Abstract
The endogenous clock that drives circadian rhythms is thought to communicate temporal information within the cell via cycling downstream transcripts. A transcript encoding a glycine-rich RNA-binding protein, Atgrp7, in Arabidopsis thaliana undergoes circadian oscillations with peak levels in the evening. The AtGRP7 protein also cycles with a time delay so that Atgrp7 transcript levels decline when the AtGRP7 protein accumulates to high levels. After AtGRP7 protein concentration has fallen to trough levels, Atgrp7 transcript starts to reaccumulate. Overexpression of AtGRP7 in transgenic Arabidopsis plants severely depresses cycling of the endogenous Atgrp7 transcript. These data establish both transcript and protein as components of a negative feedback circuit capable of generating a stable oscillation. AtGRP7 overexpression also depresses the oscillation of the circadian-regulated transcript encoding the related RNA-binding protein AtGRP8 but does not affect the oscillation of transcripts such as cab or catalase mRNAs. We propose that the AtGRP7 autoregulatory loop represents a “slave” oscillator in Arabidopsis that receives temporal information from a central “master” oscillator, conserves the rhythmicity by negative feedback, and transduces it to the output pathway by regulating a subset of clock-controlled transcripts.
An endogenous clock imposes rhythmicity on physiological processes in plants, animals, and some prokaryotes with an approximately 24-h period, reflecting the period of the Earth’s rotation (1–4). During the last two decades, the analysis of mutants affected in circadian-regulated output has provided valuable insights into the central clock machinery. A concept has been developed where a “master” clock controls individual subordinated “slave” oscillators, each of which in turn controls overt rhythms (5).
Single genes regulating conidiation rhythms in Neurospora crassa and eclosion rhythms in Drosophila melanogaster have been isolated (6–9). Current understanding of the biochemical mechanism underlying circadian timekeeping is largely based on the characteristics of the Neurospora FREQUENCY (FRQ) and Drosophila PERIOD (PER) and TIMELESS (TIM) proteins (10–12). These clock proteins negatively regulate oscillation of their own transcripts, thereby forming an autoregulatory feedback circuit involving transcription of the clock gene, translation, posttranslational protein modification, and nuclear import (2, 9, 13, 14). The analysis of the per homologue in the giant silkmoth Antheraea pernyi recently demonstrated that this clock molecule can operate in a distinctly different way in the adult brain: PER oscillations are not the result of a transcriptional autoregulatory loop but rather generated by an endogenous per antisense RNA that oscillates antiphasic to per and might block per transcript function in a cyclic manner (15). It is generally accepted but not yet proven that the clock proteins transduce temporal information to generate the overt rhythms by causing downstream transcripts to cycle (3, 16).
In higher plants, a large number of transcripts undergo circadian oscillations (17–19). Because none of the corresponding gene products has been shown to be causally involved in the generation of rhythmicity, presumably these rhythms merely reflect outputs from the clock. So far only in Arabidopsis thaliana, clock-related mutants have been identified employing luciferase reporter gene activity under control of a circadian-regulated promoter as a screenable clock-output phenotype (20).
In contrast to this strategy of clock mutant analysis we have chosen a reverse-genetics approach to investigate the potential role of a circadian-regulated RNA-binding protein in the genesis of endogenous rhythmicity in Arabidopsis thaliana. This protein previously was identified in a screen for transcripts differentially expressed as a function of time of day (19, 21, 22).
EXPERIMENTAL PROCEDURES
RNA Isolation and RNA Gel Blot Hybridization.
Isolation of total RNA, separation on 1.5% agarose-formaldehyde gels, and transfer to nylon membranes (GeneScreen, DuPont) were performed as described (21, 22). cDNA probes were radioactively labeled with the “Prime it II” kit (Stratagene). Gene- and strand-specific antisense probes that distinguish between the Arabidopsis thaliana genes Atgrp7 (23), also designated ccr2 (24), and Atgrp8 (23), also designated ccr1 (24), both encoding glycine-rich RNA-binding proteins with 75% sequence identity (23, 24), were derived from the 5′ untranslated regions. They were obtained by replacing the nonamer primers in the labeling reaction by oligonucleotides covering the respective translation start sites. Hybridization was performed according to ref. 25. The Northern blots were quantitated using a PhosphorImager (Molecular Dynamics) and associated software. Atgrp7 signals were normalized to signals obtained by hybridization with a barley 26 S rDNA probe (26).
Protein Isolation and Immunoblots.
Four-week-old plants were ground in liquid nitrogen. To minimize variation in the extraction, 100 μl of sample buffer (21, 22) was added per 10 mg of powder. Samples were boiled for 10 min and insoluble material was pelleted. The protein concentration of the supernatant was determined according to Esen (27). A test gel was stained with Coomassie blue to check for sample variation. Five micrograms of total protein was separated on 15% SDS polyacrylamide gels and electroblotted to Polyvinyliden difluoride membranes (Pierce). AtGRP7 was assayed using an antiserum against bacterially expressed SaGRP (21) at a 1:2,500 dilution. In some of the experiments, levels of plastid ATPase (α and β subunits) were assayed afterward using an antiserum raised against rye ATPase (28) at a 1:4,000 dilution, followed by chemiluminescence detection (POD chemiluminescence kit, Boehringer Mannheim). Following the immunodetection, the filters were stained with 0.1% amido black in 45% methanol/10% acetic acid and destained in 80% methanol/4% acetic acid to verify equal sample application. Scans of the immunoblot were evaluated using the nih image 1.59 program. AtGRP was normalized against loaded protein by densitometry of the amido black-stained filter.
Plasmid Construction and Transformation of Arabidopsis thaliana.
The Atgrp7 protein-coding region was amplified by PCR from the cDNA with primers 5′-GGCCATGGCGTCCGGTGAT-3′ and 5′-GGGATCCTTACCATCCTCCACC-3′ covering the translation start and stop (bold) and comprising engineered NcoI and BamHI sites (underlined), respectively. The gel-purified 540-bp amplification product was inserted into the vector pRT104 (29) between the cauliflower mosaic virus (CaMV) 35S RNA promoter with the duplicated enhancer (30) fused to the tobacco mosaic virus omega element (31) and the CaMV polyadenylation signal. The entire expression cassette was transferred as a HindIII fragment to the binary vector pBin19 (32) and introduced into Arabidopsis thaliana strain C24 by Agrobacterium-mediated root-transformation (33). Calli resistant to 50 μg/ml kanamycin were regenerated, rooted, and allowed to set seeds.
For Northern blot kinetics, kanamycin-resistant F2 seedlings were grown on ½ MS medium (34) in light/dark cycles, as indicated in the figure legends.
RESULTS
As a way to understand the molecular mechanisms underlying clock control of rhythmic endogenous processes, we previously isolated oscillating transcripts differentially expressed as a function of time of day in the long-day plant Sinapis alba (white mustard) by subtractive hybridization (21, 22). One of these transcripts, which reaches its maximal concentration in the evening, codes for a glycine-rich protein (SaGRP) with an N-terminal RNA recognition motif. Based on the homology to RNA-binding proteins such as nucleolin (35) and the splicing factor hnRNPA1 (36), and its ability to interact with RNA (unpublished observation; ref. 24) as well as its localization within the nucleus (21), it seems reasonable to assume a regulatory role for this plant protein.
As a first step toward the analysis of the RNA-binding protein in transgenic Arabidopsis thaliana plants, we have isolated the Arabidopsis counterpart of Sagrp1, which corresponds to both Atgrp7 (23) and ccr2 (24). As in Sinapis alba, an additional derivative of this cDNA was isolated. It contains a 300-bp insertion within the RNA recognition motif and corresponds to the unspliced 1-kb pre-mRNA (21).
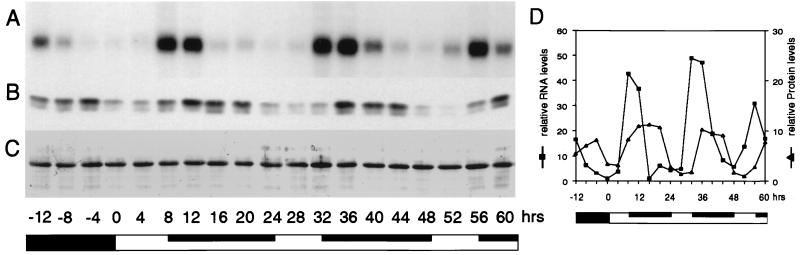
Northern blot analysis using a gene-specific probe confirmed that the Atgrp7 transcript, like Sagrp1, undergoes oscillations in young plants grown in light/dark cycles, with the highest levels occurring 8 to 12 h after onset of illumination and persisting thereafter in constant light (Fig. 1A) (21, 24). Peak quantities of Atgrp7 transcript are about 40 times the amount at the trough (Fig. 1D). Immunoblot analysis demonstrated that the AtGRP7 steady state concentration also oscillates (Fig. 1B). Most notably, AtGRP7 protein peaks are delayed relative to the Atgrp7 transcript peaks by about 4 h. When the AtGRP7 protein has accumulated to high levels, the Atgrp7 transcript level declines and does not rise again until the AtGRP7 protein has reached its trough (Fig. 1D). This delayed oscillation of the RNA-binding protein relative to its transcript could reflect a translational control or could be the result of an autoregulatory feedback loop in which an elevated level of the protein negatively affects the accumulation of its own transcript. In the latter case, constitutive overexpression of this protein in transgenic plants should eliminate the detectable oscillation of its endogenous transcript.
Figure 1.
Atgrp7 mRNA and protein cycling in Arabidopsis thaliana: Delay of the protein peak relative to the transcript peak. Plants were first entrained to light/dark cycles (LD 8:16), harvested at 4-h intervals and subsequently left in LL for the indicated number of hours after “lights on” on the final day in LD. (A) RNA gel blot with 10 μg of total RNA. Atgrp7 transcript was detected with a gene-specific probe derived from the 5′ untranslated region. (B) Protein gel blot of the same plants. Immunodetection of AtGRP (B) and amido black staining of the filter (C). (D) Relative Atgrp7 transcript and AtGRP protein levels. Atgrp7 RNA levels were normalized to controls obtained with rDNA hybridization. Densitometric evaluation of the stained filter in C was used to normalize protein levels in B. Values are expressed relative to the minimal level defined as 1. The solid and open bars represent dark and light periods, respectively. The inserted dark bars indicate subjective night. hrs, hours before and after onset of illumination on the last day in LD.
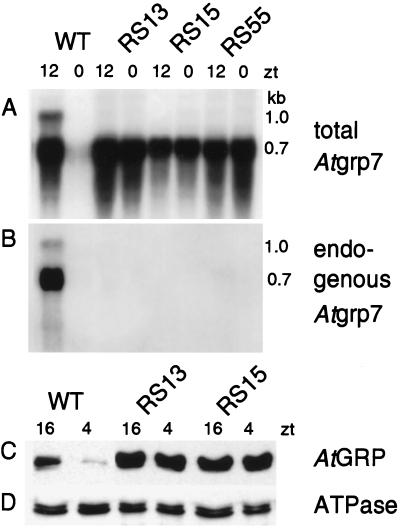
To obtain high, constitutive AtGRP7 expression, the coding sequence under control of the CaMV promoter with the duplicated enhancer (30) was introduced into Arabidopsis thaliana. Atgrp7 steady-state transcript concentrations were assayed in F2 seedlings grown in light/dark cycles (Fig. 2A). Three lines (RS13, RS15, and RS55) were identified that show equally high Atgrp7 transcript levels in the morning (zt0; zt, zeitgeber time) as well as in the evening (zt12), in contrast to the wild-type plants. Immunoblot analysis confirmed a high AtGRP7 protein level at the minimum (zt4) as well as at the maximum (zt16) due to transgene expression in contrast to the wild-type plants (shown for lines RS13 and RS15 in Fig. 2C).
Figure 2.
AtGRP7 overexpression suppresses the endogenous Atgrp7 transcript. RNA was isolated from three independent transgenic lines and wild-type controls grown in light/dark cycles at zt0 and zt12. The Northern blot with 20 μg of RNA was hybridized with the Atgrp7 cDNA to measure total Atgrp7 transcript levels (A). Note that due to the higher loading the 1-kb pre-mRNA is clearly detectable in the wild-type plants in contrast to Fig. 1A. After stripping of the membrane, expression of the endogenous Atgrp7 transcript was monitored by hybridization with a gene-specific probe derived from the 5′ untranslated region not contained within the transgene (B). (C and D) Immunoblot analysis of wild-type plants and the transgenic lines RS13 and RS15, harvested at the time of the protein trough (zt4) and the protein peak (zt16) with antibodies against SaGRP (C) and the α and β subunits of ATPase (D).
In the wild-type plants, an additional 1.0-kb Atgrp7 transcript species is visible (Fig. 2A). It corresponds to the unspliced pre-mRNA which, due to inefficient removal of the intron, accumulates to levels that can be detected on blots with high amounts of RNA (21, 24). In the Arabidopsis lines overexpressing AtGRP7, this 1-kb pre-mRNA is absent. The high AtGRP7 level from the CaMV-AtGRP7 construct might thus have either selectively decreased the level of the unspliced pre-mRNA or repressed the level of all of the endogenous Atgrp7 transcript species. To discriminate between these two alternatives, the blot was reprobed with the Atgrp7 5′ untranslated region that distinguishes between the transcripts of the endogenous gene and the transgene (Fig. 2B). Almost no endogenous Atgrp7 transcript could be detected in the transgenic lines at zt12, whereas both the fully spliced Atgrp7 transcript and the pre-mRNA were detectable in the wild type. Thus, overexpression of AtGRP7 greatly suppresses the abundance of all of the endogenous Atgrp7 transcripts.
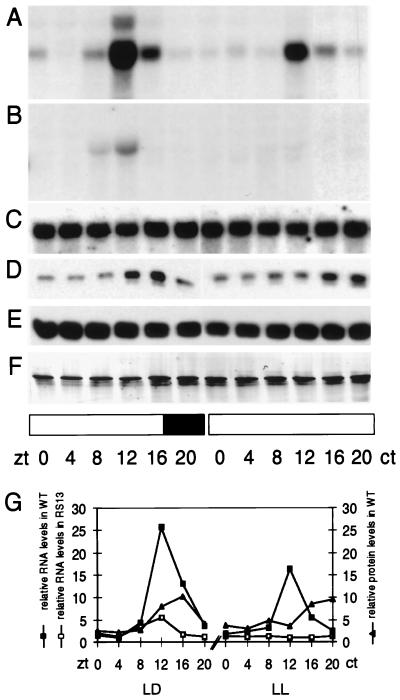
To examine this molecular phenotype in greater detail, a time course of Atgrp7 expression in transgenic and control plants was made over the entire circadian cycle (Fig. 3). The Atgrp7 transcript showed a high, relatively uniform level in light/dark cycles (LD) as well as in continuous light (LL) in the overexpressing line RS13 (Fig. 3C). Similarly, the AtGRP protein level did not show circadian oscillations in the transgenic line (Fig. 3E) in contrast to wild-type plants (Fig. 3D). Whereas normal Atgrp7 cycling was observed both in LD and in LL in wild-type plants (Fig. 3A), oscillations of the endogenous Atgrp7 transcript in transgenic plants were severely depressed in LD (Fig. 3 B Left and G) and no longer detectable in LL (Fig. 3 B Right and G).
Figure 3.
Atgrp7 transcript and AtGRP protein time course in wild-type plants and the transgenic line RS13. Plants were harvested at 4-h intervals during one light/dark cycle and on the second day after transfer to LL. Expression of the endogenous Atgrp7 transcript was monitored with a gene-specific probe derived from the 5′ untranslated region, which also included part of the promoter to increase specific activity of the hybridization probe, in wild-type plants (A) and in the representative line RS13 (B). (C) Total Atgrp7 transcript level in the transgenic line RS13 measured by hybridization with the Atgrp7 cDNA. (D) Immunoblot analysis of wild-type plants with the antibody against SaGRP. (E) Immunoblot analysis of the transgenic line RS13 with the antibody against SaGRP and amido black staining of the filter (F). (G) Quantitation of the endogenous Atgrp7 transcript profile in wild-type plants, shown in A, and the transgenic line RS13, shown in B, and AtGRP protein oscillations in wild-type plants, shown in D. Atgrp7 RNA levels were normalized to controls obtained with rDNA hybridization. Protein levels were normalized to the densitometric evaluation of the amido black-stained blot. Identical results were obtained with the transgenic line RS15. The solid and open bars represent dark and light periods, respectively.
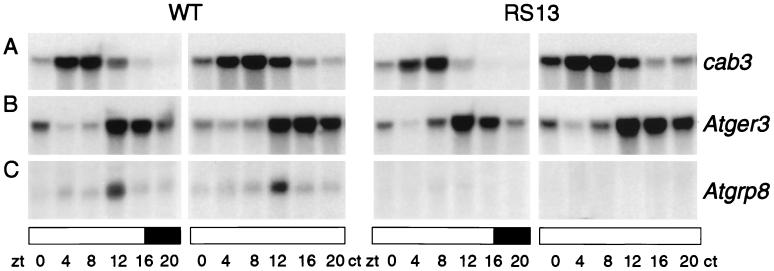
The presence of an RNA recognition motif suggests that AtGRP7 might interact with other transcripts and in this way may confer circadian rhythmicity on them. Therefore we compared the steady-state concentrations of selected oscillating transcripts, representing different circadian phases, in wild-type plants with those in the AtGRP7 overexpressing lines. No significant differences between control plants and the transgenic line RS13 were observed for the cab (chlorophyll a/b binding protein) transcripts that peak around noon (Fig. 4A) (37), for a germin-like protein, Atger3, that peaks in the late evening (Fig. 4B) (38), and for catalase3 (39) (not shown) in light–dark cycles, as well as in constant light.
Figure 4.
Influence of AtGRP7 overexpression on other circadian-regulated transcripts. RNA was isolated from wild-type plants and the representative transgenic line RS13, which were harvested at 4-h intervals in a light/dark cycle (LD 16:8) and on the second day after transfer to continuous illumination. The Northern blot with 10 μg of RNA was hybridized with a cab3 probe (37), which recognizes all the cab transcripts (A), a probe for a germin-like protein, Atger3, (38) (B), and a gene-specific probe derived from the 5′ untranslated region of the Atgrp8 gene (23) (C), respectively. Note that transcript peaks are delayed due to the LD 16:8 conditions used in this experiment compared with the phase of maximal transcript accumulation observed in LD 8:16 (cf. Fig. 1 and our unpublished observation). Identical results were obtained for the transgenic line RS15. The solid and open bars represent dark and light periods, respectively.
However, circadian oscillations of Atgrp8, a transcript encoding a related glycine-rich RNA-binding protein (23, 24) that cycles in phase with Atgrp7 in the wild-type plants (Fig. 4C Left), were almost fully suppressed in the transgenic plants in LD and not detectable any more in LL (Fig. 4C Right).
DISCUSSION
In the present study, we identify the RNA-binding protein AtGRP7 as the first component of a circadian-regulated feedback loop in higher plants. Our data indicate that AtGRP7 serves a 2-fold role both as target and modulator of circadian regulation, because it also influences cycling of a heterologous transcript.
We show that the robust Atgrp7 transcript oscillation leads to circadian cycling of the AtGRP7 protein. Increasing AtGRP7 protein concentration coincides with the decay of Atgrp7 mRNA quantity, and, conversely, the Atgrp7 mRNA level does not rise again before AtGRP7 has reached its trough level. These findings are consistent with Atgrp7 and AtGRP7 being part of a negative autoregulatory circuit. Moreover, the observed phase difference of about 4 h might provide a delay to generate a stable oscillation. Without such a delay the oscillation in the loop would damp out rather quickly and come to equilibrium (4, 40). To explain the phase of AtGRP7 protein expression, an as yet undetermined posttranscriptional mechanism in addition to mRNA cycling has to be assumed. Persistent 24-h oscillations of the Drosophila clock components PER and TIM, for example, are assumed to depend on delayed nuclear entry of the PER and TIM proteins. In this manner, per and tim expression could proceed for several hours until PER and TIM repress accumulation of their cognate transcripts within the nucleus (11, 14, 41, 42).
Constitutive AtGRP7 overexpression in transgenic Arabidopsis leads to a dramatic depression of Atgrp7 transcript oscillations, proving that AtGRP7 indeed exerts a negative feedback onto its own transcript. In LD-grown plants, the residual low-level Atgrp7 oscillation in LD suggests that there is input from an unknown external factor. The level of AtGRP7 repressor activity obtained in the transgenic plants is not sufficient to completely stop the oscillation of Atgrp7 and can still be overridden by this positively acting factor. In LL, Atgrp7 expression can no longer be detected, very likely reflecting the damping effect of these light conditions on Atgrp7 oscillations that is also evident in wild-type plants (Fig. 3A). Recently, in the short-period toc1 (timing of cab expression) mutant, the ccr2 transcript corresponding to Atgrp7 has been demonstrated to oscillate with a period that is significantly shorter than in wild-type plants (43). Therefore, the toc1 gene product could be one candidate for such an external factor that affects the AtGRP7 feedback loop.
Atgrp7 mRNA cycling is mainly generated at the transcriptional level, as the Atgrp7 promoter confers circadian rhythmicity. In transgenic Arabidopsis plants carrying the β-glucuronidase gene under control of a 1.5-kb fragment upstream of the transcription start site, the β-glucuronidase mRNA peaks in the subjective evening, whereas it is barely detectable in the subjective morning (D.S. and M.N., unpublished observation). Therefore, in wild-type plants Atgrp7 transcript levels seem to be elevated through rhythmic transcriptional activation by the clock in the subjective evening. After a lag phase, Atgrp7 mRNA accumulation seems to be repressed by the AtGRP7 protein, resulting in a stable high-amplitude oscillation. The molecular basis of the AtGRP7 autoregulatory circuit remains to be determined. On the one hand, AtGRP7 could interfere with transcriptional activation of its own gene by a central oscillator. This may occur indirectly via association of AtGRP7 with a transcription factor. Such a mechanism has been proposed to account for feedback inhibition of per transcription by the Drosophila clock protein PER (2, 3). Direct action of AtGRP7 as a transcriptional repressor is also conceivable, because a potential interaction of an RNA-binding protein with DNA is not without precedent: hnRNPK has been shown to interact with a polypyrimidine tract in the c-myc promoter and to act as a transcription factor (44). Also, the RNA recognition motif-containing protein mRNP4/FRG Y2 from Xenopus oocytes stimulates transcription from specific promoters (45). Alternatively, AtGRP7 might limit transcript accumulation by influencing transcript stability. In this case, the oscillatory feedback loop cannot solely be described by molecules involved in activation and repression of Atgrp7 transcription. Measuring the effect of a high, constitutive AtGRP7 level on reporter gene constructs with various cis-regulatory parts of the AtGRP7 gene will allow us to discriminate between these different mechanisms.
Whereas direct proof for a function in the generation of circadian physiological processes will require the generation of Atgrp7 mutants, its homology to splicing factors suggests that AtGRP7 might transfer its rhythmic activity to other transcripts by means of RNA maturation processes. We demonstrate that AtGRP7 negatively regulates another circadian-regulated transcript encoding the RNA-binding protein AtGRP8 (23), although other investigated circadian-regulated transcripts, such as cab, a germin-like protein, or catalase mRNAs, are not affected by AtGRP7 overexpression.
Because there is precedent for the existence of more than one master oscillator in plants (46), at present it is conceivable that AtGRP7 might be part of one of such central oscillators that controls a limited set of rhythmic phenomena. Based on Pittendrigh’s (5, 47) concept that the temporal organization within a cell is established by a central “master” pacemaker governing multiple “slave” oscillators, another interpretation arises: The circadian AtGRP7 feedback loop could also represent one of several “slave” oscillators in Arabidopsis that acquires a circadian period by receiving impulses from the “master” clock. The suboscillator would conserve the rhythmicity by feedback regulation and would transduce it to the output pathway, thus controlling a subset of circadian-regulated processes.
Consistent with this view is the observation that in the overexpressing line, despite a high AtGRP7 repressor concentration, there is external input into the feedback loop, allowing for a residual low-level oscillation in LD. Furthermore, the Arabidopsis toc1 mutant shortens the period of cycling of the cab2 promoter activity, leaf movement rhythms, as well as Atgrp7 oscillations (20, 43), indicating that the AtGRP7 autoregulatory circuit receives temporal information from the toc1 gene product. Assuming that toc1 is part of an oscillator, it may govern a possibly multiple-branched pathway, one regulating cab rhythms peaking at midday and another regulating Atgrp7 rhythms peaking in the evening.
Although components of circadian output pathways that affect subsets of clock-controlled processes have been described in other organisms, there is no indication for their function as a slave oscillator, as either no rhythmicity or no feedback regulation have been demonstrated. For instance, the Drosophila lark factor, a putative RNA-binding protein, negatively regulates eclosion but does not affect adult locomoter activity rhythms. Its mRNA does not oscillate in abundance, and it is not known whether the encoded protein displays rhythmic activity (48). In the cyanobacterium Synechococcus, a mutation in a sigma70-like transcription factor results in a low-amplitude rhythm phenotype. This factor seems to be part of a clock output pathway, because its loss affects the rhythmic transcription of a subset of clock-controlled genes (49). However, no negative autoregulation has been demonstrated yet for this gene.
The search for additional genes whose expression is affected by AtGRP7 should allow us to define more precisely the physiological role of this oscillatory feedback loop within the circadian network in Arabidopsis.
Acknowledgments
We thank R. Töpfer and J. Schell for the expression plasmid pRT104, A. Bachmair for the duplicated CaMV enhancer, and J. Feierabend for the ATPase antibody. We thank S. Crosthwaite, Y. Liu, and G. Armstrong for critical comments on the manuscript and D. Rubli and U. Sperling for help in preparing the figures. This work was supported by grants from the Swiss National Foundation and the Swiss Federal Institute of Technology to D.S.
ABBREVIATIONS
- CaMV
cauliflower mosaic virus
- zt
zeitgeber time
- ct
circadian time
- LD
light/dark cycles
- LL
continuous light
- SaGRP
Sinapis alba glycine-rich protein
- AtGRP
Arabidopsis thaliana glycine-rich protein
References
- 1.Sweeney B M. Rhythmic Phenomena in Plants. San Diego: Academic; 1987. [Google Scholar]
- 2.Dunlap J C. Annu Rev Genet. 1996;30:579–601. doi: 10.1146/annurev.genet.30.1.579. [DOI] [PubMed] [Google Scholar]
- 3.Hall J C. Trends Neurosci. 1995;18:230–240. doi: 10.1016/0166-2236(95)93908-g. [DOI] [PubMed] [Google Scholar]
- 4.Kay S A, Millar A J. Cell. 1995;83:361–364. doi: 10.1016/0092-8674(95)90113-2. [DOI] [PubMed] [Google Scholar]
- 5.Pittendrigh C S. In: Handbook of Behavioral Neurobiology: Biological Rhythms. Aschoff J, editor. Vol. 4. New York: Plenum; 1981. pp. 57–80. [Google Scholar]
- 6.McClung C R, Fox B A, Dunlap J C. Nature (London) 1989;339:558–562. doi: 10.1038/339558a0. [DOI] [PubMed] [Google Scholar]
- 7.Bargiello T A, Jackson F R, Young M W. Nature (London) 1984;312:752–754. doi: 10.1038/312752a0. [DOI] [PubMed] [Google Scholar]
- 8.Reddy P, Zehring W A, Wheeler D A, Pirrotta V, Hadfield C, Hall J C, Rosbash M. Cell. 1984;38:701–710. doi: 10.1016/0092-8674(84)90265-4. [DOI] [PubMed] [Google Scholar]
- 9.Myers M P, Wager-Smith K, Wesley C S, Young M W, Sehgal A. Science. 1995;270:805–808. doi: 10.1126/science.270.5237.805. [DOI] [PubMed] [Google Scholar]
- 10.Hardin P E, Hall J C, Rosbash M. Nature (London) 1990;343:536–540. doi: 10.1038/343536a0. [DOI] [PubMed] [Google Scholar]
- 11.Zeng H, Hardin P E, Rosbash M. EMBO J. 1994;13:3590–3598. doi: 10.1002/j.1460-2075.1994.tb06666.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aronson B D, Johnson K A, Loros J J, Dunlap J C. Science. 1994;263:1578–1584. doi: 10.1126/science.8128244. [DOI] [PubMed] [Google Scholar]
- 13.Vosshall L B, Price J L, Sehgal A, Saez L, Young M W. Science. 1994;263:1606–1609. doi: 10.1126/science.8128247. [DOI] [PubMed] [Google Scholar]
- 14.Curtin K D, Huang Z J, Rosbash M. Neuron. 1995;14:365–372. doi: 10.1016/0896-6273(95)90292-9. [DOI] [PubMed] [Google Scholar]
- 15.Sauman I, Reppert S M. Neuron. 1996;17:889–900. doi: 10.1016/s0896-6273(00)80220-2. [DOI] [PubMed] [Google Scholar]
- 16.van Gelder R N, Krasnow M A. EMBO J. 1996;15:1625–1631. [PMC free article] [PubMed] [Google Scholar]
- 17.Beator J, Kloppstech K. Mol Biol (Life Sci Adv) 1994;13:203–219. [Google Scholar]
- 18.Johnson C H. Semin Cell Biol. 1994;5:355–362. doi: 10.1006/scel.1994.1042. [DOI] [PubMed] [Google Scholar]
- 19.Staiger D. In: Vistas on Biorhythmicity. Greppin H, Degli Agosti R, Bonzon M, editors. Geneva: Imprimerie Nationale; 1996. pp. 119–133. [Google Scholar]
- 20.Millar A J, Carre I A, Strayer C A, Chua N-H, Kay S A. Science. 1995;267:1161–1163. doi: 10.1126/science.7855595. [DOI] [PubMed] [Google Scholar]
- 21.Heintzen C, Melzer S, Fischer R, Kappeler S, Apel K, Staiger D. Plant J. 1994;5:799–813. doi: 10.1046/j.1365-313x.1994.5060799.x. [DOI] [PubMed] [Google Scholar]
- 22.Heintzen C, Fischer R, Melzer S, Kappeler S, Apel K, Staiger D. Plant Physiol. 1994;106:905–915. doi: 10.1104/pp.106.3.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Nocker S, Vierstra R. Plant Mol Biol. 1993;21:695–699. doi: 10.1007/BF00014552. [DOI] [PubMed] [Google Scholar]
- 24.Carpenter C D, Kreps J A, Simon A E. Plant Physiol. 1994;104:1015–1025. doi: 10.1104/pp.104.3.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang H, McLesse J, Weisbart M, Dionne J-L, Lemaire I, Aubin R A. Nucleic Acids Res. 1993;21:3337–3338. doi: 10.1093/nar/21.14.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Forde B G, Kreis M, Bahramian M B, Mattews J A, Miflin B J, Thompson R D, Bartels D, Flavell R. Nucleic Acids Res. 1981;9:6689–6707. doi: 10.1093/nar/9.24.6689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Esen A. Anal Biochem. 1978;89:264–273. doi: 10.1016/0003-2697(78)90749-2. [DOI] [PubMed] [Google Scholar]
- 28.Batschauer A, Mösinger E, Kreuz K, Dörr I, Apel K. Eur J Biochem. 1986;154:625–634. doi: 10.1111/j.1432-1033.1986.tb09444.x. [DOI] [PubMed] [Google Scholar]
- 29.Töpfer R, Matzeit V, Gronenborn B, Schell J, Steinbiss H-H. Nucleic Acids Res. 1987;15:5890. doi: 10.1093/nar/15.14.5890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bachmair A, Becker F, Masterson R V, Schell J. EMBO J. 1990;9:4543–4549. doi: 10.1002/j.1460-2075.1990.tb07906.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gallie D R, Sleat D E, Watts J W, Turner P C, Wilson T M A. Nucleic Acids Res. 1987;15:3257–3273. doi: 10.1093/nar/15.8.3257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bevan M. Nucleic Acids Res. 1984;12:8711–8721. doi: 10.1093/nar/12.22.8711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Valvekens D, van Montagu M, van Lijsebettens M. Proc Natl Acad Sci USA. 1988;85:5536–5540. doi: 10.1073/pnas.85.15.5536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murashige T, Skoog F. Physiol Plant. 1962;15:473–497. [Google Scholar]
- 35.Lapeyre B, Bourbon H, Amalric F. Proc Natl Acad Sci USA. 1987;84:1472–1476. doi: 10.1073/pnas.84.6.1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cobianchi F, Karpel R L, Williams K R, Notario V, Wilson S H. J Biol Chem. 1988;263:1063–1071. [PubMed] [Google Scholar]
- 37.Leutweiler L S, Meyerowitz E M, Tobin E M. Nucleic Acids Res. 1986;14:4051–4064. doi: 10.1093/nar/14.10.4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Membre, N., Berna, A., Neuteling, G., David, A., David, H., Staiger, D., Vasquez, J. S., Raynal, M., Delseney, M., Bernier, F. (1997) Plant Mol. Biol., in press. [DOI] [PubMed]
- 39.Zhong H H, McClung C R. Mol Gen Genet. 1996;251:196–203. doi: 10.1007/BF02172918. [DOI] [PubMed] [Google Scholar]
- 40.Friesen W O, Block G D, Hocker C G. Annu Rev Physiol. 1993;55:661–681. doi: 10.1146/annurev.ph.55.030193.003305. [DOI] [PubMed] [Google Scholar]
- 41.Sehgal A, Rothenfluh-Hilfiker A, Hunter-Ensor M, Chen Y, Myers M P, Young M W. Science. 1995;270:808–810. doi: 10.1126/science.270.5237.808. [DOI] [PubMed] [Google Scholar]
- 42.Saez L, Young M W. Neuron. 1996;17:911–920. doi: 10.1016/s0896-6273(00)80222-6. [DOI] [PubMed] [Google Scholar]
- 43.Kreps J A, Simon A E. Plant Cell. 1997;9:297–304. doi: 10.1105/tpc.9.3.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Michelotti E F, Michelotti G A, Aronsohn A I, Levens D. Mol Cell Biol. 1996;16:2350–2360. doi: 10.1128/mcb.16.5.2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Deschamps S, Viel A, Garrigos M, Denis H, le Maire M. J Biol Chem. 1992;267:13799–13802. [PubMed] [Google Scholar]
- 46.Hennessey T L, Field C B. J Biol Rhythms. 1992;7:105–113. doi: 10.1177/074873049200700202. [DOI] [PubMed] [Google Scholar]
- 47.Pittendrigh C S. Cold Spring Harbor Symp Quant Biol. 1960;25:159–184. doi: 10.1101/sqb.1960.025.01.015. [DOI] [PubMed] [Google Scholar]
- 48.Newby L M, Jackson F R. J Neurobiol. 1996;31:117–128. doi: 10.1002/(SICI)1097-4695(199609)31:1<117::AID-NEU10>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 49.Tsinoremas N F, Ishiura M, Kondo T, Andersson C R, Tanaka K, Takahashi H, Johnson C H, Golden S S. EMBO J. 1996;15:2488–2495. [PMC free article] [PubMed] [Google Scholar]