Abstract
Two high copy suppressors of temperature-sensitive TATA-binding protein (TBP) mutants were isolated. One suppressor was TIF51A, which encodes eukaryotic translation initiation factor 5A. The other high copy suppressor, YGL241W, also known as KAP114, is one of 14 importin/karyopherin proteins in yeast. These proteins mediate the transport of specific macromolecules into and out of the nucleus. Cells lacking Kap114 partially mislocalize TBP to the cytoplasm. Kap114 binds TBP in vitro, and binding is disrupted in the presence of GTPγS. Therefore, Kap114 is an importer of TBP into the nucleus, but alternative import pathways must also exist.
TATA-binding protein (TBP) is essential for RNA polymerase I, II, and III transcription (1). TBP, along with its associated factors (the TAFs), recruits the basal transcription factors and RNA polymerase to promoters to mediate transcription (1). The yeast TBP gene (SPT15) is essential, and the protein localizes to the nucleus (2).
The compartmentalization of the eukaryotic cell into the nucleus and cytoplasm necessitates the existence of mechanisms to transport macromolecules into and out of the nucleus. The nuclear membrane is punctuated by aqueous channels termed nuclear pores, through which some molecules can diffuse. However, most macromolecules must be transported through these channels in a protein-mediated fashion (3). The core components of such transport pathways consist of a transport receptor (termed an importin or karyopherin), a signal in the transport cargo that allows receptor recognition, and accessory factors that aid in directing the receptor/cargo complex from one compartment to another in a vectorial manner. These accessory factors include the nucleoporin proteins of the nuclear pore and the small GTPase Ran.
The most extensively characterized importin/karyopherin is importin-β (Kap95 in yeast). Importin-β, together with its adapter protein importin-α (Srp1 in yeast), mediates the nuclear import of proteins bearing a basic, lysine-rich nuclear localization sequence. On entry into the nucleus, this transport complex is most likely dissociated by the presence of Ran in its GTP-bound form (4). The compartmentalization of Ran’s GTPase activating protein to the cytoplasm and its guanine nucleotide exchange factor to the nucleus suggests that Ran is primarily in its GTP-bound form in the nucleus and in its GDP-bound form in the cytoplasm (4). This Ran GTP/GDP gradient is believed to be essential for directional transport (4).
Many other importins/karyopherins in addition to the importin-α/importin-β complex have now been identified and characterized (5 and 6). Importins/karyopherins are most similar to one another in their first 150 amino acids (5). This N-terminal domain of importin-β contains the core of its Ran binding domain (5). Completion of the yeast Saccharomyces cerevisiae genome project revealed that there are 14 potential importins/karyopherins in yeast (7). To date, a function in nuclear transport has been identified for all but three of these (8).
The rapid expansion of knowledge of importins and their cargo has broadened the repertoire of known transport signals beyond the canonical nuclear localization sequence. For example, the importin Crm1/Xpo1 mediates the export of proteins out of the nucleus by recognizing a leucine-rich nuclear export signal (6). The identification of various transport signals allows one to hypothesize and test what transport pathways a particular cargo may be using. However, it remains unknown how most proteins are transported.
A high copy suppressor screen of an spt15 temperature-sensitive allele resulted in isolation of BRF1/PCF4/TDS4 [TFIID (TBP) suppressor], which encodes a pol III-specific TAF (9). Two genes that were weak, allele-nonspecific suppressors also were isolated and are described in this report. One of these high copy suppressors (TDS2) was identified as the putative importin YGL241W. YGL241W was recently renamed in the Saccharomyces Genome Database as KAP114, a karyopherin of 114 kDa. The TDS3 suppressor gene was found to be identical to TIF51A, which encodes eukaryotic translation initiation factor 5A (eIF-5A). We now present data that Kap114, a previously uncharacterized importin/karyopherin, is a mediator of TBP import into the nucleus.
Materials and Methods
Screen for High Copy Suppressors of Mutant spt15 Alleles.
A 2μ/URA3 plasmid-based yeast genomic library (10) was transformed into yeast strains containing temperature-sensitive alleles of the TBP gene SPT15. Two different spt15 alleles were used: one carrying the mutation Y94C (YSB66: Mata ura3–52 leu2Δ1 trp1Δ63 spt15-Y94C) and the other K133, 138L (9). Transformants that could grow at 37°C (for Y94C) or 38°C (for K133, 138L) were isolated. Library plasmids were recovered in Escherichia coli and were retested for suppression of temperature sensitivity. Suppressing plasmids were subcloned and sequenced by using standard methods. One library plasmid containing the Kap114/TDS2 gene was designated YEpTDS2.
Plasmid and Strain Construction.
To replace the KAP114 gene with a gene encoding a Kap114-green fluorescent protein (GFP) fusion, the 3′ end of the KAP114 gene was PCR-amplified by using the 5′ SacII-linked primer HM79 (TCCCCGCGGCTCACCCCTGCTATCATTAG) and the 3′ XhoI-linked primer HM80 (CCGCTCGAGCCAAAAGGGCTTCTGAAAGAAC). The PCR product was digested with SacII and AvaI and was ligated in-frame into a SacII/XhoI fragment of pPS1857. pPS1857 (11) is a nonreplicating plasmid derived from pRS306 (12) and contains the S65T V163A mutant of GFP and the NUF2 3′ untranslated region. The resulting plasmid, pPS1858, was linearized with XbaI and was transformed into FY23 (13). Integration into the kap114 locus was verified by PCR. Expression of Kap114-GFP in URA+ transformants was determined by α-GFP immunoblotting and fluorescence microscopy. The resulting Kap114-GFP strain PSY1783 was used in our studies. The Kap95-GFP strain (PSY1786) used in these studies was made by integration of linearized pJK169 into FY23 (11). pRS425-TDS2 was generated by cloning a SalI fragment containing TDS2/KAP114 from YEpTDS2 into SalI linearized pRS425 (12).
A PCR-based method was used to replace the KAP114 gene with the HIS3 marker in the diploid strain PSY902 (ura3–52/ura3–52 leu2Δ1/leu2Δ1 his3Δ200/his3Δ200 ade2/ADE2 ade3/ADE3 lys2/LYS2 trp1/TRP1) (14). On sporulation, four-spore tetrads grew, and the HIS3 marker segregated 2:2. Correct integration of the marker into the KAP114 locus was verified by PCR and Southern blotting. One Δkap114 haploid (PSY1784: Matα kap114Δ∷HIS3 ura3–52 leu2Δ1 his3Δ200 trp1) and its wild-type sister spore (PSY1785: Mata ura3–52 leu2Δ1 his3Δ200) were used in our studies. For some experiments, the strain YSB103 (MATα ura3–52 leu2–3, 112 his3Δ200 kap114Δ∷URA3) and its wild-type counterpart YSB105 were used.
Immunoblots.
Whole cell lysates (except those described below in protein binding experiments) were prepared from ≈1 × 108 freeze-thawed cells in ≈200 μl PBSMT (PBS/5 mM MgCl2/0.5% Triton X-100) plus protease inhibitors (0.5 mM phenylmethylsulfonyl fluoride; 3 μg/ml each pepstatin, leupeptin, aprotinin, and chymostatin) by glass bead lysis and were centrifuged for 10 min at 4°C. Samples were run on SDS/PAGE gels and were transferred to nitrocellulose. After incubation with primary antibody, blots were incubated in a 1:5,000 dilution of horseradish peroxidase-conjugated secondary antibody (Jackson Immunoresearch), and protein was visualized via enhanced chemiluminescence (Renaissance kit, NEN). TBP was detected by using a 1:2,000 dilution of a polyclonal antibody (S.B., unpublished work); GFP fusions were detected with a 1:5,000 dilution of a polyclonal antibody (11); Srp1 was detected by using a 1:3,000 dilution of a polyclonal antibody (a gift from D. Görlich, Zentrum für Molekular Biologie der Universität Heidelberg); Gsp1 was detected by using a 1:1,000 dilution of a polyclonal antibody (no Tween) (11); glutathione S-transferase (GST) was detected by using a 1:3,000 dilution of a mouse monoclonal antibody (Santa Cruz Biotechnology).
Visualization of Kap114-GFP and TBP-GFP in Living Cells.
Cells were grown to mid-log phase. A Nikon fluorescence microscope equipped with a GFP filter set and a 100× DIC objective were used to observe the yeast. Images were captured by using metamorph imaging software (Universal Imaging, West Chester, PA).
Immunofluorescence.
Immunofluorescence was performed as described (11). Hemagglutinin (HA)–TBP was detected by using a 1:300 dilution of 12CA5 anti-HA monoclonal antibody (Boehringer Mannheim) and a 1:1,000 dilution of FITC-conjugated mouse-specific antibodies (Jackson Immunoresearch).
Protein Binding Experiments.
Recombinant GST and GST-TBP fusion protein were expressed in E. coli using standard techniques. GST proteins were bound to glutathione-agarose beads at ≈3–5 mg of protein per milliliter of beads. Beads were equilibrated in buffer T (20 mM Tris⋅HCl, pH 7.5/10% glycerol/1 mM EDTA) containing 150 mM potassium acetate (KAc). Yeast extracts were made by resuspending pelleted cells in an equal volume of buffer T containing 0.5 KAc. Cells were lysed by adding an equal volume of glass beads (450 microns) and vortexing for five 1-min cycles with cooling on ice in between. Cell debris and glass beads were pelleted by centrifugation. The clear supernatant was diluted with buffer T to a final KAc concentration of 150 mM.
Binding reactions contained 300 μg of yeast protein and 25 μl of GST or GST-TBP beads. Binding reactions were incubated for 10 min at room temperature in the presence of 1 mM guanosine 5′-O-(2-thiodiphosphate) (GDPβS) (Sigma) or 1 mM guanosine 5′-O-(3-thiotriphosphate) (GTPγS) (Sigma) or no analogue. Reactions then were incubated at 4°C overnight with constant rotation. Beads were pelleted for 5 sec in a microfuge and were washed five times in 1 ml of buffer T plus 150 mM KAc. Bound proteins were resuspended in gel loading buffer and were electrophoresed on a 10% SDS-polyacrylamide protein gel. Immunoblotting with α-GFP antibodies was used to detect GFP fusion proteins.
Results
Overexpression of Kap114/TDS2 Suppresses the Temperature Sensitivity of a TBP Mutation.
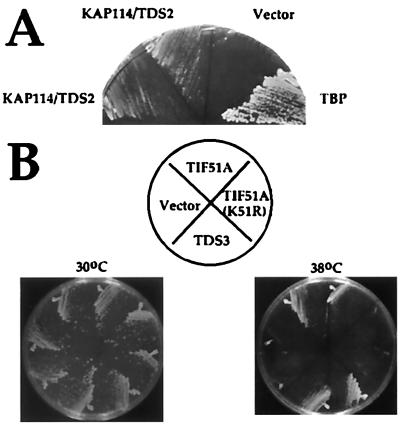
Two genetic screens were performed to identify genes that, when overexpressed, suppress the temperature sensitivity of mutant spt15 alleles. Transformation of YSB66 (carrying the TBP mutant Y94C) with a YEp24-based genomic library (10) resulted in the isolation of TDS2 (Fig. 1A). Subcloning and sequencing of the genomic insert revealed that TDS2 is the uncharacterized importin YGL241W (from now on designated KAP114). Kap114 overexpression also suppressed the temperature sensitivity of several other TBP mutants, including K133, 138L (data not shown). Suppression was attributable to the importin KAP114 rather than a nonspecific effect on nuclear transport because overexpression of the other importin family members KAP95, PSE1, NMD5, MTR10, KAP104, SXM1, or KAP123 failed to suppress (data not shown). Furthermore, overexpression of Kap114 did not suppress temperature-sensitive mutants in several other RNA polymerase II transcription factors (data not shown). One possible explanation for suppression is that Kap114 is involved in TBP transport and that overexpression allows the cell to live by providing the nucleus with increased TBP levels.
Figure 1.
Overexpression of Kap114 or eIF-5A suppresses the temperature sensitivity of TBP mutant strains. (A) YSB66, a strain bearing the temperature-sensitive mutation Y94C in TBP, was transformed with a 2μ plasmid containing the KAP114/TDS2 gene (YEpTDS2a), with the 2μ vector YEp24 alone (vector) or with a plasmid containing the wild-type TBP gene (pRS316-IID). Individual transformants were streaked onto selective media and were grown at 30°C (not shown) and 37°C for 3 days. (B) YSB67, a temperature-sensitive strain bearing the mutations K133L, K138L in TBP (9, 15), was transformed with a 2μ plasmid containing the TDS3 clone, YEp24 alone (vector), a 2μ TIF51A (eIF-5A) plasmid (YEp352T-5A) (16), or a 2μ plasmid encoding a nonhypusinated mutant of TIF51A [YEp352T-5A(K51R)] (16). Individual transformants were streaked onto selective media and were grown at 30°C and 38°C for 3 days.
In a similar screen for high copy suppressors of mutations in the basic region of TBP, TDS4/BRF1 was isolated (9). Tds4/Brf1 is a component of the RNA polymerase III basal transcription factor TFIIIB. This screen also resulted in the isolation of TDS3 (Fig. 1B). TDS3 is identical to TIF51A, one of two yeast genes encoding eIF-5A. High copy eIF-5A suppresses the temperature sensitivity of strain YSB67 (9, 15), in which lysines 133 and 138 TBP were changed to leucine (Fig. 1B) as well as that of the Y94C mutant in YSB66 (data not shown). eIF-5A is essential for viability and must be modified by the postranslational addition of hypusine to a specific lysine to function (16). Suppression of the TBP mutant phenotype also requires hypusination of eIF-5A because the mutant K51R, which cannot be hypusinated (16), does not suppress (Fig. 1B).
Overexpression of Kap114 Increases TBP Levels.
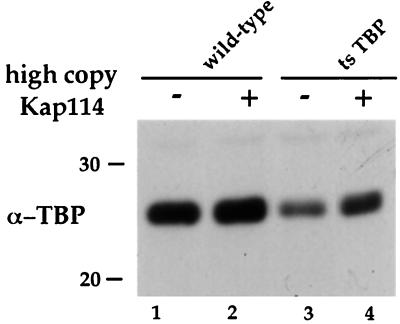
One mechanism by which overexpression of Kap114 might suppress TBP temperature-sensitive mutants is by increasing TBP levels. This hypothesis was tested by performing SDS/PAGE followed by α-TBP immunoblotting of equal amounts of total protein prepared from wild-type (FY23) and TBP temperature-sensitive cells (YSB66) that had been transformed with either empty vector or a high copy plasmid encoding for Kap114 (Fig. 2). In both wild-type and TBP temperature-sensitive cells, TBP levels are increased when Kap114 is overexpressed (Fig. 2). High copy Kap114 does not boost TBP levels in the mutant strain to that of wild-type levels (Fig. 2, compare lanes 1 and 4). However, it is possible that the increase seen in TBP levels is sufficient to confer viability. Kap114 may increase TBP levels by interacting with and thus stabilizing TBP. Alternatively, high Kap114 levels may increase transport of TBP into the nucleus, where it may be stabilized by associations with TAFs and the chromosomes.
Figure 2.
Overexpression of Kap114 increases TBP levels. Lysates of the TBP temperature-sensitive (ts) strain YSB66 and the isogenic wild-type strain (FY23) bearing either an empty vector (pRS425) or a high copy plasmid encoding Kap114 (pRS425-TDS2) were prepared from cells grown to mid-log phase. Equal amounts of protein were resolved by SDS/PAGE and were immunoblotted using α-TBP antibodies. Lanes: 1 and 2, wild-type cells; 3 and 4, temperature-sensitive TBP cells; 1 and 3, + vector; 2 and 4, +high copy Kap114.
Kap114 Is a Nonessential Member of the Importin/Karyopherin Family of Proteins.
YGL241W/KAP114 was identified as a putative importin because of the similarity of its amino terminus to the amino terminus of other importins (7). Kap114 is most similar to the importin Cse1 (Fig. 3). Cse1 is responsible for the export of Srp1 out of the nucleus (17–19). Kap114 is 18% identical to and 43% similar to Cse1 (Fig. 3). Over its first 150 amino acids, Kap114 is 24% identical to Cse1, 23% identical to Nmd5, 21% identical to Kap95, and 18% identical to Sxm1.
Figure 3.
The importin Kap114 is most similar to the yeast protein Cse1. The amino acid sequence of Kap114 (S. cerevisiae ORF YGL241W) was aligned with its most similar protein, the importin Cse1, by using the Clustal method (DNAstar, Madison, WI). Identical amino acids are shaded black, and similar amino acids are shaded gray.
KAP114 is not essential for viability. The KAP114 gene was disrupted in diploid cells by replacing the coding region of the gene with a marker gene. On sporulation and tetrad dissection, four-spore tetrads grew. All four spores grew with equal efficiency and did not show any cold or heat sensitivity (data not shown). Therefore, any transport mechanism mediated by Kap114 must be at least partially redundant with another TBP nuclear localization mechanism.
Kap114 Localizes Primarily to the Nucleus.
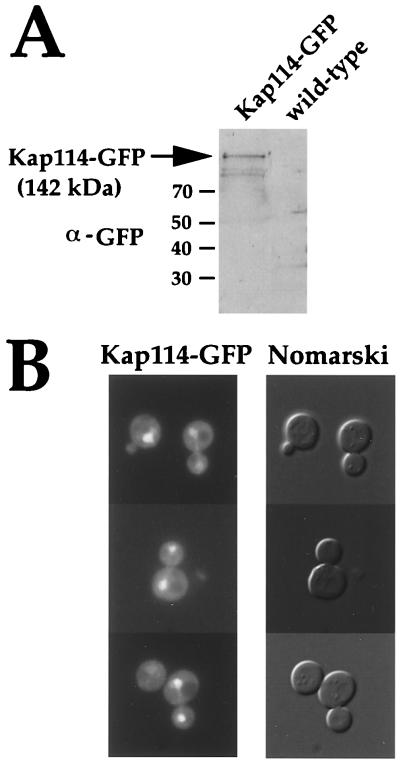
Studies done thus far indicate that most of the importins are localized primarily at the nuclear pore and/or in the nucleus (11, 19–24). To determine the subcellular localization of Kap114, a yeast strain in which the Kap114 ORF was fused to GFP was constructed. An α-GFP immunoblot showed that the Kap114-GFP fusion protein is expressed and that it runs at its predicted molecular weight of ≈142 kDa (Fig. 4A). Kap114-GFP localized primarily to the nucleus in ≈75% of cells (Fig. 4B). Kap114-GFP also was seen, but to a lesser extent, in the cytoplasm (Fig. 4B). The nuclear localization of Kap114 is consistent with its being an importin. Based on immunoblotting and fluorescence microscopy, Kap114 does not appear to be as abundant as other importin proteins (data not shown).
Figure 4.
Kap114-GFP localizes primarily to the nucleus. (A) Lysates of wild-type cells (FY23) and cells bearing an integrated Kap114-GFP gene (PSY1783) were prepared from cells grown to mid-log phase. Equal amounts of protein were resolved by SDS/PAGE and were immunoblotted using α-GFP antibodies. (B) Kap114-GFP cells (PSY1783) were grown in selective media at 25°C to mid-log and were analyzed by fluorescence microscopy. (Left) Cells under a GFP filter set. (Right) The corresponding cells by Nomarski optics.
TBP Is Partially Mislocalized to the Cytoplasm in Δkap114 Cells.
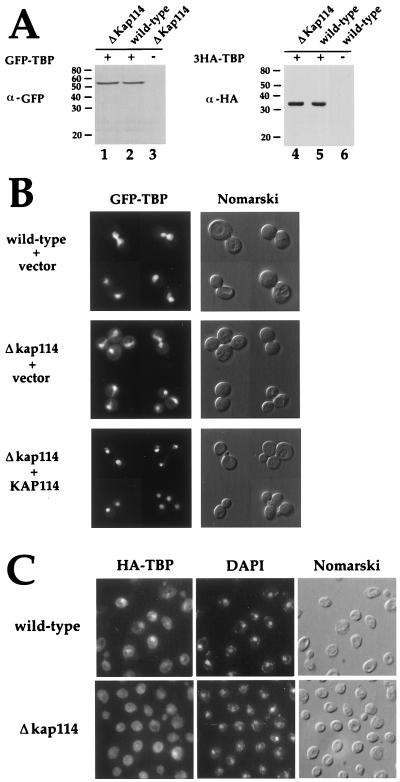
With TBP as a candidate substrate for Kap114-mediated transport, we set out to determine whether TBP was mislocalized in cells lacking Kap114. Wild-type and Δkap114 strains were transformed with a plasmid encoding either GFP-TBP or an epitope-tagged triple-hemagglutinin-TBP (3HA-TBP) expressed from the TBP promoter (2). Patterson et al. have shown that GFP-TBP is fully functional (2). Immunoblotting showed that GFP-TBP and 3HA-TBP run at their predicted molecular masses (54 and 33 kDa, respectively), were intact, and were expressed at equal levels in both wild-type and ΔKap114 cells (Fig. 5A).
Figure 5.
TBP is partially mislocalized to the cytoplasm in cells lacking Kap114. (A) A centromeric plasmid encoding GFP-TBP (pDP15GFPTBP) (2) was transformed into a Δkap114 strain (PSY1784) and the isogenic wild-type strain (PSY1785). A centromeric plasmid encoding 3HA-TBP (3HA-TBP/YCplac111) was transformed into a Δkap114 strain (YSB103) and the isogenic wild-type strain (YSB105). Whole cell lysates were prepared from cells grown at 25°C in selective media. Equal amounts of protein were resolved by SDS/PAGE and then were analyzed by immunoblot using α-GFP antibodies (lanes 1–3) or 12CA5 α-HA antibodies (lanes 4–6). Lanes: 1, Δkap114 cells + GFP-TBP; 2, wild-type cells + GFP-TBP; 3, Δkap114 cells; 4, Δkap114 + 3HA-TBP; 5, wild-type cells + 3HA-TBP; 6, wild-type cells + vector (YCplac111). (B) The strains expressing GFP-TBP described in A also were transformed with either vector alone (YEp24) or with a 2μ plasmid expressing Kap114 (YEpTDS2a). Cells were grown to mid-log phase at 25°C in selective media and then were analyzed by fluorescence microscopy. Cells were visualized by using a GFP filter set (Left) and by Nomarski optics (Right). (Top, wild-type + vector) The localization of GFP-TBP in wild-type cells (PSY1785) that have been transformed with pDP15GFPTBP and YEp24 vector. (Middle, Δkap114 + vector) The localization of GFP-TBP in Δkap114 cells that have been transformed with pDP15GFPTBP and YEPp24 vector. (Bottom, Δkap114 + KAP114) The localization of GFP-TBP in Δkap114 cells that have been transformed with pDP15GFPTBP and the 2μ KAP114/TDS2 plasmid YEpTDS2. (C) The strains expressing 3HA-TBP in A were grown to mid-log phase at 25°C in selective media and were fixed, and immunofluorescence was performed by using 12CA5 α-HA antibodies and FITC-conjugated secondary antibodies. Cells were visualized by using a FITC filter set (Left), a DAPI filter set (Center), and Nomarski optics (Right). (Upper) The localization of 3HA-TBP in wild-type cells (YSB105). (Lower) The localization of 3HA-TBP in Δkap114 cells (YSB103).
The localization of GFP-TBP was examined in wild-type and in Δkap114 cells by fluorescence microscopy. As predicted, GFP-TBP was localized to the nucleus in wild-type cells (Fig. 5B; ref. 2). GFP-TBP was partially mislocalized to the cytoplasm in ≈70% of Δkap114 cells (Fig. 5B). Similar results were obtained by immunofluorescence microscopy of cells expressing 3HA-TBP (Fig. 5C). Expression of Kap114 from a plasmid corrected the mislocalization of GFP-TBP in cells missing Kap114, indicating that the cytoplasmic localization was attributable to loss of Kap114 and not an independent mutation (Fig. 5B).
Kap114 Binds to TBP, and This Interaction Is Disrupted by GTPγS.
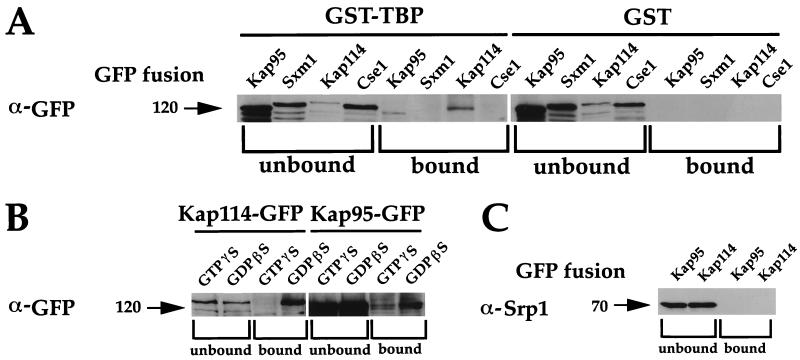
If Kap114 can import TBP into the nucleus, one would predict a physical interaction between the proteins. To determine whether Kap114 interacts with TBP, whole cell lysates of yeast expressing Kap114-GFP were incubated with GST-TBP and GST-bound glutathione-agarose beads. Supernatants and bound material were resolved by SDS/PAGE and were immunoblotted with α-GFP antibodies. Kap114 bound to GST-TBP but not to GST alone (Fig. 6A). As a test for specificity, several other importin-GFP fusions also were tested for binding to GST-TBP. Cse1-GFP and Sxm1-GFP did not bind to GST-TBP (Fig. 6A). Kap95-GFP exhibited some binding to GST-TBP but not to GST alone. It is interesting to note that, although Kap95 was several times more abundant than Kap114 in whole cell lysate, Kap95 binding to GST-TBP appears to be weaker than Kap114 binding (Fig. 6 A and B; data not shown).
Figure 6.
Kap114 binds to TBP. (A) Whole cell lysates were prepared from cells bearing GFP fusions to either Kap95 (PSY1786), Sxm1 (PSY1267) (11), Kap114 (PYS1783), or Cse1 (PSY1262) (19). Lysates were incubated overnight with GST-TBP or GST bound to glutathione-agarose beads. Pellets (bound) and ≈10 μg of supernatants (unbound) were resolved by SDS/PAGE and were immunoblotted by using α-GFP antibodies. (B) The TBP/Kap114 interaction is disrupted by a nonhydrolyzable GTP analogue. Lysates were incubated in the presence and absence of 1 mM GTPγS or 1 mM GDPβS before binding as in A. (C) The blots in A also were probed with α-Srp1 antibodies (Kap114 and Kap95 lanes shown).
A common feature of importin/cargo interactions is their sensitivity to GTP (4). Artificially increasing the concentration of RanGTP both in vivo and in vitro has been shown to disrupt importin/cargo binding (4). As would be predicted of an importin/cargo interaction, binding of Kap114-GFP to GST-TBP was completely disrupted by the addition of the nonhydrolyzable analogue GTPγS to the cell lysates (Fig. 6B). The Kap114-GFP/GST-TBP interaction was not altered by the addition of GDPβS to the cell lysates (Fig. 6B; data not shown). The Kap95-GFP/GST-TBP interaction was decreased, but not completely disrupted, by the addition of GTPγS to cell lysates (Fig. 6B). The addition of GDPβS to the cell lysates did not increase the amount of Kap95-GFP binding to GST-TBP (Fig. 6B; data not shown).
These blots also were probed with other antibodies to determine whether other transport factors were a part of this Kap114-GFP/GST-TBP complex. Blotting with α-Srp1 revealed no significant binding of Srp1 to GST-TBP (Fig. 6C). In addition, no Gsp1 (the essential yeast Ran) was seen bound to GST-TBP (data not shown). However, we cannot rule out the possibility that some Srp1 or Gsp1 was bound but below our levels of detection. Probing of blots with α-GST antibodies indicated that GST and GST-TBP were present and intact (data not shown).
Discussion
We have identified a transport pathway in S. cerevisiae. Kap114, a member of the importin/karyopherin family of proteins in yeast, is a mediator of TBP nuclear import. Kap114 overexpression suppresses spt15 mutant alleles in an allele-nonspecific manner, and TBP is partially mislocalized in cells lacking Kap114. Furthermore, Kap114 binds to GST-TBP, and this binding is disrupted by the addition of a nonhydrolyzable GTP analogue. We note that, while this paper was in review, another group using a different approach also found that Kap114 could interact with and transport TBP (25).
The allele-nonspecific nature of the Kap114 suppression is consistent with Kap114 being an importer of TBP into the nucleus; overexpression of Kap114 may boost nuclear levels of TBP in these mutant strains, thus compensating for partially defective TBP function. We demonstrate that at least one mechanism by which overexpression of Kap114 suppresses TBP temperature-sensitive alleles is by increasing total intracellular TBP levels. This increase in levels might be attributable to slower degradation kinetics of TBP in the nucleus than in the cytoplasm.
The observation that the interaction of Kap114 and TBP is completely disrupted by GTPγS is also consistent with our conclusion that Kap114 imports TBP. It has been shown that RanGTP drives the dissociation of the importin-α/cargo complex from importin-β and the dissociation of the import receptor transportin from its cargo (4). The current model is that the relatively high concentration of GTP-bound Ran in the nucleus, compared with the relatively high levels of GDP-bound Ran in the cytoplasm, drives the dissociation of nuclear import receptors and their cargo (4).
Kap114-mediated import is clearly not the only pathway by which TBP can enter the nucleus. TBP was still localized primarily to the nucleus in cells missing Kap114. Furthermore, TBP is essential for cell viability whereas Kap114 is not. Other mechanisms by which TBP can enter the nucleus remain to be determined. TBP may diffuse into the nucleus, as it is theoretically small enough to pass through the nuclear pores. Alternatively, TBP may enter the nucleus in a complex with other proteins. In fact, it remains a possibility that the Kap114/GST-TBP interactions are indirect. For example, TBP may enter the nucleus via a piggy-back mechanism in which Kap114 does not recognize TBP itself but, rather, a protein with which it is complexed, such as a TAF. However, Pemberton et al. recently demonstrated that Kap114 and TBP directly interact (25). Furthermore, overexpression of Kap114 did not suppress several different temperature-sensitive alleles of pol II and pol III TAFs (data not shown). However, it remains possible that distinct pathways could exist for transporting, and perhaps independently regulating, the TBP-containing complexes necessary for pol I, II, and III transcription. Regulated nuclear localization of gene-specific transcription factors is a common mechanism for controlling gene expression, and it would be interesting if basal transcription factors were similarly regulated.
Kap95 bound to GST-TBP, and this binding was significantly decreased by the presence of GTPγS. These data suggest that Kap95 may be another TBP import receptor. However, Kap95 overexpression did not suppress the temperature sensitivity of the TBP mutant strain YSB66. TBP does not appear to have a nuclear localization signal that would normally be recognized by the importin-α/importin-β heterodimer. Furthermore, we did not detect any Srp1 binding to the Kap95/GST-TBP complex. These observations suggest that, if Kap95 is involved in TBP transport, it may be doing so in an importin-α-independent manner. It has been shown that importin-β mediates the transport of several different cargoes in an importin-α independent manner (26–31).
It is also possible that Kap95p and Kap114 may act together to mediate TBP transport. We previously reported that Kap95 and the importin Pse1 could be co-immunoprecipitated from yeast whole cell lysates (11). One interpretation of this result is that Kap95 and Pse1 work together in a complex to mediate transport. It also has been recently shown that two vertebrate importins, importin-β and importin 7 (RanBP7), act together in the binding and import of linker histone H1 (31). Perhaps many importins work in combination to mediate the transport of particular proteins or protein complexes.
We also identified eIF-5A as an allele-nonspecific suppressor of mutations in TBP. This result is particularly interesting considering that eIF-5A has been reported to be involved in the transport of the HIV-1 protein rev and its bound Rev-responsive element-containing transcript (32–34). However, the role of eIF-5A in transport remains controversial (26, 35). Other studies indicate that eIF-5A may be involved translation (36–39), raising the possibility that the mechanism of eIF-5A suppression involves increasing translation levels to compensate for reduced mRNA, tRNA, or rRNA levels in TBP mutant strains.
We have defined a transport pathway by demonstrating that the previously uncharacterized importin Kap114 is a mediator of TBP import into the nucleus. These studies open exciting possibilities to further define the components of this transport pathway and to find other cargoes that may be imported via this pathway.
Acknowledgments
We are very grateful to Hong Zhou for technical assistance in the early stages of this work. We thank George Patterson and David Piston for GFP-TBP plasmids (pDP15GFPTBP), Laurent Kuras and Kevin Struhl for 3HA-TBP plasmids, John Hershey for YEp352T-5A and YEp352T-5A(K51R), Dave Chao for the GST-TBP plasmid, and members of the Silver and Buratowski labs for helpful discussions. This work was supported by the National Institutes of Health (grants to P.A.S. and S.B.). S.B. is supported by a Pew Scholar in Biomedical Sciences Award and a Junior Faculty Research Award from the American Cancer Society.
Abbreviations
- TBP
TATA-binding protein
- TAF
TBP-associated factor
- GFP
green fluorescent protein
- eIF-5A
eukaryotic translation initiation factor 5A
- GST
glutathione S-transferase
- HA
hemagglutinin
References
- 1.Hernandez N. Genes Dev. 1993;7:1291–1308. doi: 10.1101/gad.7.7b.1291. [DOI] [PubMed] [Google Scholar]
- 2.Patterson G H, Schroeder S C, Bai Y, Weil A, Piston D D. Yeast. 1998;14:813–825. doi: 10.1002/(SICI)1097-0061(19980630)14:9<813::AID-YEA280>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 3.Mattaj I W, Englmeier L. Annu Rev Biochem. 1998;67:265–306. doi: 10.1146/annurev.biochem.67.1.265. [DOI] [PubMed] [Google Scholar]
- 4.Görlich D. EMBO J. 1998;17:2721–2727. doi: 10.1093/emboj/17.10.2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Görlich D, Dabowski M, Bischoff F R, Kutay U, Bork P, Hartmann E, Prehn S, Izaurralde E. J Cell Biol. 1997;138:65–80. doi: 10.1083/jcb.138.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moroianu J. J Cell Biochem. 1998;70:231–239. [PubMed] [Google Scholar]
- 7.Pennisi E. Science. 1998;279:1129–1131. doi: 10.1126/science.279.5354.1129. [DOI] [PubMed] [Google Scholar]
- 8.Albertini M, Pemberton L F, Rosenblum J S, Blobel G. J Cell Biol. 1998;143:1447–1455. doi: 10.1083/jcb.143.6.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buratowski S, Zhou H. Cell. 1992;71:221–230. doi: 10.1016/0092-8674(92)90351-c. [DOI] [PubMed] [Google Scholar]
- 10.Carlson M, Botstein D. Cell. 1982;28:145–154. doi: 10.1016/0092-8674(82)90384-1. [DOI] [PubMed] [Google Scholar]
- 11.Seedorf M, Damelin M, Kahana J, Taura T, Silver P A. Mol Cell Biol. 1999;19:1547–1557. doi: 10.1128/mcb.19.2.1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sikorski R S, Hieter P. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Winston F, Dollard C, Ricuperso-Hovasse S L. Yeast. 1995;11:53–55. doi: 10.1002/yea.320110107. [DOI] [PubMed] [Google Scholar]
- 14.Baudin A, Ozier-Kalogeropoulou O, Denouel A, Lacroute F, Cullin C. Nucleic Acids Res. 1993;21:3329–3330. doi: 10.1093/nar/21.14.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buratowski S, Zhou H. Science. 1992;255:1130–1132. doi: 10.1126/science.1546314. [DOI] [PubMed] [Google Scholar]
- 16.Schnier J, Schwelberger H G, Smit-McBride Z, Kang H A, Hershey J W. Mol Cell Biol. 1991;11:3105–3114. doi: 10.1128/mcb.11.6.3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kunzler M, Hurt E C. FEBS Lett. 1998;433:185–190. doi: 10.1016/s0014-5793(98)00892-8. [DOI] [PubMed] [Google Scholar]
- 18.Solsbacher J, Maurer P, Bischoff F R, Schlenstedt G. Mol Cell Biol. 1998;18:6805–6815. doi: 10.1128/mcb.18.11.6805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hood J K, Silver P A. J Biol Chem. 1998;273:35142–35146. doi: 10.1074/jbc.273.52.35142. [DOI] [PubMed] [Google Scholar]
- 20.Aitchison J D, Blobel G, Rout M P. Science. 1996;274:624–627. doi: 10.1126/science.274.5287.624. [DOI] [PubMed] [Google Scholar]
- 21.Koepp D M, Wong D H, Corbett A H, Silver P A. J Cell Biol. 1996;133:1163–1176. doi: 10.1083/jcb.133.6.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rout M P, Blobel G, Aitchison J D. Cell. 1997;89:715–725. doi: 10.1016/s0092-8674(00)80254-8. [DOI] [PubMed] [Google Scholar]
- 23.Schlenstedt G, Smirnova E, Deane R, Solsbacher J, Kutay U, Görlich D, Ponstingl H, Bischoff F. EMBO J. 1997;16:6237–6249. doi: 10.1093/emboj/16.20.6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ferrigno P, Posas F, Koepp D, Saito H, Silver P A. EMBO J. 1998;17:5606–5614. doi: 10.1093/emboj/17.19.5606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pemberton L F, Rosenblum J S, Blobel G. J Cell Biol. 1999;145:1407–1417. doi: 10.1083/jcb.145.7.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Henderson B R, Percipalle P. J Mol Biol. 1997;274:693–707. doi: 10.1006/jmbi.1997.1420. [DOI] [PubMed] [Google Scholar]
- 27.Truant R, Fridell R A, Benson R E, Bogerd H, Cullen B R. Mol Cell Biol. 1998;18:1449–1458. doi: 10.1128/mcb.18.3.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Truant R, Cullen B R. Mol Cell Biol. 1999;19:1210–1217. doi: 10.1128/mcb.19.2.1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Palmeri D, Malim M H. Mol Cell Biol. 1999;19:1218–1225. doi: 10.1128/mcb.19.2.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lam M H, Briggs L J, Hu W, Martin T J, Gillespie M T, Jans D A. J Biol Chem. 1999;274:7391–7398. doi: 10.1074/jbc.274.11.7391. [DOI] [PubMed] [Google Scholar]
- 31.Jäkel S, Albig W, Kutay U, Bischoff F R, Schwamborn K, Doenecke D, D G. EMBO J. 1999;18:2411–2423. doi: 10.1093/emboj/18.9.2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ruhl M, Himmelspach M, Bahr G M, Hammerschmid F, Jaksche H, Wolff B, Aschauer H, Farrington G K, Probst H, Bevec D, et al. J Cell Biol. 1993;123:1309–1320. doi: 10.1083/jcb.123.6.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bevec D, Jaksche H, Oft M, Wohl T, Himmelspach M, Pacher A, Schebesta M, Koettnitz K, Dobrovnik M, Csonga R, et al. Science. 1996;271:1858–1860. doi: 10.1126/science.271.5257.1858. [DOI] [PubMed] [Google Scholar]
- 34.Schatz O, Oft M, Dascher C, Schebesta M, Rosorius O, Jaksche H, Dobrovnik M, Bevec D, Hauber J. Proc Natl Acad Sci USA. 1998;95:1607–1612. doi: 10.1073/pnas.95.4.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shi X P, Yin K C, Waxman L. Biol Signals. 1997;6:143–149. doi: 10.1159/000109120. [DOI] [PubMed] [Google Scholar]
- 36.Benne R, Brown-Luedi M L, Hershey J W. J Biol Chem. 1978;253:3070–3077. [PubMed] [Google Scholar]
- 37.Benne R, Hershey J W. J Biol Chem. 1978;253:3078–3087. [PubMed] [Google Scholar]
- 38.Park M H, Wolff E C, Folk J E. Biofactors. 1993;4:95–104. [PubMed] [Google Scholar]
- 39.Kang H A, Hershey J W. J Biol Chem. 1994;269:3934–3940. [PubMed] [Google Scholar]