Abstract
Thyroid hormone is a critical mediator of central nervous system (CNS) development, acting through nuclear receptors to modulate the expression of specific genes. Transcription of the rat hairless (hr) gene is highly up-regulated by thyroid hormone in the developing CNS; we show here that hr is directly induced by thyroid hormone. By identifying proteins that interact with the hr gene product (Hr), we find that Hr interacts directly and specifically with thyroid hormone receptor (TR)—the same protein that regulates its expression. Unlike previously described receptor-interacting factors, Hr associates with TR and not with retinoic acid receptors (RAR, RXR). Hr can act as a transcriptional repressor, suggesting that its interaction with TR is part of a novel autoregulatory mechanism.
Many factors, both genetic and environmental, contribute to the formation and function of the mammalian central nervous system (CNS). An essential component of these processes is thyroid hormone (TH); if thyroid hormone levels are perturbed, abnormal development ensues, resulting in neurological deficits that include severe mental retardation (1–4). The effects of TH are mediated through the action of specific nuclear receptor proteins (5, 6). Thyroid hormone receptors (TR) act by binding to specific DNA sequences and subsequently activating or repressing the transcription of nearby genes in response to hormone binding (7, 8). Several proteins that interact with TR and other nuclear hormone receptors, including both coactivators and corepressors, have been identified (9, 10).
Although much is known about the mechanism of action of TR and other nuclear receptors, far less is known about the genes regulated by these receptors. We have shown that the rat hairless (hr) gene is highly up-regulated (>10-fold) by thyroid hormone in developing brain (11). The rapid induction (<4 hr) occurs even in the absence of protein synthesis, suggesting that hr is directly regulated by TR. Direct target genes are particularly important because such genes likely constitute the first step in the genetic program responsible for TH-mediated aspects of neural development. We show here that the upstream regulatory region of the hr gene includes a potent thyroid hormone response element (TRE), indicating that hr is indeed a direct target of TR.
The murine hr locus was originally identified as a spontaneous mutation caused by an endogenous retrovirus (12). The hr gene is expressed predominantly in skin and brain; the mutant phenotype in skin is progressive hair loss and increased susceptibility to cancer, while the neurological phenotype has not yet been described (11, 13). The hr gene encodes a putative protein of approximately 127 kDa that lacks homology to known structural motifs other than a cluster of cysteine residues proposed to form a zinc finger (13). Toward defining the function of the hr gene product (Hr), we identified proteins that interact with Hr. Surprisingly, we found that Hr interacts with TR. Previously identified proteins that interact with TR have been shown to interact with multiple nuclear receptors (9, 10, 14–21). In contrast, of the retinoid and steroid receptors tested here, Hr interacts only with TR. The interaction of Hr with TR suggests that Hr is part of a novel autoregulatory mechanism by which Hr may influence the expression of downstream TH-responsive genes.
MATERIALS AND METHODS
TRE Mapping/DNA Binding Assay.
A rat genomic library (Stratagene) was screened using a probe from the 5′ end of the hr cDNA. Four overlapping clones were isolated. A 15-kb NotI fragment was digested with KspI, and the resulting 6- and 9-kb fragments were subcloned into pBluescript (Stratagene). The subcloned fragments were digested with AluI and used for gel shift analysis. Proteins were synthesized by coupled in vitro transcription/translation (Promega) in the presence of [35S]methionine (NEN). Synthesis was analyzed by running a fraction of the radiolabeled products on a gel followed by autoradiography. DNA-binding reactions contained approximately 200 ng of digested DNA mixed with [35S]TR and retinoid X receptor (RXR). Samples were run on 5% polyacrylamide gels in 0.5× TBE, fixed, dried, and exposed to x-ray film. Fragments that gave shifted bands were restriction mapped, and smaller fragments were subcloned. The subcloned fragments were digested with AluI and used for DNA binding. This process was repeated until the smallest binding fragment was determined to be a 106-bp HinfI–EagI fragment. After sequencing the 106-bp fragment, overlapping oligonucleotides spanning the fragment were synthesized and used as competitors for DNA binding. For transfection experiments, oligonucleotides were cloned upstream of a minimal thymidine kinase (TK) promoter by digesting TK-luc with HindIII, then ligating the annealed, phosphorylated oligonucleotides. Constructs were sequenced to confirm the sequence, orientation, and number of oligonucleotides present.
Cell Culture/Transfection.
GH1 and CHO cells were obtained from American Type Culture Collection and maintained in DMEM supplemented with 10% fetal calf serum. For induction experiments, serum was depleted of thyroid and steroid hormones by treatment with AG-1-X8 resin (Bio-Rad) and charcoal (Sigma) as described (22). Cells were grown for 1 day in hormone-depleted media before transfection. Transfection was by lipofection (lipofectamine, Life Technologies, Gaithersburg, MD) in 6-well plates. After transfection, thyroid hormone (L-T3) was added to 10−7 M. Cells were transfected (per well of a 6-well plate) with 167 ng of reporter plasmid, 50 ng of expression plasmid, and 80 ng of CMX–β-gal. pCMX-GAL-Hr was constructed by inserting the BamHI fragment from pLexA-hr into pCMX-GAL4. pCMX-GAL4, GALpx3- tkluc, and pCMX GAL-RXR were kindly provided by K. Umesono (Kyoto University). Cells were harvested using 1× reporter lysis buffer (Promega) and assayed for β-galactosidase (β-gal) and luciferase activity.
Two-Hybrid Screen.
To construct pLexA-hr, a 2.2-kb HindIII fragment corresponding to amino acids 568-1207 of Hr was isolated, the ends were filled in with Klenow, BamHI linkers were ligated, then the fragment was cloned into the BamHI site of pLexA (23). The resulting plasmid was transformed into yeast strain L40 (23). The resulting strain was used to screen a human brain cDNA library constructed as a fusion with the activation domain of VP16. DNA was isolated from HIS+, lacZ+ colonies (24), propagated in Escherichia coli, purified, and sequenced. Cells were tested for β-galactosidase activity as described (25). To test the hormone dependence of interaction, Triac (Sigma) was added to the media and assay buffer (final concentration, 10−6 M).
Far Western Assay.
TrpE-hr was constructed by insertion of a 2.2-kb HindIII fragment corresponding to amino acids 568-1207 of hr into pATH21 (kindly provided by N. Patel, University of Chicago). Glutathione S-transferase (GST)-hr was constructed by insertion of the 2.2-kb BamHI fragment from pLexA-hr into pGEX3X (Pharmacia). Purified GST-hRXRβ (amino acids 198–462) and GST-cTRα (amino acids 1–408) were obtained from Santa Cruz Biotechnology. Extracts from bacteria expressing fusion proteins were separated by SDS/PAGE and transferred to nitrocellulose. After transfer, filters were prepared for far Western blotting as described (26) except that 35S-labeled proteins were used as probes. To test the hormone dependence of interaction, Triac was added to the hybridization buffer (final concentration, 10−6 M). pTZ18 (rTRβ1) was kindly provided by H. Towle (University of Minnesota, Minneapolis); pCMX TRα1, pCMX hRARα, and pCMX hRXRα were kindly provided by K. Umesono.
Immunohistochemistry.
An epitope for myc was appended to the amino terminus of Hr at amino acid 200 by subcloning a 3.1-kb BamHI–XbaI fragment of hr into the vector pBluescript KS(+) myc (kindly provided by M. Bellini, Carnegie Institution of Washington, Baltimore). The resulting myc-hr fusion was excised by digestion with XbaI and partial digestion with HindIII and inserted downstream of the Rous sarcoma virus (RSV) long terminal repeat. GH1 cells were grown on coverslips and transfected by lipofection. Cells were fixed with 1.6% paraformaldehyde for 20 min, blocked for 30 min in PBS with 5% normal goat serum, and incubated with a mouse monoclonal antibody to myc (9E10, kindly provided by Z. Wu, Carnegie Institution of Washington, Baltimore) for 1 hr. Detection was with Cy3 anti-mouse antibody (Jackson ImmunoResearch). Cells were mounted in 50% glycerol with 0.25 μg/ml 4′,6-diamidino-2-phenylindole (DAPI). Cells transfected in parallel with RSV–β-gal were used as a negative control for the anti-myc antibody and stained with 5-bromo-4-chloro-3-indolyl β-d-galactoside (X-gal) as a positive control for transfection. Identical results were obtained using an Hr-specific antisera raised to the GST-hr fusion protein described above.
RESULTS
Expression of hr Is Directly Regulated by Thyroid Hormone.
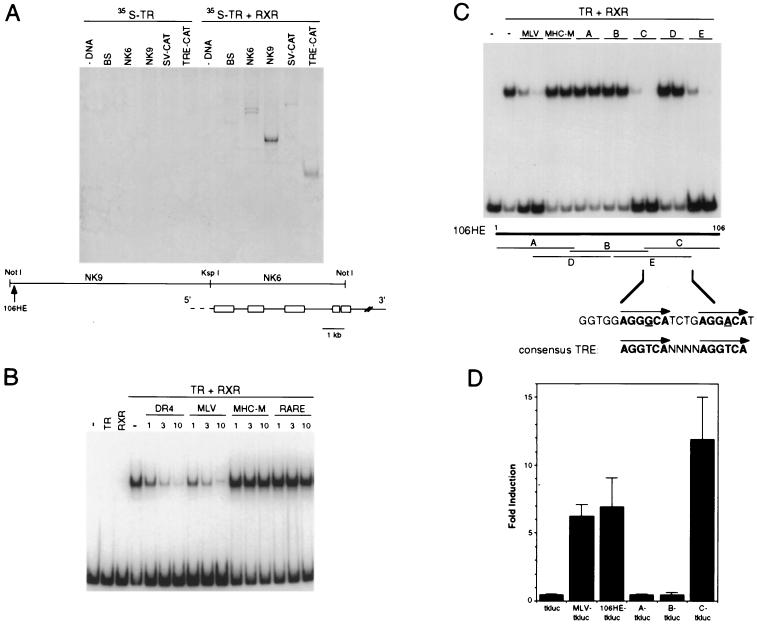
To determine whether hr is a direct target of TR, we examined whether cis-acting sequences controlling its expression include a binding site for TR and/or TR/RXR heterodimers. Genomic sequences from the hr gene were digested with restriction enzymes and used as probes in a gel retardation assay using [35S]TR. No binding was observed when fragments were incubated with [35S]TR alone, but binding was detected when both TR and RXR (unlabeled) were present (Fig. 1A). A high-affinity TR/RXR binding site was detected within a 9-kb NotI–KspI fragment immediately upstream of the hr transcription unit. By subcloning and testing progressively smaller restriction fragments, the TR/RXR binding site was mapped to within 106 bp located approximately 9 kb upstream of the first exon. The isolated, 32P-labeled 106-bp sequence bound specifically to TR/RXR heterodimers, as binding was reduced TRE-containing oligonucleotides (DR4, MLV) but not by a mutated TRE (MHC-M) or a retinoic acid response element (RARE) (Fig. 1B).
Figure 1.
Hr is directly regulated by thyroid hormone. (A) A high-affinity TR/RXR binding site was mapped in the hr gene. Genomic fragments from the hr gene were digested with frequent-cutting restriction enzymes, then incubated with 35S-labeled thyroid hormone receptor (TR) in the absence (Left) or presence (Right) of unlabeled retinoid X receptor (RXR). Positions of genomic fragments are shown relative to the published mouse hr genomic structure (13). DNA used for binding reactions: - DNA, no DNA; BS, Bluescript; NK6, 6-kb NotI–KspI genomic fragment from hr; NK9, 9-kb NotI–KspI genomic fragment from hr; ΔSV-CAT, negative control plasmid; TRE-CAT, positive control plasmid. The faint bands in lanes 9 and 11 (negative control) were low-affinity or nonspecific sites and were not mapped further. Arrow indicates position of the smallest genomic fragment that bound to TR/RXR, a 106-bp HinFI–EagI fragment (106HE). (B) The hr gene has a high-affinity TRE. Binding of TR/RXR in a gel retardation assay with 32P-labeled 106-bp HinFI–EagI fragment. Lanes: 1, no protein; 2, TR only; 3, RXR only; 4–16, TR + RXR. Competitor DNA: (-), none; DR4, synthetic direct repeat TRE; MLV, Moloney leukemia virus TRE; MHC-M, mutated TRE from α-myosin heavy chain gene; RARE, retinoic acid response element (DR5) (27). 1, 3, 10 indicates fold molar excess of competitor DNA. (C) The hr TRE is an imperfect direct repeat of the DR+4 class. Overlapping oligonucleotides encompassing the 106-bp HinFI-EagI fragment were used as competitors for DNA binding. Competitor DNAs: (-), none; MLV, Moloney leukemia virus TRE; MHC-M, mutated TRE from α-myosin heavy chain gene (negative control); A–E, overlapping oligonucleotides of potential TRE from hr. First lane of each pair is approximately 3-fold molar excess; second lane is approximately 10-fold molar excess of competitor DNA. (Middle) Relative positions of putative TRE oligonucleotides. (Bottom) Sequence common to the two oligonucleotides that compete for binding (C and E); an imperfect direct repeat spaced by four nucleotides is indicated in bold with mismatched bases underlined. Consensus TRE is idealized DR+4 (27). (D) The hr TRE confers thyroid hormone responsiveness. The 106-bp HinFI-EagI fragment and putative TRE oligonucleotides (A–C) were inserted individually upstream of a minimal TK promoter driving expression of luciferase. Fold induction is luciferase activity in the presence of thyroid hormone (TH) divided by activity in the absence of TH; baseline activity (−TH) was approximately the same for all constructs. Data are the mean of three independent experiments done in triplicate.
To more precisely define the TR/RXR binding site, overlapping oligonucleotides encompassing the 106-bp sequence were synthesized and used as competitors for binding to the 106-bp fragment (Fig. 1C). Only oligonucleotides C and E were effective competitors and, thus, contained the TR/RXR binding site. These oligonucleotides share a 23-bp sequence that includes an imperfect direct repeat (ggtggAGGGCATCTGAGGACAtc) separated by four nucleotides. TREs often consist of half-sites spaced by four nucleotides (DR+4), with an optimal half-site of AGGTCA (27, 28). Both half-sites of the hr TRE match the optimal half-site in five of six positions. Thus, the hr gene has a potential TRE of the type DR+4.
To test whether the direct repeat sequence motif indeed confers thyroid hormone responsiveness, the 106-bp fragment and putative TRE oligonucleotides were individually placed upstream of a minimal TK promoter driving expression of a luciferase reporter gene. Introduction of these constructs into GH1 (rat pituitary) cells, which express endogenous thyroid hormone receptors, showed that transcription is activated in the presence of thyroid hormone only when the direct repeat sequence is present (Fig. 1D) (106HE-tkluc, C-tkluc). Therefore, the direct repeat sequence in the hr gene acts as a TRE. Together with previous data showing that up-regulation of hr by thyroid hormone occurs rapidly (<4 hr) and without the need for new protein synthesis (11), these results demonstrate that hr is a direct response gene for thyroid hormone, the first such gene identified in the mammalian CNS.
Hr Interacts with TR.
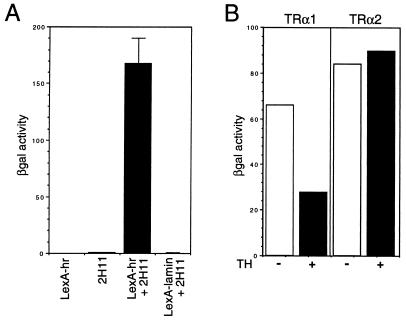
To begin to understand the function of the hr gene product (Hr), we identified proteins that interact with Hr by using a two-hybrid assay (23). The C-terminal 639 amino acids of Hr (amino acids 568–1207; includes the putative zinc finger) were fused to the lexA DNA-binding domain and used as “bait” (LexA-hr) to screen a human brain cDNA library. One cDNA that was isolated multiple times (2H11) was characterized (Fig. 2A). Remarkably, clone 2H11 encodes a thyroid hormone receptor (TRα2) (29). Interaction is not specific for the TRα2 isoform, as TRα1 was found to interact as well (Fig. 2B). Interaction was influenced by hormone binding, because interaction of TRα1 but not TRα2 was reduced 2-fold by thyroid hormone (Fig. 2B). Thus, it appears that the product of the hr gene, a direct target of transcriptional regulation by TR, interacts with the same factor that regulates its expression.
Figure 2.
Hr interacts with TR. (A) Positives isolated in a two-hybrid screen were assayed for β-galactosidase activity. Yeast strains carrying the indicated plasmids were tested. LexA-hr (bait) only, 2H11 (TRα2-VP16) only, LexA-hr + 2H11, LexA-lamin + 2H11. Results are representative of at least three individual transformants. 2H11 encodes TRα2 (amino acids 14–490) (29). TRα1 is a functional TR, whereas TRα2, which has a divergent C terminus, lacks the ability to bind thyroid hormone (29, 30); the preference for isolating TRα2 is likely because the mRNA for TRα2 is more abundant than that for TRα1 (31, 32). (B) Effect of hormone on interaction of Hr and TR. Solution β-galactosidase assay for yeast-carrying LexA-hr and TRα1-VP16 (Left) and TRα2-VP16 (Right) in the presence (+) and absence (−) of thyroid hormone. Open bars, − thyroid hormone; solid bars, + thyroid hormone.
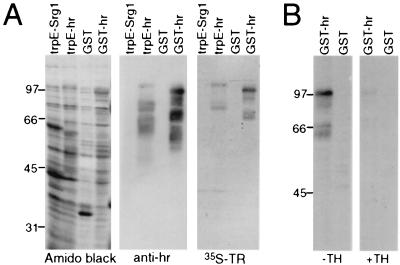
To confirm the direct interaction between Hr and TR, a far Western assay was used. Hr (amino acids 568–1207) was expressed in bacteria as a fusion protein with either GST or trpE. Extracts from bacteria expressing Hr fusion proteins were separated by SDS/PAGE and transferred to nitrocellulose. The immobilized, renatured proteins were incubated with [35S]TRα1 (Fig. 3A). TRα1 detected a protein the size of the Hr fusion proteins, which was recognized by Hr-specific antisera. Therefore, Hr interacts specifically with TRα1. These data also show that no other factors (for example, other proteins in yeast) are required for this interaction. The effect of hormone on interaction was tested using the far Western assay; hormone binding drastically reduced interaction of Hr and TR (Fig. 3B).
Figure 3.
Hr interacts directly with TR. (A) Far Western assay for interaction of Hr with TR. Extracts from bacteria expressing trpE-Srg1 (negative control), trpE-hr, GST only, and GST-hr were separated by SDS/PAGE and transferred to nitrocellulose. Total protein was detected by amido black staining (Left); Hr fusion proteins were detected by Western blot analysis with anti-hr antisera (Center); interaction of Hr with TR was detected by far Western blot analysis with 35S-labeled TRβ1 (Right). Detection of multiple lower molecular mass bands by [35S]TR is from binding to fusion protein degradation products. Sizes of molecular mass standards are in kilodaltons. (B) Effect of hormone on TR/Hr interaction. Far Western assay was as described above with 35S-labeled TRβ1 in the absence (Left) and presence (Right) of thyroid hormone.
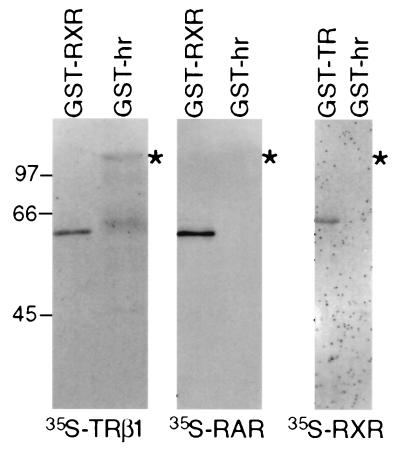
Previously identified factors that interact with TR have been shown to also associate with retinoid receptors and, in some cases, other nuclear receptors as well (9, 10, 14–21). To examine the specificity of interaction between TR and Hr, Hr was tested for interaction with TRβ1, retinoic acid receptor (RAR) and RXR (Fig. 4). TRβ1 bound to the Hr fusion protein, indicating that interaction is not TRα-specific. In contrast, binding was not detected with RAR or RXR. [35S]RAR interacted with GST-RXR, and [35S]RXR interacted with GST-TR, verifying that RAR and RXR are functional in this assay. Consistent with these results, LexA-RAR [ligand binding domain (LBD)] did not interact with Hr in the two-hybrid assay, whereas the TR LBD did interact (data not shown). Two steroid hormone receptors (glucocorticoid and mineralocorticoid) were tested for interaction using the far Western assay and also failed to interact with Hr (data not shown). Therefore, of the receptors tested, binding is specific for TR. These results are particularly important because they indicate that unlike previously identified receptor-interacting factors, Hr appears to distinguish between TR and RAR.
Figure 4.
Hr interacts specifically with TR. GST fusion proteins with retinoid X receptor (GST-RXR), thyroid hormone receptor (GST-TR), and Hr (GST-hr) were probed with 35S-labeled TRβ1 (Left), 35S-labeled RARα (Center), or 35S-labeled RXRα (Right). Asterisk (∗) indicates position of GST-hr.
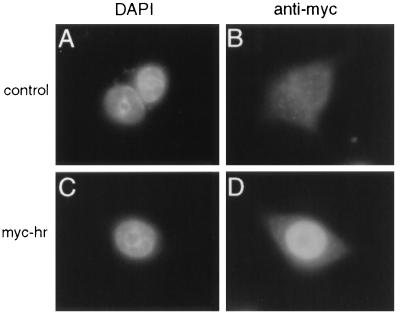
In addition to their interaction in vitro, Hr and TR are expressed in the same cell types in vivo. We have shown previously that hr is expressed in tissues that express TR; in situ hybridization analyses have shown that hr and TR transcripts are present in the same cell populations in the brain (11, 33, 34). For interaction of Hr and TR to occur in vivo, both must occupy the same subcellular compartment. TR resides in the nucleus; to determine whether Hr is also nuclear (it lacks a discernible nuclear localization sequence), sequences encoding an epitope (myc) detected by a specific monoclonal antibody were appended to the hr cDNA (myc-hr). The resulting cDNA was transfected into GH1 cells, and the myc epitope was detected by immunofluorescence. Nuclear staining was observed in cells transfected with myc-hr but not in control cells (Fig. 5). Identical results were obtained using Hr-specific antisera (data not shown). Thus, like TR, Hr is a nuclear protein.
Figure 5.
Hr is a nuclear protein. Myc-hr was expressed in GH1 cells by transient transfection and detected using a mouse monoclonal antibody against the myc epitope. (A and B) Cells transfected with RSV–β-gal (control). (C and D) Cells transfected with RSVmyc-hr. DAPI staining (A and C) indicates nuclei. Magnification, ×1,000.
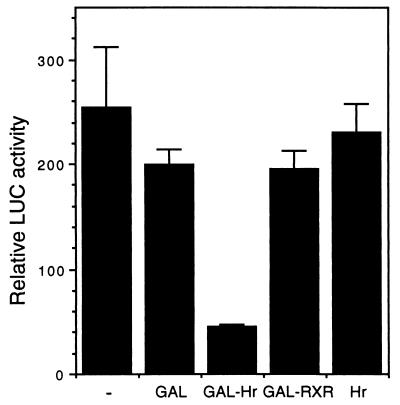
Because Hr is a nuclear protein that interacts with a known transcription factor, Hr was tested for a role in transcriptional regulation. When Hr is coexpressed with TRα1 and a TRE-containing reporter (MLV-tkluc), thyroid hormone-induced transcription is reduced 2-fold (data not shown), suggesting that Hr acts as a repressor. To assay for endogenous repressor function, Hr (amino acids 568-1207) was fused to the GAL4 DNA-binding domain (DBD). When tested for the ability to repress transcription from a GAL4-dependent reporter, the GAL4 DBD-Hr fusion protein reduced basal level transcription by about 5-fold (Fig. 6). This effect is specific to Hr sequences because the GAL4 DBD alone or a GAL4 DBD-RXR fusion protein did not affect activity. Hr alone did not affect activity, indicating that repression by Hr required tethering to DNA. The ability of Hr to repress transcription indicates that when bound to thyroid hormone receptors, Hr may function as a transcriptional modulator.
Figure 6.
Hr can act as a repressor. Hr/GAL4 plasmids were cotransfected with a GAL4-regulated promoter (GALpx3-tkluc) into CHO cells. All transfections included CMX–β-gal as an internal control. LUC activity = light units normalized to β-gal activity. Experiment was repeated three times in triplicate with similar results; shown is a representative experiment. (−), expression vector alone; GAL, GAL4 DNA-binding domain (DBD); GAL-Hr, GAL4 DBD fused to Hr (amino acids 568–1207); GAL-RXR, GAL4 DBD fused to the ligand-binding domain of hRXRα; Hr, full-length Hr.
DISCUSSION
Together with previous evidence that the hr gene is rapidly up-regulated by thyroid hormone even in the absence of protein synthesis, the mapping of a high-affinity TRE in the hr gene demonstrates that hr is a direct target of thyroid hormone receptors in the developing mammalian CNS. Although a handful of genes whose expression is influenced by thyroid hormone in the CNS have been identified, induction of these genes has not been shown to be rapid (>24 hr) nor resistant to inhibitors of protein synthesis (35, 36). Thus, hr is the first direct response gene for thyroid hormone identified in the developing mammalian nervous system. Given that postnatal CNS development is extremely sensitive to thyroid hormone levels (1–4), expression of hr likely constitutes a key step in the genetic program responsible for TH-dependent aspects of CNS development. The TRE found in the hr gene is similar in sequence to previously described TREs (DR+4) (27, 28), although it is unusually distant (9 kb) from the transcription start site.
Our screen for proteins that interact with Hr led to the startling discovery that the product of this thyroid hormone-responsive gene interacts directly and specifically with TR—the same protein that induces its expression. Equally important is the finding that although Hr binds to TR, it does not appear to bind to RAR or to their common partner, RXR. Though many proteins that interact with nuclear hormone receptors have been identified, all have been shown to be widely expressed and to bind to multiple receptors (9, 10). In contrast, hr is predominantly expressed in brain and skin (11, 13), and Hr binds to only one (TR) of several steroid and retinoid receptors tested. Hr and TR are both nuclear proteins and are coexpressed in various regions of the brain (11, 33, 34), suggesting that the interaction observed in vitro also occurs in vivo.
Transcriptional repression and hormone-dependent interaction with TR are properties that Hr shares with corepressors (17, 18), suggesting that Hr has a similar function. In addition, the induction of hr expression by thyroid hormone, coupled with the interaction of Hr protein with TR, suggests a novel regulatory pathway. Once induced by thyroid hormone, Hr likely binds to TR and modulates the expression of downstream genes. The existence of a similar autoregulatory mechanism for other nuclear receptors is hinted at by the product of an estrogen-responsive gene (efp) that shows homology to receptor-interacting proteins (37, 38). As a direct target gene, together with its ability to interact with TR, hr likely serves a dual role—as a downstream target as well as upstream regulator of thyroid hormone action.
Acknowledgments
We thank S. Hollenberg for two-hybrid vectors, yeast strains, and advice; J. Arriza for the human brain cDNA library; K. Umesono for providing multiple plasmids; H. Towle for pTZ18; O. Martin for technical assistance; our colleagues at the Carnegie Institution for helpful reagents, advice, and comments on the manuscript; and the Carnegie Institution of Washington, where these studies were initiated. This work was supported by the John Merck Fund at Community Trust and the National Institutes of Health (National Institute of Diabetes and Digestive and Kidney Diseases).
ABBREVIATIONS
- CNS
central nervous system
- β-gal
β-galactosidase
- TH
thyroid hormone
- TR
thyroid hormone receptor
- TRE
thyroid hormone response element
- GST
glutathione S-transferase
References
- 1.Schwartz H L. In: Molecular Basis of Thyroid Hormone Action. Oppenheimer J H, Samuels H H, editors. New York: Academic; 1983. pp. 413–444. [Google Scholar]
- 2.Morreale de Escobar G, Ruiz-Marcos A, Escobar del Rey F. In: Congenital Hypothyroidism. Dussault J H, Walker P, editors. New York: Dekker; 1983. pp. 85–126. [Google Scholar]
- 3.DeLong G R, Adams R D. In: The Thyroid. Braverman L E, Utiger R D, editors. New York: Lippincott–Raven; 1991. pp. 1027–1039. [Google Scholar]
- 4.Bernal J, Nunez J. Eur J Endocrinol. 1995;133:390–398. doi: 10.1530/eje.0.1330390. [DOI] [PubMed] [Google Scholar]
- 5.Oppenheimer J H. In: The Thyroid. Braverman L E, Utiger R D, editors. New York: Lippincott–Raven; 1991. pp. 204–222. [Google Scholar]
- 6.Samuels H H. In: Molecular Basis of Thyroid Hormone Action. Oppenheimer J H, Samuels H H, editors. New York: Academic; 1983. pp. 1–34. [Google Scholar]
- 7.Evans R M. Science. 1988;240:889–895. doi: 10.1126/science.3283939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tsai M-J, O’Malley B W. Annu Rev Biochem. 1994;63:451–486. doi: 10.1146/annurev.bi.63.070194.002315. [DOI] [PubMed] [Google Scholar]
- 9.Horwitz K B, Jackson T A, Bain D L, Richer J K, Takimoto G S, Tung L. Mol Endocrinol. 1996;10:1167–1177. doi: 10.1210/mend.10.10.9121485. [DOI] [PubMed] [Google Scholar]
- 10.Beato M, Sánchez-Pacheco A. Endocr Rev. 1996;17:587–609. doi: 10.1210/edrv-17-6-587. [DOI] [PubMed] [Google Scholar]
- 11.Thompson C C. J Neurosci. 1996;16:7832–7840. doi: 10.1523/JNEUROSCI.16-24-07832.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stoye J P, Fenner S, Greenoak G E, Moran C, Coffin J M. Cell. 1988;54:383–391. doi: 10.1016/0092-8674(88)90201-2. [DOI] [PubMed] [Google Scholar]
- 13.Cachon-Gonzalez M B, Fenner S, Coffin J M, Moran C, Best S, Stoye J P. Proc Natl Acad Sci USA. 1994;91:7717–7721. doi: 10.1073/pnas.91.16.7717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baniahmad A, Ha I, Reinberg D, Tsai S, Tsai M-J, O’Malley B W. Proc Natl Acad Sci USA. 1993;90:8832–8836. doi: 10.1073/pnas.90.19.8832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oñate S A, Tsai S Y, Tsai M-J, O’Malley B W. Science. 1995;270:1354–1357. doi: 10.1126/science.270.5240.1354. [DOI] [PubMed] [Google Scholar]
- 16.Lee J W, Ryan F, Swaffield J C, Johnston S A, Moore D D. Nature (London) 1995;374:91–94. doi: 10.1038/374091a0. [DOI] [PubMed] [Google Scholar]
- 17.Chen J D, Evans R M. Nature (London) 1995;377:454–457. doi: 10.1038/377454a0. [DOI] [PubMed] [Google Scholar]
- 18.Hörlein A J, Näär A M, Heinzel T, Torchia J, Gloss B, Kurokawa R, Ryan A, Kamei Y, Söderström M, Glass C K, Rosenfeld M G. Nature (London) 1995;377:397–404. doi: 10.1038/377397a0. [DOI] [PubMed] [Google Scholar]
- 19.Zeiner M, Gehring U. Proc Natl Acad Sci USA. 1995;92:11465–11469. doi: 10.1073/pnas.92.25.11465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.L’Horset F, Dauvois S, Heery D M, Cavaillès V, Parker M G. Mol Cell Biol. 1996;16:6029–6036. doi: 10.1128/mcb.16.11.6029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.vom Baur E, Zechel C, Heery D, Heine M, Garnier J M, Vivat V, LeDouarin B, Gronemeyer H, Chambon P, Losson R. EMBO J. 1996;15:110–124. [PMC free article] [PubMed] [Google Scholar]
- 22.Samuels H H, Stanley F, Casanova J. Endocrinology. 1979;105:80–85. doi: 10.1210/endo-105-1-80. [DOI] [PubMed] [Google Scholar]
- 23.Hollenberg S M, Sternglanz R, Cheng P F, Weintraub H. Mol Cell Biol. 1995;15:3813–3822. doi: 10.1128/mcb.15.7.3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robzyk K, Kassir Y. Nucleic Acids Res. 1992;20:3790. doi: 10.1093/nar/20.14.3790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reynolds A, Lundblad V. In: Short Protocols in Molecular Biology. Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. New York: Wiley; 1992. pp. 13–27. [Google Scholar]
- 26.Cavaillès V, Dauvois S, Danielian P S, Parker M G. Proc Natl Acad Sci USA. 1994;91:10009–10013. doi: 10.1073/pnas.91.21.10009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Umesono K, Murakami K K, Thompson C C, Evans R M. Cell. 1991;65:1255–1266. doi: 10.1016/0092-8674(91)90020-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Näär A M, Boutin J-M, Lipkin S M, Yu V C, Holloway J M, Glass C K, Rosenfeld M G. Cell. 1991;65:1267–1279. doi: 10.1016/0092-8674(91)90021-p. [DOI] [PubMed] [Google Scholar]
- 29.Lazar M A. Endocr Rev. 1993;14:184–193. doi: 10.1210/edrv-14-2-184. [DOI] [PubMed] [Google Scholar]
- 30.Schueler P A, Schwartz H L, Strait K A, Mariash C N, Oppenheimer J H. Mol Endocrinol. 1990;4:227–234. doi: 10.1210/mend-4-2-227. [DOI] [PubMed] [Google Scholar]
- 31.Thompson C C, Weinberger C W, Lebo R, Evans R M. Science. 1987;237:1610–1614. doi: 10.1126/science.3629259. [DOI] [PubMed] [Google Scholar]
- 32.Murray M B, Zilz N D, McCreary N L, MacDonald M J, Towle H C. J Biol Chem. 1988;263:12770–12777. [PubMed] [Google Scholar]
- 33.Bradley D J, Towle H C, Young W S., III J Neurosci. 1992;12:2288–2302. doi: 10.1523/JNEUROSCI.12-06-02288.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mellström B, Naranjo J R, Santos A, Gonzalez A M, Bernal J. Mol Endocrinol. 1991;5:1339–1350. doi: 10.1210/mend-5-9-1339. [DOI] [PubMed] [Google Scholar]
- 35.Farsetti A, Mitsuhashi T, Desvergne B, Robbins J, Nikodem V. J Biol Chem. 1991;266:23266–23232. [PubMed] [Google Scholar]
- 36.Strait K A, Zou L, Oppenheimer J H. Mol Endocrinol. 1992;6:1874–1880. doi: 10.1210/mend.6.11.1282672. [DOI] [PubMed] [Google Scholar]
- 37.Inoue S, Orimo A, Hosoi T, Kondo S, Toyoshima H, Kondo T, Ikegami A, Ouichi Y, Orimo H, Muramatsu M. Proc Natl Acad Sci USA. 1993;90:11117–11121. doi: 10.1073/pnas.90.23.11117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.LeDouarin B, Zechel C, Garnier J-M, Lutz Y, Tora L, Poerrat B, Heery D, Gronemeyer H, Chambon P, Losson R. EMBO J. 1995;14:2020–2033. doi: 10.1002/j.1460-2075.1995.tb07194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]