Abstract
Using systematic evolution of ligands by exponential enrichment (SELEX), an RNA molecule was isolated that displays a 1,000-fold higher affinity for guanosine residues that carry an N-7 methyl group than for nonmethylated guanosine residues. The methylated guanosine residue closely resembles the 5′ terminal cap structure present on all eukaryotic mRNA molecules. The cap-binding RNA specifically inhibited the translation of capped but not uncapped mRNA molecules in cell-free lysates prepared from either human HeLa cells or from Saccharomyces cerevisiae. These findings indicate that the cap-binding RNA will also be useful in studies of other cap-dependent processes such as pre-mRNA splicing and nucleocytoplasmic mRNA transport.
The cap structure that is present at the 5′ end of eukaryotic mRNAs consists of a methylated guanosine residue that is linked to the penultimate nucleotide of the RNA through an inverted 5′–5′ triphosphate linkage (1, 2). This unusual linkage of the cap structure to the 5′ end of the mRNA contributes to the stability of the RNA, protecting the 5′ end against attack by exonucleases and phosphatases present in the cell (3).
Both in the nucleus and in the cytoplasm, distinct cap-binding protein complexes assemble at the cap structure. In the nucleus, cap-binding protein complexes are required for efficient splicing of pre-mRNAs in mammalian cell extracts (4–7) and in Xenopus oocyte nuclei (8). Specifically, one nuclear cap-binding complex is required for the recognition of the 5′ splice site of the 5′ proximal intron by U1 small nuclear ribonucleoprotein (9, 10). Nuclear cap-binding complexes have also been implicated in the export of mRNAs from the nucleus into the cytoplasm (11–13). In the cytoplasm, cap-binding complexes, distinct in their composition from nuclear cap-binding complexes, have long been known to have important roles in the initiation of mRNA translation (2, 14). For example, there is considerable evidence that eukaryotic initiation factor 4G (eIF-4G), a component of the cytoplasmic cap-binding protein complex, interacts both with factor eIF-4E, which binds to the 5′ terminal mRNA cap structure, and with factor eIF-3, which is associated with the small 40S ribosomal subunit. It has been proposed that the simultaneous association of eIF-4G with eIF-4E and eIF-3 allows the recruitment of 40S subunits onto the mRNA (15, 16).
Although these biochemical approaches have revealed substantial information about the interactions of cap-binding complexes with both the cap structure and with other proteins known to be required for cap-dependent processes in vitro, the interactions that govern cap-dependent mechanisms inside cells have been difficult to study. This has been due to the inability to introduce cap analogs or other inhibitors that interfere with cap-dependent processes into eukaryotic cells, with the exception of Xenopus oocytes (see above). Here, we report the isolation of a short RNA molecule that binds with high affinity to the 5′ terminal cap structure on mRNAs. Interaction of the RNA with the mRNA cap results in the selective inhibition of cap-dependent translation, probably by competition with the cytoplasmic cap-binding protein complex for binding to the cap structure. Selected cap-binding RNAs could be expressed in eukaryotic cells and used to inhibit cap-dependent processes.
MATERIALS AND METHODS
Selection of RNAs That Bind to 7-Methyl GTP (m7-GTP).
The template DNA used for the synthesis of the initial random RNA population was constructed with oligonucleotides S5P1 (5′-CTGAATTCTAATACGACTCACTATAGGGCGAATTGGAGCTCGCTAGCCTT-3′), S40N1 (5′-TGGGTACCGTCGACATCCGAATGCC(N40)AAGGCTAGCGAGCTCCAATTCGCCC-3′), and S3P1 (5′-TGGGTACCGTCGACATCCGAATGCC-3′) (17). S40N1 contains 40 nt of randomized sequence (N40) flanked on either side by 25 nt of fixed sequence. Oligonucleotide S3P1 contains the 5′ terminal fixed sequences present in S40N1. Oligonucleotide S5P1 contains sequences for a promoter of T7 RNA polymerase (shown in italics) followed by fixed sequences that are complementary to the 3′ terminal 25 nt in S40N1. The fixed sequence elements further harbor restriction endonuclease sites for SacI and KpnI (underlined) that were used to clone individual amplified DNA sequences. All three oligonucleotides were annealed, and DNA sequences were amplified by PCR. Only three PCR cycles were used initially, so as not to reduce the sequence complexity of the random pool due to preferential amplification of sequences by the Taq DNA polymerase. A pool of ≈1013 DNA molecules was then transcribed by T7 RNA polymerase (see below) to generate a random pool of 90-nt RNAs. Prior to rounds one, two, and three, the systematic evolution of ligands by exponential enrichment (SELEX) RNAs were first passed through a 2 ml Sepharose 4B (Sigma) column to remove RNA species with affinity for the resin. Unbound RNAs were then incubated with 0.1 ml m7-GTP Sepharose-4B (Pharmacia), equilibrated in binding buffer (100 mM Hepes-KOH, pH 7.0/5 mM MgCl2/5 mM KCl/300 mM NaCl) for 1 hr at 4°C. The resin was then washed with 40 column volumes of binding buffer, and the bound RNAs were eluted with 16 mM m7-GTP (Sigma) in binding buffer. In all subsequent cycles, SELEX RNA-bound columns were eluted with 16 mM GTP (counter-SELEX) prior to elution with m7-GTP. The eluted RNAs were reverse transcribed by avian myeloblastosis virus reverse transcriptase (80 units) (GIBCO/BRL) using primer S3P1 in 50 mM Tris⋅HCl (pH 8.3), 6 mM MgCl2, 40 mM KCl, 10 mM DTT, and 0.5 mM dNTPs for 3 hr at 43°C. The cDNA molecules were then amplified by PCR, purified by electrophoresis through polyacrylamide gels, eluted, and transcribed in vitro using T7 RNA polymerase to synthesize the SELEX RNA pool for the next round of selection. After the eighth selection cycle, the PCR-generated 115-bp cDNA fragment was isolated from polyacrylamide gels and digested with KpnI and SacI. The fragments were ligated to a 2.8-kb pGEM 3 vector-derived DNA fragment which had been digested with KpnI and SacI. C600 Escherichia coli cells were transformed and plasmids from individual bacterial clones were subjected to dideoxynucleotide sequencing (Sequenase kit; GIBCO/BRL).
In Vitro RNA Synthesis.
Approximately 3–5 μg of the SELEX cDNA, linearized with BamHI, were incubated in transcription buffer {8 mM Hepes-KOH, pH 7.5/1.2 mM MgCl2/0.2 mM spermidine/40 mM DTT/100 units RNasin/1.8 mM NTPs/30 μCi [α-32P]UTP (800 Ci/mmol; 1 Ci = 37 GBq)/2.5 units yeast inorganic pyrophosphatase (Sigma)/10 μg/ml T7 RNA polymerase (gift from B. Burnett, University of Colorado Health Sciences Center)} at 37°C for 4 hr. The reactions were then incubated with 3 units of DNase RQ1 at 37°C for 15 min. After extraction with phenol/chloroform and precipitation with ethanol, RNA was resuspended and fractionated through a G25-Sephadex column to remove unincorporated nucleotides. Similarly, large quantities of selected RNAs R8-35 and R0-1 were made in this way, except that those RNAs were further purified by electrophoresis on a 5% polyacrylamide/7 M urea gel as described (18). The isolated RNAs were quantified by measuring the A260 (40 μg/ml per A260 unit).
The luciferase (LUC) reporter mRNAs were generated from the plasmid pT7LUCpA (19), linearized with HpaI or BamHI such that the 3′ terminal RNA sequences contained or lacked polyadenosine residues, respectively. Capped mRNAs were synthesized in the presence of cap analog m7GpppG (New England Biolabs) that was added at a 5-fold higher concentration than GTP to the transcription reactions. Unincorporated cap analog was removed by precipitation with ethanol and fractionating the reaction mixture twice with G25-Sephadex columns. The bicistronic mRNA m7-CAT/Bip/LUCpA was expressed from plasmid pT7-CAT-Bip-LUCpA (20) linearized with HpaI. The integrity of the mRNAs was monitored by electrophoresis on agarose or denaturing polyacrylamide gels.
Determination of Binding Constants.
Five micrograms of 32P-labeled R8-35 SELEX RNA was incubated with 100 μl of m7-GTP-Sepharose 4B at 4°C for 2 hr. The unbound RNAs were removed from the reaction by washing the resin with 40 column volumes of binding buffer. The R8-35 RNA-bound Sepharose resin was then divided into five equal aliquots. Increasing amounts of various nucleosides and nucleotide analogs (see Table 2) were added to the RNA-bound resin in a total volume of 200 μl of binding buffer. After incubation on a rotator at 4°C for 12 hr, the Sepharose beads were sedimented, and the radioactivity in the supernatant was determined by liquid scintillation counting. The RNA concentration needed to elute 50% of the bound RNA was determined (i.e., IC50).
Table 2.
IC50 values of nucleotides and nucleotide analogs for R8-35 RNA binding to m7-GTP-Sepharose
| Competitor | IC50, μM | |
|---|---|---|
| m7-GTP | 0.5 | |
| GTP | 1200.0 | |
| m7GpppG | 1.0 | |
| GpppG | >2800.0 | |
| m7GpppA | 2.2 | |
| m7-GDP | 1.5 | |
| dGTP | 2900.0 | |
| GMP | 1800.0 | |
| Guanosine | 90.0 | |
| ITP | 4700.0 | |
| CTP | 6500.0 | |
| UTP | 6000.0 | |
| ATP | 4800.0 |
Enzymatic Probing of RNA Structures.
Structural probing of the R8-35 SELEX RNA was performed with 5′ end-labeled RNA as described (21), except that tRNA (1 μg/μl) was added to the reaction mixtures. RNases A, T1, T2 and cobra venom RNase 1 were employed in these experiments.
Preparation of Translation Lysates.
The HeLa cell S10 and yeast S30 translation lysates were prepared as described (19, 22).
In Vitro Translation Assays.
Increasing amounts of various SELEX RNAs were pre-incubated with capped or uncapped LUC mRNA on ice for 10 min. The translation extracts and buffers were added and the incubations were continued for 45 min either at 30°C (for HeLa lysates) or at room temperature (for yeast lysates). The concentrations of LUC reporter mRNAs in the HeLa lysate reactions (40% vol/vol) (22) and the yeast S30 lysate (50% vol/vol) reactions (19) were 40 μM and 25 μM, respectively, in 15 μl reaction mixtures. The reactions were stopped by placing on ice. Polypeptide synthesis was monitored by measuring LUC activity (23).
Ribosome Binding Assays.
Twenty-five micrograms of capped LUC transcripts lacking poly(A) tails were 3′ end-labeled using 60 μCi of [32P]pCp (3,000 Ci/mmol) and 100 units of T4 RNA ligase (New England Biolabs) and an incubation period of 30 min at 37°C according to the manufacturer’s recommendation. The end-labeled RNAs were then extracted with phenol/chloroform and precipitated with ethanol. Unincorporated pCp was removed using G25-spin columns. Ribosome binding assays were performed as described using yeast S30 lysates and labeled RNA in a total volume of 50 μl (24). The buffer conditions were the same as in the translation reactions, except that 1.2 mM cycloheximide, 8 mM GMP-PNP, 5 mM m7-GTP, or 5 mM EDTA was added as indicated. Briefly, reactions were pre-incubated without mRNA for 5 min at room temperature. Then reporter mRNAs, which were pre-incubated with or without the SELEX RNAs for 10 min on ice, were added to yield a 25 μM final concentration. The incubation was continued for 20 min at room temperature. Finally, 100 μl of cold buffer A [30 mM Hepes-KOH, pH 7.4/100 mM KOAc/23 mM Mg(OAc)2/2 mM DTT] containing 0.25% glutaraldehyde was added, and the reactions were incubated on ice for 5 min. The reactions were then layered on 10 ml linear 10–30% sucrose gradients in buffer A. The gradients were harvested after centrifugation in a Beckman SW41Ti rotor at 40,000 rpm for 130 min at 4°C. Fractions (0.65 ml) were collected from the bottom of the gradient, and the radioactivity in the samples was determined by liquid scintillation counting.
RESULTS
Rationale.
The SELEX procedure is a powerful approach to isolate RNA ligands with high affinities for a variety of molecular ligands (25, 26), including nucleotides (27). The finding that an RNA molecule could be isolated that bound with 10,000-fold higher affinity to theophylline than to caffeine (28), which differs from theophylline only by the presence of a methyl group at nitrogen atom N-7, prompted our efforts to isolate an RNA molecule that could bind with higher affinity to m7-GTP than to nonmethylated GTP. The goal was to identify an RNA species that could bind to the 5′ end of methyl-7-guanosine-containing mRNAs in eukaryotic cells. In principle, such an RNA could sterically interfere with cap-dependent processes such as pre-mRNA processing, nucleocytoplasmic mRNA transport, and translation of mRNAs.
Selection of a Cap-Binding RNA.
m7-GTP-Sepharose 4B was chosen as the target for the selection because it resembles the 5′ cap structure and has been successfully used to isolate eIF-4E, the cap-binding protein, from rabbit reticulocyte lysates (29). Table 1 summarizes the ligand-binding properties of 90-nt RNAs that originally contained 40 nt of random sequence, through eight rounds of selection. Less than 1% of the RNA molecules in the pool bound to the resin in the first round of selection. The RNAs that bound to the resin in the second round of selection were first eluted with GTP; this process, known as counter-SELEX, is used to increase the specificity of the selection process (28). In this way, RNA species that recognize determinants of the GTP moiety of m7-GTP should have been removed. The RNA population that remained bound to the resin was then eluted with m7-GTP and processed for the next round of selection. More than 80% of the in vitro-synthesized RNA pool bound to the resin after six rounds of selection, suggesting that the transcribed pool of RNA was enriched for RNAs that interacted specifically with m7-GTP. After eight rounds of selection, individual cDNAs were cloned from the PCR-amplified cDNA pool and the nucleotide sequences of 52 individual inserts were determined. RNAs were transcribed, and the effects on translation of uncapped and capped mRNAs were determined (see below). This analysis revealed that all selected RNAs inhibited translation of a capped reporter mRNA by 30–60% (not shown). Two RNAs, R8-35 and R8-50, which differed only by a single nucleotide (nucleotide number 38 in R8-50 is a cytidine; Fig. 1) inhibited cap-dependent translation by 90% (see below). RNA R0-1 and R0-8 were cloned from the unselected pool; their effects on translation is shown below. First, R8-35 was chosen for a more detailed examination of its biochemical and biological properties.
Table 1.
Increase in affinity and specificity of RNA pools through rounds of selection
| Cycle of selection | RNA bound, % | m7-GTP elution, % |
|---|---|---|
| 1 | — | — |
| 2 | 38 | — |
| 3 | 71 | 6.3 |
| 4 | 77 | 18 |
| 5 | 80 | 25 |
| 6 | 87 | 29 |
| 7 | 88 | 27 |
| 8 | 78 | 40 |
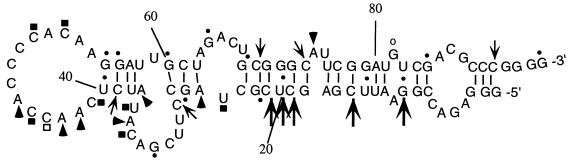
Figure 1.
Predicted secondary structure of R8-35 RNA. The computer-predicted secondary structure of the cap-binding RNA is shown. Susceptibilities of residues to various single- and double-stranded specific nucleases are indicated. Strong reactivities are marked by filled-in symbols and weak reactivities are denoted with open symbols. RNase A (□ and ▪) displays specificity for single-stranded U and C residues. RNase T1 (○ and •) cleaves 3′ of single-stranded G residues. RNase T2 (▵ and ▴) cleaves 3′ of A residues. RNase V1 cleaves helical regions (a strong hit is indicated by a large arrow, a weaker hit is marked by a small arrow). The nucleotide numbers are indicated on the RNA molecule. The sequences from nt 1 to 16 and from nt 89 to 95 are derived from the cloning vector pGEM3 (Promega). Nucleotides 17–30 and 71–88 are derived from the fixed 5′-end and 3′-end sequences, respectively. The variable region extends from nt 31 to 70.
Predicted RNA Structure of the Cap-Binding SELEX R8-35 RNA.
The Wisconsin GCG RNA fold program was used to predict the secondary structure of R8-35 RNA. Fig. 1 shows that R8-35 could be folded into a long hairpin structure with several internal bulges and loops and a predicted free energy of ΔG = −22.5 kcal/mol. To test the validity of the predicted structure, radiolabeled R8-35 RNA was treated with a number of different nucleases that are predicted to cleave either single-stranded (RNases A, T1, T2) or helical (cobra venom RNase 1) regions in RNA. The sites in the RNA that were susceptible to nuclease cleavage were determined; the susceptible nucleotides are marked in the computer-predicted RNA structure shown in Fig. 1. Overall, the data obtained from nuclease digestions supported the computer-predicted structure. Curiously, cytidines 47, 48, and 49, predicted to reside in a large single-stranded loop, were not cleaved by RNase A, suggesting that the large single-stranded loop contains secondary or higher order RNA structure. However, substitution of these residues did not affect the biochemical and biological properties of R8-35 RNA (data not shown). Also, the observed susceptibilities of guanosines 55 and 56 to single-stranded nucleases suggest that alternative RNA conformations exist.
Affinity of SELEX RNA R8-35 for the Cap Structure.
Competition binding analysis was used to determine the relative affinities of various nucleotides for SELEX RNA R8-35. The IC50, or concentration of competitor at which the binding of R8-35 to m7-GTP-Sepharose is 50% competed, should be equal to the dissociation constant if the inhibitor binds competitively to one site on m7-GTP. The IC50 of R8-35 RNA for m7-GTP, a compound closely resembling the cap structure, was 0.5 μM (Table 2). In contrast, the IC50 of R8-35 for GTP was ≈1,000-fold higher. Thus, the R8-35 RNA can distinguish at the molecular level between two very similar molecules, m7-GTP and GTP, which differ only by the presence of a methyl group and a positive charge at the N-7 atom of guanosine. Similarly, the R8-35 RNA bound with apparently higher affinity to m7GpppG, m7GpppA, and m7-GDP than to nonmethylated compounds such as GpppG, dGTP, GMP, ITP, CTP, UTP and ATP (Table 2). Curiously, the base guanosine by itself appeared to have higher affinity to R8-35 than the various nucleoside tri-, di-, and monophosphates (Table 2).
The dissociation constant of the cap-binding protein eIF-4E for m7GpppG is ≈5 μM (30). Because SELEX RNA R8-35 displays a similar affinity for the cap structure, it is predicted that SELEX R8-35 RNA would compete with eIF-4E for occupancy at the cap structure. The outcome of this competition could lead to interference of cap-dependent translation.
The Cap-Binding RNA Inhibits Translation of Capped mRNAs.
The effect of Selex RNA R8-35 on the translation of capped and uncapped LUC-encoding reporter mRNAs was tested in a translation-competent lysate prepared from human HeLa cells (22).
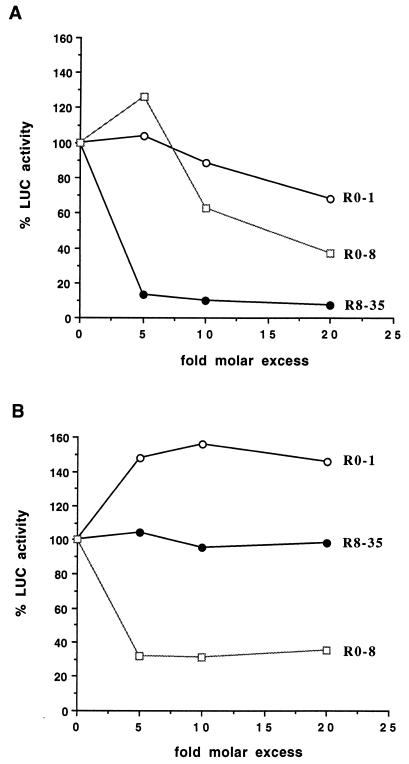
Fig. 2 shows that the addition of a 5-fold molar excess of R8-35 RNA (i.e., 200 μM) over the reporter mRNA inhibited the translation of capped mRNA by >80% (Fig. 2A) but had no effect on the translation of uncapped reporter mRNAs (Fig. 2B). Control RNAs were chosen from cDNAs cloned randomly from the starting pool of unselected clones. Selex RNA R0-1 did not inhibit translation of capped mRNAs (Fig. 2A) and slightly stimulated translation of uncapped mRNAs (Fig. 2B). This stimulation was specific for R0-1 and was not observed with several other RNAs. In contrast, Selex RNA R0-8 inhibited the translation of both capped (Fig. 2A) and uncapped (Fig. 2B) mRNAs.
Figure 2.
Effects of various SELEX RNAs on translation of capped and uncapped reporter mRNAs in a HeLa cell-free lysate. (A) Effects of SELEX RNAs R8-35, R0-1, and R0-8 on the translation of capped, polyadenylated LUC reporter mRNAs. LUC activity in the presence of various concentrations of SELEX RNAs is shown. (B) Effects of Selex RNAs on the translation of uncapped, polyadenylated LUC mRNAs. LUC activity in the presence of various concentrations of SELEX RNAs is shown.
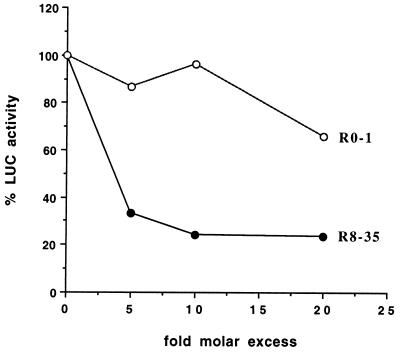
Next, the effects of R8-35 and R0-1 on translation of capped mRNAs were also tested in cell-free S30 lysates prepared from Saccharomyces cerevisiae (19). The translation of capped mRNAs that lacked 3′ terminal polyadenosine residues was monitored to exclude the effects of polyadenosine binding protein on translational initiation (24, 31). Fig. 3 shows that translation of capped reporter mRNA was inhibited by ≈70% in the presence of R8-35 RNAs at a 5-fold molar excess over mRNA (i.e., 125 μM). At similar concentrations, the control R0-1 RNA did not affect the translation of capped LUC mRNA. R8-35 RNAs displayed a small inhibitory effect (≈10–20% inhibition) on the translation of uncapped mRNAs (data not shown). Again, this suggests that R8-35 exerted its effects by binding to the cap structure and not by antisense inhibition.
Figure 3.
Effects of SELEX RNAs on translation of capped, nonpolyadenylated LUC reporter mRNAs in a cell-free lysate from S. cerevisiae. LUC activity in the presence of various concentrations of SELEX RNAs is shown.
The Initiation Step of Translation Is Inhibited by the Cap Structure-Binding RNA.
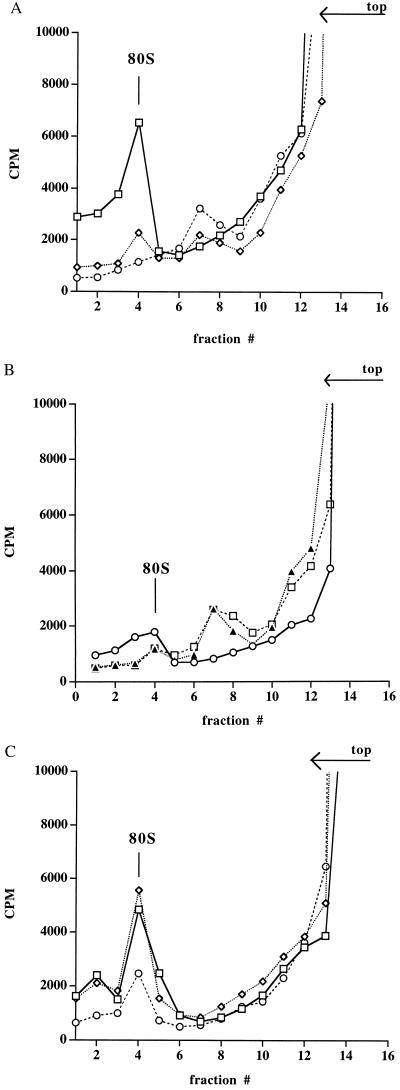
To determine which step in cap-dependent translation was inhibited by R8-35 RNA, ribosome binding assays were performed to observe any effect of the cap-binding RNA on the formation of 80S ribosomes. Incubation of 3′ end-labeled capped Luc mRNA with yeast S30 lysate in the presence of the elongation inhibitor cycloheximide resulted in the formation of 80S complexes that could be visualized after sedimentation through sucrose gradients (Fig. 4A, fraction 4) (24, 32). Addition of EDTA or m7-GTP to the cycloheximide-treated reaction resulted in the disappearance of the 80S complex (Fig. 4A), strongly suggesting that the 80S complex contains a ribosome (24, 32). The inhibition of formation of 80S complexes by m7-GTP (Fig. 4A) was likely due to the binding of m7-GTP to the cap-binding protein eIF-4E, thereby reducing the concentration of 40S complexes that are available to mediate loading of ribosomal subunits onto the mRNA (2). The presence of ribosomes in the 80S peak (Fig. 4A) is further supported by the observation that few 80S complexes were observed in the absence of cycloheximide (Fig. 4B). Furthermore, formation of 80S complexes was also abrogated in the presence of GMP-PNP, a nonhydrolyzable GTP analog that prevents joining of the 40S and 60S ribosomal subunits (32) (Fig. 4B). Instead, the radiolabeled RNA sedimented at a lower S value (Fig. 4B, fraction 7). Fraction 7 may contain both 43S ribosomal complexes and ribonucleoprotein particles, because addition of m7-GTP to GMP-PNP-treated reactions had only a small effect on the sedimentation of the radiolabeled RNA (Fig. 4B).
Figure 4.
Effects of cap analogs and SELEX RNAs on the formation of 80S ribosomal complexes on capped LUC mRNA molecules. (A) Formation of 80S ribosomal complexes in the presence of 1.2 mM cycloheximide (□), 1.2 mM cycloheximide and 5 mM m7-GTP (◊), or 1.2 mM cycloheximide and 5 mM EDTA (○) after sucrose gradient centrifugation is shown. The radioactivity in collected fractions is shown. The top of the gradient and the position of 80S ribosomes are indicated. (B) Formation of 80S ribosomal complexes in the absence of any inhibitor (○), in the presence of 8 mM GMP-PNP (▴), or 8 mM GMP-PNP and 5 mM m7-GTP (□) is depicted. (C) Formation of 80S complexes in the presence of 1.2 mM cycloheximide (◊), 1.2 mM cycloheximide and 1.5 mM (60-fold molar excess) of R8-35 (○), or 1.2 mM cycloheximide and 1.5 mM R0-1 (□) is shown. The conditions for translation in yeast extracts are the same as described in Fig. 3, except for the addition of translation inhibitors.
Addition of R8-35 RNAs to cycloheximide-treated ribosome-binding reactions decreased the formation of 80S ribosomes (Fig. 4C) to a similar extent as the addition of m7-GTP (see Fig. 4A). Addition of the control RNA R0-1 to the binding reactions did not significantly inhibit the formation of 80S ribosomal complexes. These data show that the cap structure-binding RNA R8-35 affects an initiation step of translation.
DISCUSSION
Employing the SELEX method, a small RNA (R8-35) was selected that binds with high affinity to the cap structure located at the 5′ end of eukaryotic mRNAs. The selected RNA binds with 1,000-fold higher affinity to m7-GTP than to GTP. In vitro translation studies showed that R8-35 RNA specifically inhibited translation of capped but not uncapped reporter mRNAs in lysates prepared from either human cells or S. cerevisiae. In addition, the translation of a reporter mRNA preceded by the internal ribosome entry site located in the human immunoglobulin heavy-chain binding protein, BiP (33), mRNA was not inhibited in the presence of R8-35 RNAs (data not shown). These data indicate that the cap-binding RNA R8-35 is a specific inhibitor of cap-dependent translation and does not affect cap-independent translation by internal initiation. Ribosome binding studies carried out in the presence of R8-35 RNAs showed a decrease in 80S ribosome complex formation. It is unlikely that R8-35 inhibited the movement of 40S subunits on the mRNA because uncapped mRNAs were translated with equivalent efficiency in the presence or absence of R8-35 RNA. Rather, this finding suggests that R8-35 prevents entry of 40S subunits onto the mRNA by interfering with the association of the cytoplasmic cap-binding protein eIF-4E with the 5′ cap.
At least three classes of RNA molecules existed in the initial unselected SELEX sequence pool—(i) RNAs such as R8-35, which inhibit translation of capped mRNAs; (ii) R0-1-like RNAs which do not inhibit translation of either capped or uncapped mRNAs; and (iii) R0-8-like RNAs which inhibit translation of both capped and uncapped mRNAs—probably by inhibiting the function of a canonical translation factor or by interacting with the mRNA.
Several studies have suggested that the 5′ terminal cap structure and associated binding proteins have a role in the assembly of splicing complexes at the 5′ splice sites of 5′ proximal introns in pre-mRNAs (9, 10). Preliminary experiments designed to monitor the effects of R8-35 and R0-1 RNAs on the efficiency of intron removal in β-globin pre-mRNA, which contains a single intron, showed that addition of R8-35 RNAs to in vitro splicing reactions prevented splicing of this transcript. These data suggest that R8-35 displaces not only the human cytoplasmic cap-binding complex, composed of factors eIF-4E/eIF-4A/eIF-4G, from the 5′ terminal cap structure, but also the human nuclear cap-binding protein complex, composed of factors CBP20 and CBP80 (34).
Indirect evidence has been provided that CBP80 is required for nucleocytoplasmic mRNA transport (12). It remains to be tested whether the function of CBP80 complexes is disrupted in the presence of R8-35. The cap structure-binding RNA offers a novel approach to study both cytoplasmic and nuclear cap-dependent processes that occur in eukaryotic cells. The use of such RNA molecules provides an alternative approach to the use of cap analogs and should be useful for both in vitro and in vivo analysis of cap-dependent processes.
Acknowledgments
We thank Simon Hambidge for laying the groundwork for this project and members of Barry Polisky’s group at NEXSTAR in Boulder, CO, for helpful suggestions. We are grateful to Karla Kirkegaard for critical reading of the manuscript. This work was supported by National Institutes of Health Grant R01 GM55979 and in part by National Institutes of Health Grant AG07347. A.A.H. is a recipient of a fellowship from the Jane Coffin Childs Memorial Fund for Medical Research, and P.S. is a recipient of a faculty research award from the American Cancer Society.
ABBREVIATIONS
- SELEX
systematic evolution of ligands by exponential enrichment
- eIF
eukaryotic initiation factor
- m7-GTP
7-methyl GTP
- LUC
luciferase
References
- 1.Shatkin A J. Cell. 1976;9:645–653. doi: 10.1016/0092-8674(76)90128-8. [DOI] [PubMed] [Google Scholar]
- 2.Banerjee A K. Microbiol Rev. 1980;44:175–205. doi: 10.1128/mr.44.2.175-205.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Furuichi Y, LaFinandra A, Shatkin A J. Nature (London) 1977;266:235–239. doi: 10.1038/266235a0. [DOI] [PubMed] [Google Scholar]
- 4.Konarska M, Padgett R, Sharp P. Cell. 1984;38:731–736. doi: 10.1016/0092-8674(84)90268-x. [DOI] [PubMed] [Google Scholar]
- 5.Edery I, Sonenberg N. Proc Natl Acad Sci USA. 1985;82:7590–7594. doi: 10.1073/pnas.82.22.7590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Krainer A, Maniatis T, Ruskin B, Green M. Cell. 1984;36:993–1005. doi: 10.1016/0092-8674(84)90049-7. [DOI] [PubMed] [Google Scholar]
- 7.Patzelt E, Tahlmann E, Hartmuth K, Blaas D, Kuechler E. Nucleic Acids Res. 1987;15:1387–1399. doi: 10.1093/nar/15.4.1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Inoue K, Ohno M, Sakamoto H, Shimura Y. Genes Dev. 1989;3:1472–1479. doi: 10.1101/gad.3.9.1472. [DOI] [PubMed] [Google Scholar]
- 9.Lewis J D, Izaurralde E, Jarmolowski A, McGuigan C, Mattaj I W. Genes Dev. 1996;10:1683–1698. doi: 10.1101/gad.10.13.1683. [DOI] [PubMed] [Google Scholar]
- 10.Ohno M, Sakamoto H, Shimura Y. Proc Natl Acad Sci USA. 1987;84:5187–5191. doi: 10.1073/pnas.84.15.5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hamm J, Mattaj I W. Cell. 1990;63:109–118. doi: 10.1016/0092-8674(90)90292-m. [DOI] [PubMed] [Google Scholar]
- 12.Izaurralde E, Stepinski J, Darzynkiewicz E, Mattaj I W. J Cell Biol. 1992;118:1287–1295. doi: 10.1083/jcb.118.6.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Izaurralde E, Lewis J, Gamberi C, Jarmolowski A, McGuigan C, Mattaj I W. Nature (London) 1995;376:709–712. doi: 10.1038/376709a0. [DOI] [PubMed] [Google Scholar]
- 14.Sonenberg N, Rupprecht K M, Hecht S M, Shatkin A J. Proc Natl Acad Sci USA. 1979;74:4345–4349. doi: 10.1073/pnas.76.9.4345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hentze M W. Science. 1997;275:500–501. doi: 10.1126/science.275.5299.500. [DOI] [PubMed] [Google Scholar]
- 16.Sachs A B, Sarnow P, Hentze M W. Cell. 1997;89:831–838. doi: 10.1016/s0092-8674(00)80268-8. [DOI] [PubMed] [Google Scholar]
- 17.Pata J D. Mechanistic Studies of Poliovirus RNA-Dependent RNA Polymerase. Boulder: Univ. of Colorado; 1993. [Google Scholar]
- 18.Wyatt J R, Sontheimer E J, Steitz J A. Genes Dev. 1992;6:2542–53. doi: 10.1101/gad.6.12b.2542. [DOI] [PubMed] [Google Scholar]
- 19.Iizuka N, Najita L, Franzusoff A, Sarnow P. Mol Cell Biol. 1994;14:7322–7330. doi: 10.1128/mcb.14.11.7322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Macejak D G, Hambidge S J, Najita L M, Sarnow P. In: EIF-4F-Independent Translation of Poliovirus RNA and Cellular mRNA Encoding Glucose-Regulated Protein 78/Immunoglobulin Heavy-Chain Binding Protein. Brinton M A, Heinz F X, editors. Washington, DC: Am. Soc. for Microbiol.; 1990. pp. 152–157. [Google Scholar]
- 21.Jacobson S J, Konings D A M, Sarnow P. J Virol. 1993;67:2961–2971. doi: 10.1128/jvi.67.6.2961-2971.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McBratney S, Sarnow P. Mol Cell Biol. 1996;16:3523–3534. doi: 10.1128/mcb.16.7.3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.De Wet J R, Wood K V, DeLuca M, Helinski D R, Subramani S. Mol Cell Biol. 1987;7:725–737. doi: 10.1128/mcb.7.2.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tarun S Z, Sachs A B. Genes Dev. 1995;9:2997–3007. doi: 10.1101/gad.9.23.2997. [DOI] [PubMed] [Google Scholar]
- 25.Ellington A D, Szostak J W. Nature (London) 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 26.Tuerk C, Gold L. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 27.Sassanfar M, Szostak J W. Nature (London) 1993;364:550–553. doi: 10.1038/364550a0. [DOI] [PubMed] [Google Scholar]
- 28.Jenison R D, Gill S C, Pardi A, Polisky B. Science. 1994;263:1425–1429. doi: 10.1126/science.7510417. [DOI] [PubMed] [Google Scholar]
- 29.Webb N R, Chari R V, DePillis G, Kozarich J W, Rhoads R E. Biochemistry. 1984;23:177–81. doi: 10.1021/bi00297a001. [DOI] [PubMed] [Google Scholar]
- 30.Goss D J, Carberry S E, Dever T E, Merrick W C, Rhoads R E. Biochemistry. 1990;29:5008–12. doi: 10.1021/bi00473a002. [DOI] [PubMed] [Google Scholar]
- 31.Tarun S Z, Sachs A B. EMBO J. 1996;15:7168–7177. [PMC free article] [PubMed] [Google Scholar]
- 32.Gray N K, Hentze M W. EMBO J. 1994;13:3882–3891. doi: 10.1002/j.1460-2075.1994.tb06699.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Macejak D G, Sarnow P. Nature (London) 1991;353:90–94. doi: 10.1038/353090a0. [DOI] [PubMed] [Google Scholar]
- 34.Izaurralde E, Lewis J, McGuigan C, Jankowska M, Darzynkiewicz E, Mattaj I. Cell. 1994;78:657–668. doi: 10.1016/0092-8674(94)90530-4. [DOI] [PubMed] [Google Scholar]