is most appealing. In this context, a possible function for the C terminus might be to transduce the Ca2+ signal to the peroxisomal or mitochondrial matrix by the transport of a solute.
The C-terminal half of a Ca2+-dependent mitochondrial transporter shows 78% homology with Grave disease carrier protein (30, 36, 37) and 67% homology with human ADP/ATP carrier protein. Adrian et al. (38) determined amino acid sequences from the ADP/ATP carrier that are required for the delivery and location of the transporter in the inner mitochondrial membrane. In this region the ADP/ATP transporter and our protein exhibit 50% identity on the amino acid level. This level of identical amino acids reflects the requirement that a membrane-spanning domain of a protein has to fit a specific membrane (39). Because the difference between the inner-mitochondrial and the peroxisomal membrane is only marginal, solute carriers for both organelles can exhibit a similar membrane-spanning amino acid sequence. A more important aspect of the location of a protein are organelle-specific targeting sequences. The C-terminal serine-lysine-leucine (SKL) sequence (40) was the first recognized peroxisomal targeting sequence. It is not present in the primary sequence of our transporter. The same applies for the yeast Pmp47 protein (15), another solute carrier localized in the peroxisome, although an internal SKL sequence exists at position 316–318. Earlier work on Pmp47 from C. boidinii points to the existence of an internal peroxisomal targeting sequence other than the internal SKL triplet (41), which is contained within a short hydrophilic loop (42). Amino acids in this loop from Pmp47 and from our protein share 50% similarity, which is the same degree of similarity for the entire protein sequence. Interestingly the single conserved triplet in this loop is LKS, the reverse sequence of the known SKL peroxisomal targeting sequence and therefore a promising candidate for future studies on the translocation of peroxisomal membrane proteins. The other peroxisomal targeting sequence of Pmp47 is located at the N terminus and is in agreement with the signal peptide of the peroxisomal 3-ketoacyl-CoA thiolase precursor (43). In our case only the arginine at position 3 is conserved whereas at position 10 there is a lysine instead of a histidine. If our protein followed the rules established by Tsukamoto et al. (44) on the N-terminal peroxisomal targeting sequence, our protein should be located in the cytosol and not in the peroxisome. The need for a special peroxisomal targeting sequence for the protein could be explained by its special topology; the 25-kDa N terminus has to face the cytosol while the rest of the protein is embedded in the peroxisomal membrane. A similar topology applies to the peroxisomal ATP-binding cassette transporters that also lack both known peroxisomal targeting sequences (45). One could speculate that integral membrane proteins of the peroxisome with extensive portions facing the cytosol need a special targeting sequence for proper translocation.
Another interesting aspect for the targeting of this protein is its apparent dual localization in mitochondria and peroxisomes. A similar distribution has been described for the α subunit of the F1-ATPase (46). In the context of a possible transduction of a Ca2+ signal to the inside of peroxisomes and mitochondria this could lead to coordinated action of both organelles. However, the question as to whether the protein exists in one form or in two isoforms with different targeting sequences will be the subject of future work.
The mainly hydrophobic C-terminal half of our solute carrier is related to Grave disease carrier, for which no function has yet been assigned (30). The most unique feature of the solute carrier is the Ca2+ dependency. To date only a single Ca2+-dependent mitochondrial carrier has been described (47) but it still awaits purification or molecular cloning.
One of the aspects of peroxisomal function is the ability of this compartment to be induced by so-called peroxisomal proliferators. Peroxisomal proliferators form a diverse group of chemicals, including hypolipidemic drugs, industrial plasticizers, and pesticides (for a review, see ref. 48). Characteristic of their effects in the liver is the induction of fatty acid metabolizing enzymes (49, 50) in association with hepatomegaly (51, 52). The cellular signal pathway activated by peroxisome proliferators depends, at least in part, on Ca2+ signaling (53). For example clofibrate-induced peroxisome proliferation can be suppressed by calcium antagonists (54, 55). The situation is complicated by the fact that different peroxisome proliferators mobilize Ca2+ in different ways (56). To our knowledge the Ca2+-dependent peroxisomal solute carrier is the best candidate for a Ca2+ sensor that could transduce either generally or specifically the Ca2+ signals induced by peroxisomal proliferators, to the peroxisome.
The other main effector in the induction of peroxisomes is the peroxisome proliferator activated receptor, which belongs to the steroid hormone receptor superfamily (57–59). This receptor is activated by long-chain fatty acids and regulates the β-oxidation of fatty acids (60). In the colon, long chain fatty acids are suspected to be tumor promoters. The γ-isoform of this receptor and our solute carrier are both expressed in the colonocytes facing the lumen of the colon (data not shown). One could speculate that the coordinated action of both proteins to induce peroxisome proliferation via a Ca2+ signal and the onset of transcription prevents the accumulation of long-chain fatty acids in colonocytes and thus prevents colon carcinogenesis (61).
Acknowledgments
We thank Drs. Christoph Richter and Paulo Gazzotti for reading the manuscript and for helpful discussions. We are indebted to Dr. Karel Wirtz for sending us the SCP-2 cDNA. This work was supported by the Swiss National Science Foundation (Grants 31-32441.91 and 32-46810.96) and by the Eidgenössiche Technische Hochschule (Zurich).
ABBREVIATIONS
- CD
circular dichroism
- EF
elongation factor
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF004161).
References
- 1.Dickerson R E. Nature (London) 1980;283:210–212. doi: 10.1038/283210a0. [DOI] [PubMed] [Google Scholar]
- 2.De Duve C. Sci Am. 1983;248:52–62. doi: 10.1038/scientificamerican0583-74. [DOI] [PubMed] [Google Scholar]
- 3.Lazarow P B, Fujiki Y. Annu Rev Cell Biol. 1985;1:489–530. doi: 10.1146/annurev.cb.01.110185.002421. [DOI] [PubMed] [Google Scholar]
- 4.Schatz G. J Biol Chem. 1996;271:31763–31766. doi: 10.1074/jbc.271.50.31763. [DOI] [PubMed] [Google Scholar]
- 5.Cuezva J M, Flores A I, Liras A, Santarén J F, Alconada A. Biol Cell. 1993;77:47–62. doi: 10.1016/s0248-4900(05)80174-1. [DOI] [PubMed] [Google Scholar]
- 6.Klingenberg M. J Bioenerg Biomembr. 1993;25:447–458. doi: 10.1007/BF01108402. [DOI] [PubMed] [Google Scholar]
- 7.Higgins C F. Annu Rev Cell Biol. 1992;8:67–113. doi: 10.1146/annurev.cb.08.110192.000435. [DOI] [PubMed] [Google Scholar]
- 8.Higgins C F, Hiles I D, Salmond G P C, Gill D R, Downie J A, Evans I J, I B, H, Gray L, Buckel S D, Bell A W. Nature (London) 1986;323:448–450. doi: 10.1038/323448a0. [DOI] [PubMed] [Google Scholar]
- 9.Van Veldhoven P, Just W, Mannerts G P. J Biol Chem. 1987;262:105–109. [PubMed] [Google Scholar]
- 10.van den Bosch H, Schutgens R B H, Wanders R J A, Tager J M. Annu Rev Biochem. 1992;61:157–197. doi: 10.1146/annurev.bi.61.070192.001105. [DOI] [PubMed] [Google Scholar]
- 11.Mosser J, Douar A-M, Sarde C O, Kioschis P, Feil R, Moser H, Poustka A-M, Mandel J-L, Aubourg P. Nature (London) 1993;361:726–730. doi: 10.1038/361726a0. [DOI] [PubMed] [Google Scholar]
- 12.Kamijo K, Taketani S, Yokota S, Osumi T, Hashimoto T. J Biol Chem. 1990;265:4534–4540. [PubMed] [Google Scholar]
- 13.Gärtner J, Moser H, Valle D. Nat Genet. 1992;1:16–23. doi: 10.1038/ng0492-16. [DOI] [PubMed] [Google Scholar]
- 14.Kuan L, Saier H J., Jr Crit Rev Biochem Mol Biol. 1993;28:209–233. doi: 10.3109/10409239309086795. [DOI] [PubMed] [Google Scholar]
- 15.Sakai Y, Saiganji A, Yurimoto H, Takabe K, Saiki H, Kato N. J Cell Biol. 1996;134:37–51. doi: 10.1083/jcb.134.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chomzynski P, Sacchi N. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 17.Weber F E, Vaughan K T, Reinach F C, Fischman D A. Eur J Biochem. 1993;216:661–669. doi: 10.1111/j.1432-1033.1993.tb18186.x. [DOI] [PubMed] [Google Scholar]
- 18.Okagaki T, Weber F E, Fischman D A, Vaughan K T, Mikawa T, Reinach F C. J Cell Biol. 1993;123:619–626. doi: 10.1083/jcb.123.3.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maruyama K, Mikawa T, Ebashi S. J Biochem (Tokyo) 1984;95:511–519. doi: 10.1093/oxfordjournals.jbchem.a134633. [DOI] [PubMed] [Google Scholar]
- 20.Kuznicki J, Strauss K I, Jacobowitz D M. Biochemistry. 1995;34:15389–15394. doi: 10.1021/bi00047a001. [DOI] [PubMed] [Google Scholar]
- 21.Studer D, Michell M, Müller M. Scanning Microsc Suppl. 1989;3:153–269. [PubMed] [Google Scholar]
- 22.Müller M, Marti T, Kriz S. In: Electron Microscopy, Seventh European Congress of the Electron Microscopy Foundation. Brederoo P, de Priester W, editors. Vol. 2. Leiden: Electron Microscopy Found.; 1980. pp. 720–772. [Google Scholar]
- 23.Schwarz H. In: Immunolabelling of Ultrathin Resin Sections for Fluorescence and Electron Microscopy. Joffrey B, Colliex C, editors. Vol. 3. Les Ulis, France: Les Editions de Physique; 1994. pp. 255–256. [Google Scholar]
- 24.Reynolds E S. J Cell Biol. 1963;17:208–212. doi: 10.1083/jcb.17.1.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith A L. Methods Enzymol. 1967;10:81–86. [Google Scholar]
- 26.Lipka G, Schulthess G, Thurnhofer H, Wacker H, Wehrli E, Zeman K, Weber F E, Hauser H. J Biol Chem. 1995;270:5917–5925. doi: 10.1074/jbc.270.11.5917. [DOI] [PubMed] [Google Scholar]
- 27.Kozak M. Nucleic Acids Res. 1987;15:8125–8132. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kobayashi T, Kagami O, Takagi T, Konishi K. J Biochem (Tokyo) 1989;105:823–828. doi: 10.1093/oxfordjournals.jbchem.a122752. [DOI] [PubMed] [Google Scholar]
- 29.da Silva A C R, Reinach F C. Trends Biochem Sci. 1991;16:53–57. doi: 10.1016/0968-0004(91)90024-p. [DOI] [PubMed] [Google Scholar]
- 30.Fiermonte G, Runswick M J, Walker J E, Palmieri F. DNA Sequence. 1992;3:71–78. doi: 10.3109/10425179209033999. [DOI] [PubMed] [Google Scholar]
- 31.Cozens A L, Runswick M J, Walker J E. J Mol Biol. 1989;206:261–280. doi: 10.1016/0022-2836(89)90477-4. [DOI] [PubMed] [Google Scholar]
- 32.Walker J E, Runswick M J. J Bioenerg Biomembr. 1993;25:435–446. doi: 10.1007/BF01108401. [DOI] [PubMed] [Google Scholar]
- 33.Provencher S W, Glöckner J. Biochemistry. 1981;20:33–37. doi: 10.1021/bi00504a006. [DOI] [PubMed] [Google Scholar]
- 34.Brandolin G, Boulay F, Dalbon P, Vignais P V. Biochemistry. 1989;28:1093–1100. doi: 10.1021/bi00429a025. [DOI] [PubMed] [Google Scholar]
- 35.Capobianco L, Brandolin G, Palmieri F. Biochemistry. 1991;30:4963–4969. doi: 10.1021/bi00234a018. [DOI] [PubMed] [Google Scholar]
- 36.Campbell D G, Li P, Oakeshott R D. J Bone Jt Surg Br Vol. 1996;78B:22–25. [PubMed] [Google Scholar]
- 37.Zarrilli R, Oates E L, Wesley McBride O, Lerman M I, Chan J Y, Santisteban P, Ursini M V, Notkins A L, Kohn L D. Mol Endocrinol. 1989;3:1498–1508. doi: 10.1210/mend-3-9-1498. [DOI] [PubMed] [Google Scholar]
- 38.Adrian G S, McCommon M T, Montgomery D L, Douglas M G. Mol Cell Biol. 1986;6:626–634. doi: 10.1128/mcb.6.2.626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bretscher M S, Munro S. Science. 1993;261:1280–1281. doi: 10.1126/science.8362242. [DOI] [PubMed] [Google Scholar]
- 40.Gould S J, Keller G-A, Hosken N, Wilkinson J, Subramani S. J Cell Biol. 1989;108:1657–1664. doi: 10.1083/jcb.108.5.1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McCammon M T, McNew J A, Willy P J, Goodman J M. J Cell Biol. 1994;124:915–925. doi: 10.1083/jcb.124.6.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dyer J M, McNew J A, Goodman J M. J Cell Biol. 1996;133:269–280. doi: 10.1083/jcb.133.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Swinkels B W, Gould S J, Bodnar A G, Rachubinski R A, Subramani S. EMBO J. 1991;10:3255–3262. doi: 10.1002/j.1460-2075.1991.tb04889.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tsukamoto T, Hata S, S, Y, Miura S, Fujiki Y, Hijikata M, Miyazawa S, Hashimoto T, Osumi T. J Biol Chem. 1994;269:6001–6010. [PubMed] [Google Scholar]
- 45.Shani N, Sapag A, Valle D. J Biol Chem. 1996;271:8725–8730. doi: 10.1074/jbc.271.15.8725. [DOI] [PubMed] [Google Scholar]
- 46.Cuezva J M, Santarén J F, Gonzalez P, Luis A M, Izquierdo J M. FEBS Lett. 1990;270:71–75. doi: 10.1016/0014-5793(90)81237-i. [DOI] [PubMed] [Google Scholar]
- 47.Haynes R C J, Picking R A, Zacks W J. J Biol Chem. 1986;261:16121–16125. [PubMed] [Google Scholar]
- 48.Lake B G. Annu Rev Pharmacol Toxicol. 1995;35:483–507. doi: 10.1146/annurev.pa.35.040195.002411. [DOI] [PubMed] [Google Scholar]
- 49.Sharma, R., Lake, B. G., Foster, J. & Gibson, G. G. (1988) Biochem. Pharmacol. 1203–1206. [DOI] [PubMed]
- 50.Sharma R K, Lake B G, Foster J, Gibson G G. Biochem Pharmacol. 1988;37:1193–1201. doi: 10.1016/0006-2952(88)90770-8. [DOI] [PubMed] [Google Scholar]
- 51.Reddy J K, Kumar N S. J Biochem (Tokyo) 1979;85:847–856. [PubMed] [Google Scholar]
- 52.Moody D E, Reddy J K. Am J Pathol. 1979;90:435–450. [PMC free article] [PubMed] [Google Scholar]
- 53.Bieri F. Biol Cell. 1993;77:43–46. doi: 10.1016/s0248-4900(05)80173-x. [DOI] [PubMed] [Google Scholar]
- 54.Ram P A, Waxman D J. Biochem J. 1994;301:753–758. doi: 10.1042/bj3010753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Watanabe T, T. S. FEBS Lett. 1988;232:293–297. doi: 10.1016/0014-5793(88)80756-7. [DOI] [PubMed] [Google Scholar]
- 56.Shackelton G L, Gibson G G, Sharma R K, D, H, Orrenius S, Kass G E N. Toxicol Appl Pharmacol. 1995;130:294–303. doi: 10.1006/taap.1995.1034. [DOI] [PubMed] [Google Scholar]
- 57.Tugwood J D, Isseman I, Anderson R G, Bundell K, McPheat W, Green S. EMBO J. 1992;11:293–297. doi: 10.1002/j.1460-2075.1992.tb05072.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Muerhoff A S, Griffin K J, Johnson E F. J Biol Chem. 1992;267:19051–19053. [PubMed] [Google Scholar]
- 59.Barot O, Aldridge T C, Latruffe N, Green S. Biochem Biophys Res Commun. 1993;192:37–45. doi: 10.1006/bbrc.1993.1378. [DOI] [PubMed] [Google Scholar]
- 60.Lee S S T, Pineau T, Drago J, Lee E J, Owens J W, Ktoetz D L, Fernandezsalguero P M, Westphal H, Gonzales F J. Mol Cell Biol. 1995;15:3012–3022. doi: 10.1128/mcb.15.6.3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mansén A, Guardiola-Diaz H, Rafter J, Branting C, Gustafson J A. Biochem Biophys Res Commun. 1996;222:844–851. doi: 10.1006/bbrc.1996.0832. [DOI] [PubMed] [Google Scholar]