Abstract
Processing of antigens for presentation by major histocompatibility complex (MHC) class I molecules requires the activity of the proteasome. The 20S proteasome complex is composed of 14 different subunits, 2 of which can be substituted by the interferon γ (IFN-γ)-inducible and MHC-encoded subunits LMP2 and LMP7 (low molecular mass poylpeptides 2 and 7). A third subunit, MECL-1, is inducible by IFN-γ but is encoded outside the MHC. Here we show by cotransfection experiments that the incorporation of MECL-1 into the 20S proteasome is directly dependent on the expression of LMP2 but independent of LMP7. Conversely, the uptake of LMP2 is strongly enhanced by MECL-1 expression. The expression of MECL-1 caused a replacement of the homologous subunit Z in the 20S proteasome complex. LMP2 is required for MECL-1 incorporation at the level of proteasome precursor formation that guarantees the concerted incorporation of two IFN-γ-inducible proteasome subunits encoded inside and outside the MHC. The obligatory coincorporation of MECL-1 and LMP2 is an important parameter for the interpretation of results obtained with LMP2-deficient cell lines and mice as well as for the design of experiments addressing the function of MECL-1 in antigen presentation.
The major histocompatibility complex (MHC) class I restricted pathway of antigen presentation has evolved some 450 million years ago, allowing the presentation of intracellular viral antigens to cytotoxic T lymphocytes on the cell surface (1, 2). Three components of this pathway are encoded completely or in part in the MHC locus: two subunits, LMP2 and LMP7 (for low molecular mass polypeptides 2 and 7), of the proteasome which degrades self and non-self proteins to peptides in the nucleus and cytoplasm (3), both subunits, TAP1 and TAP2 (for transporter associated with antigen presentation 1 and 2), of a peptide transporter which conveys the peptides into the endoplasmic reticulum lumen (4), and the MHC class I heavy chain itself which binds octa- or nonameric peptides at defined C-terminal and internal anchor residues in its peptide binding cleft. The known murine class I molecules and peptide transporters accommodate only peptides with hydrophobic C termini whereas human peptide transporters efficiently translocate peptides with basic and hydrophobic C termini that can be bound by different human MHC class I molecules (4–6). A trimeric complex of peptide, MHC class I heavy chain and β2-microglobulin may be transported to the cell surface where MHC plus peptide is recognized by specific antigen receptors of cytotoxic T cells.
All three of these components, LMP2/7, TAP1/2, and MHC heavy chain, are inducible by the antiviral cytokine interferon γ (IFN-γ). During the assembly of the 20S proteasomes in IFN-γ-stimulated cells LMP2 and LMP7 replace two homologous and constitutive proteasome subunits designated δ (or “Y”) and MB-1 (or “X”) in the barrel-shaped complex (7, 8). This subunit exchange has been shown to alter the cleavage priority of the 20S proteasome in vitro (9, 10), whereas an increase in MHC class I cell surface expression and an improvement in the presentation of some antigens have been demonstrated in vivo (11–14). Recently a third subunit exchange has been discovered in the 20S proteasome: the IFN-γ inducible subunit MECL-1 replaces a homolgous and constitutively expressed subunit designated “MC14” in the mouse and “Z” in the human (15–17). Interestingly, in contrast to LMP2 and LMP7, MECL-1 is not encoded in the MHC locus (18).
To characterize the impact of MECL-1 incorporation on antigen presentation and the cleavage characteristics of the 20S proteasome, we attempted to over-express human and murine MECL-1 in a number of cell lines. These experiments yielded the surprising finding that over-expressed MECL-1 proteins could only be incorporated into 20S proteasomes when the LMP2 subunit was coexpressed in transfected cell lines. Conversely, MECL-1 expression in LMP2/LMP7 double transfected mouse fibroblast cells enhanced the incorporation of LMP2. Thus two independent subunit exchange events exist for the murine and human 20S proteasomes: the coincorporation of LMP2/MECL-1 and the single incorporation of LMP7.
MATERIALS AND METHODS
Cell Lines and Transfections.
The human lymphoblastoid cell line T2 (full name 174XCEM.T2; ref. 19) is a deletion mutant lacking the MHC class II locus and hence is deficient for LMP2 and LMP7. Transfectants T2/LMP2, T2/LMP7, and T2/LMP2+7 have been obtained by transfection of T2 with LMP2 and/or LMP7 as described (9). The clone TMP16 has been generated by transfection of T2 cells with the human MECL-1 expression construct pSG5hMECL-1 (described below) and the puromycin resistance construct pLXSP (E. Palmer, personal communication). The T27MP15 clone was obtained by transfection of T2/LMP2+7 cells with pSG5hMECL-1 and pLXSP. Cotransfections of T2 cells were performed by conventional electroporation (230 V, 960 μF) using 6 × 106 cells, 20 μg of pSG5hMECL-1 DNA, and 4 μg of pLXSP DNA in a volume of 600 μl PBS. Transfected cells were plated in 96-well plates under cloning conditions and selected with 0.4 μg/ml puromycin. Puromycin-resistant clones were analyzed for MECL-1 expression by Northern blot analysis.
The mouse fibroblast line B8 (H. Hengel and U. H. Koszinowski, personal communication) has been obtained by transfection of BALB/c derived C4 cells with the IE1 gene encoding the pp89 protein of mouse cytomegalovirus. Double transfection of B8 cells with murine LMP2 and LMP7 expression constructs yielded the clone BC27H7 as detailed elsewhere (20). The BME13 cell clone has been generated by cotransfection of B8 cells with the mouse MECL-1 expression construct pSG5 mMECL-1 and pLXSP. The B27M.2 clone was obtained by transfection of BC27H7 cells with pSG5 mMECL-1 and pLXSP. Cotransfection of B8 cells were performed by standard calcium phosphate precipitation using 106 cells, 8 μg of pSG5 mMECL-1 DNA, and 2 μg of pLXSP DNA. Transfected cells were plated under cloning conditions and selected with 2.5 μg/ml puromycin, and resistant clones were screened for MECL-1 expression by Western blot analysis.
The treatment of cells with recombinant murine or human IFN-γ (Boehringer Mannheim) was performed for 3 days in culture at a concentration of 20 units/ml for all reported experiments.
Expression Vectors.
The human MECL-1 expression construct pSG5hMECL-1 was generated as follows: human cDNA of MECL-1 was obtained by retrotranscription of total mRNA from IFN-γ-treated HeLa cells and PCR using primers GAGGAATTCCCAAGATGCTGAAGCCAGCCCTGG and GAAGGATCCCTCAGCTTACTCCACCTCCATAGC according to standard protocols. The PCR product was cloned via EcoRI and BamHI sites into the eukaryotic expression vector pSG5 (Stratagene) and sequenced to exclude PCR errors. The murine MECL-1 expression vector pSG5 mMECL-1 was constructed as follows: cDNA of MECL-1 was obtained by retrotranscription of total mRNA from IFN-γ-treated B8 cells and PCR using primers GAGGAATTCCTGCGAGATGCTGAAGC and GACGAATTCACTTCATTCCACCTCCATGG according to standard protocols. The PCR product was cloned via EcoRI into the eukaryotic expression vector pSG5 (Stratagene) and sequenced to exclude PCR errors.
Antibodies.
A mouse MECL-1 reactive polyclonal antibody was generated by immunization of rabbits with the mouse MECL-1-derived and keyhole limpet hemocyanin-conjugated synthetic peptide TAGGAKLQRALSTPTEPVQ. The antibody recognizes MECL-1 on blotted two-dimensional (2D) isoelectric focusing (IEF)/PAGE gels of purified proteasomes from IFN-γ-treated B8 cells. Cross-reactions with the homologous proteasome subunit Z (or MC14) or other proteins in Western blots of total cell lysates were not observed.
Northern Blot Analysis.
Total RNA was obtained by acid guanidinum thiocyanate-phenol-chloroform extraction, and Northern blotting was performed as described (21); 15 μg of total RNA were loaded per lane. The blots were radioactively probed with the entire coding region of human or mouse MECL-1 cDNA. Autoradiographies were quantified by densitometry in the linear range of quantification and the values were normalized to the amount of 18S and 28S rRNAs.
Western Blot Analysis.
About 2 × 106 cells were lysed in 100 μl of buffer A (50 mM Tris⋅HCl, pH 7.5/5 mM MgCl2/0.2% Triton X-100/1 mM phenylmethylsulfonyl fluoride/6 μg/ml aprotinin/7 μM pepstatin/10 μM leupeptin) by 3-fold freezing and thawing. The postnuclear supernatant was quantified by optical density at 280 nm assuming that 0.1 OD corresponds to a concentration of 100 μg protein per ml. Aliquots of 100 μg were supplemented with a 1× Laemmli sample buffer and applied to a 15% PAGE. The gels were blotted onto nitrocellulose membrane, and proteins were visualized by Ponceau red staining. The blots were blocked for 1 h in blocking buffer [PBS + 10% horse serum + 5% (wt/vol) low fat dry milk + 0.4% Tween 20] at room temperature and agitated overnight at 4°C in a 500-fold dilution of mouse MECL-1 reactive polyclonal antibody in blocking buffer. Blots were washed three times in PBS + 0.4% Tween 20 and incubated for 1 h in a 20,000-fold dilution of a goat anti-rabbit IgG-peroxidase conjugate (Dianova, Hamburg. Germany) in blocking buffer. After four washes proteins were visualized on films by the enhanced chemiluminescence reaction.
For sucrose gradient analysis the postnuclear supernatant from 1 × 107 cells was loaded onto a 10–40% sucrose gradient in lysis buffer and ultracentrifuged in a SW40 rotor at 40,000 rpm for 15.50 h. Fractions of 600 μl were collected and the MECL-1 content was analyzed by Western blot analysis.
Proteasome Purification and IEF/PAGE.
20S proteasomes were purified from cell lines as described (20). A total of 60 μg of purified proteasomes were analyzed per 2D IEF/PAGE gel as described (15). The gels were stained with Coomassie stain and spots of interest where quantified by densitometry in the linear range of quantification.
RESULTS
With the plan to characterize the impact of MECL-1 incorporation on the cleavage characteristics of the 20S proteasome, we cloned the human MECL-1 cDNA into the eukaryotic expression vector pSG5 for over-expression of MECL-1 in HeLa cells. HeLa cells were initially chosen as recipients since their endogenous expression of LMP2, LMP7, and MECL-1 is extremely low. When the 20S proteasome subunit composition of two independent MECL-1 transfectants was analyzed by 2D IEF/PAGE, we noted that in spite of a very high expression of MECL-1 mRNA the amount of MECL-1 protein incorporation into the 20S proteasome was much less than found for IFN-γ-stimulated cells (data not shown).
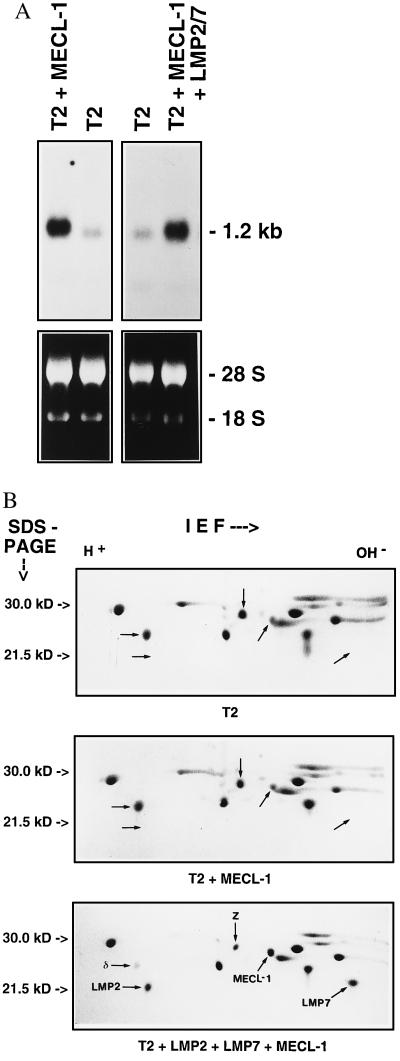
This unexpected result suggested that MECL-1 subunit incorporation may be dependent on factors that are inducible by IFN-γ and prompted us to examine whether the presence of LMP2 or LMP7 influences MECL-1 incorporation into the 20S proteasome complex. For this purpose we stably over-expressed MECL-1 in lymphoblastoid T2 cells that lack LMP2 and LMP7 expression. For comparison MECL-1 was also transfected into a previously generated T2 transfectant T2/LMP2+7 which stably expresses murine LMP2 and LMP7 (9). By quantitative Northern blot analysis of MECL-1 mRNA expression (Fig. 1A) a cell clone TMP16 was selected from a panel of 17 T2 transfectants for further experiments. TMP16 expressed 6.7-fold higher MECL-1 mRNA levels than endogenously found in T2 cells (Fig. 1A). Similarly, a clone designated T27MP15 was isolated from 15 MECL-1 transfectants of T2/LMP2+7 cells that over-expressed the MECL-1 mRNA by a factor of 4.8. To determine MECL-1 incorporation, 20S proteasome complexes were isolated from T2, TMP16, and T27MP15 cells and their subunit composition was analyzed on Coomassie-stained 2D IEF/PAGE gels (Fig. 1B). The indicated positions of the homologous human proteasome subunits Z and MECL-1 in 2D gels were confirmed by N-terminal microsequencing from blotted gels, and their relative amounts were quantified by densitometry and normalized to the amounts of an invariant proteasome subunit.
Figure 1.
(A) Northern blot analysis of human MECL-1 expression in T2 cells and transfectants TMP16 (T2 + MECL-1) and T27MP15 (T2 + MECL-1 + LMP2/7). (Upper) Autoradiography of MECL-1 Northern blot, the 1.2-kb RNA marker band is indicated. (Lower) Photography of ethidium bromide-stained total RNA before blotting; 18S and 28S rRNAs are assigned. (B) 2D IEF/PAGE gels of 20S proteasomes purified from T2 cells (T2), TMP16 cells (T2 + MECL-1), and T27MP15 cells (T2 + LMP2 + LMP7 + MECL-1). The subunits LMP2, δ, MECL-1, Z, and LMP7 were assigned according to their migratory position as previously determined by microsequencing analysis. In addition the identity of human MECL-1 and Z was confirmed by N-terminal microsequencing from a Western blot of 2D gels of T27MP15 proteasomes. The sequences obtained were as follows: TTIAGLVFQD for MECL-1 and TTIAGVVYKD for Z. The MB-1 subunit is not visible because its isoelectric point is quite basic, causing it to migrate out of the IEF gel rod under the applied conditions. A total of 60 μg of proteasome was loaded for each gel; proteins were visualized by Coomassie stain.
Strikingly, a 6.7-fold over-expression of MECL-1 mRNA in TMP16 cells did not lead to any increase in the amount of MECL-1 protein when compared with that of T2 proteasomes. In contrast, despite a slightly lower MECL-1 mRNA over-expression in T27MP15 cells a 5-fold increase of mature MECL-1 in isolated 20S proteasomes was obtained. This result demonstrates that the presence of either LMP2, LMP7, or both subunits together is a prerequisite for efficient MECL-1 incorporation into 20S proteasomes. Importantly, the increase in the amount of MECL-1 protein in T27MP15 proteasomes caused a 2-fold reduction of the proteasome subunit Z as compared with T2 cells. Thus these results show evidence that the constitutive subunit Z can be replaced by MECL-1 independently of IFN-γ-inducible factors other than LMP2 and LMP7.
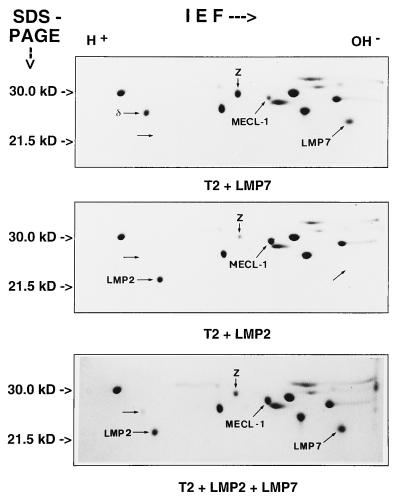
To determine whether either LMP2 or LMP7 alone are sufficient to promote MECL-1 subunit uptake into the 20S proteasome we analyzed the proteasome subunit composition of the previously generated T2 cell clones T2/LMP2, T2/LMP7, and T2/LMP2+7 that, respectively, express murine LMP2, LMP7, or LMP2 and LMP7 together (9). The IEF/PAGE analyses shown in Fig. 2 demonstrate that LMP2 was necessary and sufficient to allow incorporation of MECL-1 into 20S proteasomes whereas LMP7 did not increase the uptake of MECL-1. The observed differences in MECL-1 protein cannot be attributed to different levels in MECL-1 mRNA expression because they were identical for the examined T2 transfectants (data not shown). A comparison of the degree of MECL-1/Z subunit exchange in proteasomes from T27MP.15 cells (Fig. 1B Bottom), with that of T2/LMP2 and T2/LMP2+7 cells (Fig. 2) shows that it correlates with the degree of LMP2 expression rather than with the MECL-1 mRNA expression in these cell lines. It is important to note that the endogenous expression of MECL-1 in T2/LMP2 cells was sufficient to achieve a 4-fold reduction in the amount of subunit Z. Taken together, we conclude that LMP2 but not LMP7 is required for MECL-1 incorporation into the 20S proteasome complex.
Figure 2.
2D IEF/PAGE gels of 20S proteasomes purified from cell lines T2/LMP2 (T2 + LMP2), T2/LMP7 (T2 + LMP7), and T2/LMP2+7 (T2 + LMP2 + LMP7). The subunits LMP2, δ, MECL-1, Z, and LMP7 were assigned according to their migratory position as described in the legend of Fig. 1B. Note that the exchange of MECL-1 for proteasome subunit Z is dependent on LMP2 expression but independent of LMP7 expression. The generation of the applied transfectants has been previously reported by Kuckelkorn et al. (9). The endogenous expression of MECL-1 was identical for all cell lines according to Northern blot analysis (data not shown).
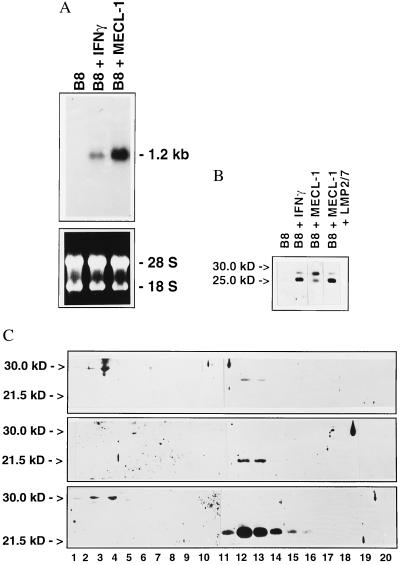
The mature 20S proteasome is assembled from precursor complexes that autocatalytically cleave the proforms of several β-type subunits like LMP2, LMP7, or MECL-1 during their assembly (22–25). This raises the question whether proteasome assembly could be the step at which LMP2 is required for MECL-1 subunit incorporation. In total lysates of B8 mouse fibroblast cells grown in the presence of IFN-γ, MECL-1 was recognized as a protein of 25 kDa by a monospecific antibody raised against this subunit. Upon longer exposure an additional band of 29 kDa became visible, which is likely to be the unprocessed MECL-1 precursor protein because neither the 25-kDa nor the 29-kDa band were detected in B8 cells grown in the absence of IFN-γ (Fig. 3B). To extend our analysis, murine MECL-1 was over-expressed in B8 cells by stable transfection and a clone, designated BME13, was isolated which expressed 2.3-fold more MECL-1 mRNA than B8 cells after stimulation with murine IFN-γ (Fig. 3A). Despite strong MECL-1 mRNA expression in BME13 cells the amount of mature 25-kDa MECL-1 protein in total lysates was far less than found in IFN-γ-treated B8 cells. Instead the 29-kDa MECL-1 precursor protein strongly accumulated in BME13 cells. This accumulation of MECL-1 must be due to the very low LMP2 expression in B8 cells (20). In agreement with this notion the coexpression of murine LMP2, LMP7, and MECL-1 in a triple transfectant (B27M.2) resulted in a much greater amount of mature MECL-1 protein as compared with MECL-1 precursor protein (Fig. 3B). When extracts of BME13 cells were analyzed on sucrose gradients, the unprocessed MECL-1 precursor proteins in BME13 remained at the top of the gradients and did not accumulate in the 13S or 16S fractions where proteasome precursor complexes are normally found (23, 24) (Fig. 3C). Only after long over-exposure a faint band of 29 kDa was found in fractions 8 and 9 corresponding to a sedimentation coefficient of 16S (data not shown). These experiments indicate that LMP2 is required already at the step of the formation of proteasome precursor complexes as a prerequisite for MECL-1 uptake, processing, and incorporation into the mature 20S proteasome.
Figure 3.
(A) Northern blot analysis of mouse MECL-1 expression in B8 mouse fibroblast cells before (B8) and after (B8 + IFN-γ) stimulation with IFN-γ and in the transfected clone BME13 (B8 + MECL-1). The blots were probed with the cDNA of the complete coding region of murine MECL-1. (Upper) Autoradiography of MECL-1 Northern blot. (Lower) Photography of ethidium bromide stained total RNA before blotting. IFN-γ treatment [20 units/ml recombinant mouse IFN-γ (Boehringer Mannheim)] was performed for 3 days before isolation of total RNA. (B) Western blot analysis of MECL-1 protein expression in untreated B8 cells (B8), in IFN-γ-treated B8 cells (B8 + IFN-γ), in BME13 cells (B8 + MECL-1) and in a B8 clone stably transfected with LMP2, LMP7, and MECL-1. The 29-kDa band corresponds to the MECL-1 precursor protein, and the 25-kDa corresponds to the mature MECL-1 protein. (C) Western blot analysis of MECL-1 (Top), LMP2 (Middle), and LMP7 (Bottom) protein distribution in BME13 cells after fractionation of cytoplasmic proteins on sucrose gradients; 30-kDa and 21.5-kDa markers in SDS/PAGE are indicated, fraction numbers are assigned below each lane. Precursor proteins are visible in fractions 1–3 and mature proteasome subunits in fractions 12 and 13. Upon longer exposure faint precursor bands of 29 kDa became also visible in fractions 8 and 9, corresponding to 16S proteasome precursor complexes. Note the much higher efficiency of LMP7 subunit incorporation into the 20S proteasome as compared with MECL-1 or LMP2 in BME13 cells.
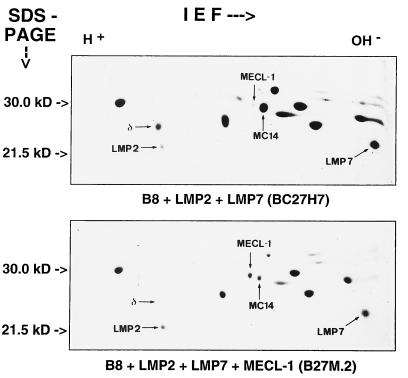
Previously we had shown that transfection of T2 cells with a LMP2 expression construct resulted in a complete replacement of the homologous proteasome subunit δ by LMP2 (9, 24) whereas the same experiment performed with B8 mouse fibroblast cells only yielded a partial exchange of LMP2 for δ. As shown in Fig. 3A, the endogenous expression of MECL-1 is very low in B8 mouse fibroblast cells in comparison to T2 human lymphoblastoid cells (Fig. 1A). This finding prompted us to test the possibility that LMP2 and MECL-1 incorporation into the 20S proteasome are mutually dependent, which could provide an explanation why a complete subunit exchange of LMP2 for δ does not occur in B8 cells. Therefore we compared 20S proteasomes from the LMP2/LMP7 double transfected line BC27H7 (see ref. 20) and the triple transfectant B27M.2, which was obtained by transfection of BC27H7 cells with a mouse MECL-1 expression construct. The analysis of the proteasome subunit composition confirmed that MECL-1 expression in B27M.2 cells induced a replacement of the murine homologue of Z (also named MC14, Fig. 4). Strikingly, MECL-1 expression and incorporation strongly promoted the subunit exchange of LMP2 for the δ subunit in B27M.2 cells when compared with BC27H7 recipient cells whereas the incorporation of LMP7 remained invariant. As the LMP2 expression in BC27H7 and B27M2 cells was identical in Northern blot analysis (data not shown), it is evident that LMP2 and MECL-1 are mutually required for incorporation into the 20S proteasome. An obligatory coincorporation of MECL-1 and LMP2 poses the difficulty in that the subunit-specific impact of MECL-1 on proteasomal peptide hydrolysis cannot be studied in any of the so far available cell line mutants or gene-deficient mice because the observed in vitro or in vivo effects could be attributed to changes of either LMP2 or MECL-1. Therefore the interdependence of LMP2 and MECL-1 is an important parameter for the interpretation of analyses employing LMP2-deficient cell lines and mice. It also sets the stage for the design of future experiments aimed at an identification of MECL-1 function involving the expression of catalytically inactivated proteasome subunits.
Figure 4.
2D IEF/PAGE gels of 20S proteasomes purified from cell lines BC27H7 (B8 + LMP2 +LMP7) and B27M.2 (B8 +LMP2 + LMP7 + MECL-1). The subunits LMP2, δ, MECL-1, MC14, and LMP7 were assigned according to their migratory position (15). Note the different relative positions of murine MECL-1 and MC14 as compared with human MECL-1 and Z (Fig. 1B). Shown are Coomassie-stained gels each containing 60 μg of 20S proteasomes.
DISCUSSION
In this study we provide evidence that the incorporation of the IFN-γ-inducible proteasome subunits MECL-1 and LMP2 are mutually dependent for their incorporation into the 20S proteasome complex. As the coincorporation of LMP2/MECL-1 and the single incorporation of LMP7 do not seem to affect each other in steady-state analysis, there exist two rather than three independent subunit exchange events in murine and human 20S proteasomes. The obligatory coincorporation of LMP2 and MECL-1 has important consequences for the interpretation of LMP2-deficient cell lines and gene targeted mice (19). A close reinspection of 2D IEF/PAGE gels of proteasomes from spleen cells of LMP2 knock out mice reported by Van Kaer et al. (12) for instance reveals besides a lack of LMP2 also a lack of a proteasome subunit in the position where mouse MECL-1 normally migrates. It is thus likely that reported in vivo or in vitro effects of LMP2 mutation are at least in part due to a concomitant lack of MECL-1. Interestingly, also shown in this analysis is that the incorporation of LMP7 remained unaffected by the deletion of LMP2. A mutual dependence of LMP2 and MECL-1 may at least in part explain discrepancies in the analyses of LMP2-deficient cell lines from several laboratories as the level of the endogenous MECL-1 expession in different cell line models will strongly influence the LMP2 incorporation (12, 13, 26). Furthermore, the different levels of endogenous MECL-1 expression in T2 and B8 cells must be the reason why LMP2 over-expression led to a complete exchange of LMP2 for the δ subunit in T2 cells (9, 24), whereas only a partial replacement could be obtained in B8 cells (20).
The LMP2 dependence of MECL-1 incorporation limits the variability in proteasome subunit replacements and raises the question whether there is a biological reason for this phenomenon. A certain cleavage activity may for instance require the presence of both MECL-1 and LMP2. Alternatively, conformational changes within the 20S proteasome possibly directing the association with proteasome activators may be dependent on both subunits. On the other hand LMP2 may be a prerequisite for MECL-1 incorporation for structural rather than functional reasons. In this respect the very recent publication of the three-dimensional structure of the 20S proteasome from Saccharomyces cerevisiae is of great interest: the yeast β-type subunits PRE3 and PUP1 which are highly homologous to δ and Z, respectively, are directly juxtaposed in the two rings of β-type subunits, and make close molecular contacts with each other whereas the yeast subunit PRE2, which is homologous to MB-1, is separated from PRE3 and PUP1 by two β-type subunits (27). Even direct subunit interactions in trans between PRE2 of one β-type ring and PRE3/PUP1 of the second β-ring were not found within the yeast 20S complex. These spatial interactions may well account for the observed two sets of independent proteasome subunits incorporation of LMP2/MECL-1 on the one hand and LMP7 on the other.
A common trait of the IFN-γ-inducible proteasome subunits LMP2, LMP7, and MECL-1 as well as their replaceable homologues δ, MB-1, and Z is the existence of a threonine residue at the N terminus and a lysine residue in position 33 of the mature β-type proteasome subunits. These two residues were found to be essential for proteolytic activity in the active centers of the complex (24, 28, 29). It is thus feasible that the immune system uses IFN-γ-inducible subunits to modulate the cleavage activities at the individual active centers of the 20S proteasome complex by means of subunit replacements. A concomitant replacement of the constitutive subunit δ by LMP2 and Z by MECL-1, for instance, has been shown to strongly reduce the peptide cleavage at the C terminus of glutamic acid (9, 10, 20). Consistent with this result a mutation in PRE3, the δ-homologue of S. cerevisiae selectively affects the hydrolysis of glutamic acid peptide substrates (30) and the P1 substrate binding pocket of PRE3 contains an arginine residue at position 45 at the base of the pocket.
We have analyzed the 20S proteasomes from cell lines reported in this study with fluorogenic peptide assays. Our preliminary analysis comparing the effect of an incorporation of human MECL-1 in T2 cells with that of murine MECL-1 in B8 cells yielded the surpirsing finding that the incorporation of human MECL-1 into 20S proteasomes enhanced the hydrolysis of the fluorogenic substrate BZ-VGR-MCA whereas murine MECL-1 induced a slight reduction of this activity (data not shown). This finding suggests that the exchange of MECL-1 for Z may modulate the trypsin-like activity of the mammalian 20S proteasome in a species-specific manner that would be consistent with the difference in specificity of human and murine peptide transporters and MHC class I molecules. Moreover, it would be in agreement with the effect of a point mutation introduced in the MECL-1 homologue of S. cerevisiae, PUP1, which causes a severe and specific reduction in the hydrolysis of arginine substrates in the mutant (W. Heinemeyer and D. Wolf, personal communication). However, it should be pointed out that the analysis of the proteasomal cleavage activity with short fluorogenic peptides may not necessarily reflect the situation with larger polypeptide substrates in vivo (1) and that a modulation of cleavages C terminal of arginine by an exchange of MECL-1 for Z is not supported by the highly resolved 20S proteasome structure from S. cerevisiae (27). Experiments involving cotransfection of murine and human MECL-1 in the same cell line employing functionally inactivated LMP2 and MECL-1 subunits as well as cross species experiments would be required to further substantiate our data on MECL-1-specific function and are presently being performed in our laboratory.
The discovery of lmp and tap genes in the MHC locus has nourished expectations that their colocalization is the consequence of a selective advantage related to their function in class I-restricted antigen presentation. It therefore came as a surprise that MECL-1, being encoded outside the MHC, is inducible by IFN-γ, and like LMP2 and LMP7 replaces a homologous and constitutively expressed proteasome subunit in the 20S proteasome complex. This atypical location of MECL-1 in the genome could be interpreted to argue against a selective advantage of colocalization. However, our finding that the incorporation of MECL-1 is dependent on LMP2 at the level of proteasome complex formation might explain why a selective pressure to maintain MECL-1 in the MHC locus was relieved.
Acknowledgments
We thank Regine Kraft and Susanne Kostka for N-terminal microsequence determination, Peter Henklein for peptide synthesis, Wolfgang Heinemeyer and Dieter H. Wolf for sharing unpublished information, and Dietmar Zaiβ and Gunter Schmidtke for critically reading the manuscript. This work was supported by the Deutsche Forschungsgemeinschaft Grant GR1517 to M.G. and Grant Kl 9-1 to P.M.K.
ABBREVIATIONS
- MHC
major histocompatibility complex
- IFN-γ
interferon γ
- LMP
low molecular mass polypeptide
- TAP
transporter associated with antigen presentation
- IEF
isoelectric focusing
- 2D
two dimensional
References
- 1.Groettrup M, Soza A, Kuckelkorn U, Kloetzel P M. Immunol Today. 1996;17:429–435. doi: 10.1016/0167-5699(96)10051-7. [DOI] [PubMed] [Google Scholar]
- 2.Heemels M-T, Ploegh H. Annu Rev Biochem. 1995;64:463–491. doi: 10.1146/annurev.bi.64.070195.002335. [DOI] [PubMed] [Google Scholar]
- 3.Coux O, Tanaka K, Goldberg A L. Annu Rev Biochem. 1996;65:801–847. doi: 10.1146/annurev.bi.65.070196.004101. [DOI] [PubMed] [Google Scholar]
- 4.Howard J. Curr Opin Immunol. 1995;7:69–76. doi: 10.1016/0952-7915(95)80031-x. [DOI] [PubMed] [Google Scholar]
- 5.Rammensee H G, Falk K, Rötzschke O. Annu Rev Immunol. 1993;11:213–244. doi: 10.1146/annurev.iy.11.040193.001241. [DOI] [PubMed] [Google Scholar]
- 6.Engelhard V H. Curr Opin Immunol. 1994;6:13–23. doi: 10.1016/0952-7915(94)90028-0. [DOI] [PubMed] [Google Scholar]
- 7.Akiyama K-y, Yokota K-Y, Kagawa S, Shimbara N, Tamura T, Akioka H, Nothwang H G, Noda C, Tanaka K, Ichihara A. Science. 1994;265:1231–1234. doi: 10.1126/science.8066462. [DOI] [PubMed] [Google Scholar]
- 8.Belich M P, Glynne R J, Senger G, Sheer D, Trowsdale J. Curr Biol. 1994;4:769–776. doi: 10.1016/s0960-9822(00)00174-3. [DOI] [PubMed] [Google Scholar]
- 9.Kuckelkorn U, Frentzel S, Kraft R, Kostka S, Groettrup M, Kloetzel P-M. Eur J Immunol. 1995;25:2605–2611. doi: 10.1002/eji.1830250930. [DOI] [PubMed] [Google Scholar]
- 10.Gaczynska M, Rock K L, Spies T, Goldberg A L. Proc Natl Acad Sci USA. 1994;91:9213–9217. doi: 10.1073/pnas.91.20.9213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fehling H J, Swat W, Laplace C, Kuehn R, Rajewsky K, Mueller U, von Boehmer H. Science. 1994;265:1234–1237. doi: 10.1126/science.8066463. [DOI] [PubMed] [Google Scholar]
- 12.Van Kaer L, Ashton-Rickardt P G, Eichelberger M, Gaczynska M, Nagashima K, Rock K L, Goldberg A L, Doherty P C, Tonegawa S. Immunity. 1994;1:533–541. doi: 10.1016/1074-7613(94)90043-4. [DOI] [PubMed] [Google Scholar]
- 13.Sibille C, Gould K G, Willard-Gallo K, Thomson S, Rivett A J, Powis S, Butcher G W, De Baetselier P. Curr Biol. 1995;5:923–930. doi: 10.1016/s0960-9822(95)00182-5. [DOI] [PubMed] [Google Scholar]
- 14.Cerundolo V, Kelly A, Elliot T, Trowsdale J, Townsend A. Eur J Immunol. 1995;25:554–562. doi: 10.1002/eji.1830250238. [DOI] [PubMed] [Google Scholar]
- 15.Groettrup M, Kraft R, Kostka S, Standera S, Stohwasser R, Kloetzel P. Eur J Immunol. 1996;26:863–869. doi: 10.1002/eji.1830260421. [DOI] [PubMed] [Google Scholar]
- 16.Hisamatsu H, Shimbara N, Saito Y, Kristensen P, Hendil K B, Fujiwara T, Takahashi E-I, Tanahashi N, Tamura T, Ichihara A, Tanaka K. J Exp Med. 1996;183:1–10. doi: 10.1084/jem.183.4.1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nandi D, Jiang H, Monaco J J. J Immunol. 1996;156:2361–2364. [PubMed] [Google Scholar]
- 18.Larsen F, Solheim J, Kristensen T, Kolsto A-B, Prydz H. Hum Mol Gen. 1993;2:1589–1595. doi: 10.1093/hmg/2.10.1589. [DOI] [PubMed] [Google Scholar]
- 19.Salter R D, Cresswell P. EMBO J. 1986;5:943–949. doi: 10.1002/j.1460-2075.1986.tb04307.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Groettrup M, Ruppert T, Kuehn L, Seeger M, Standera S, Koszinowski U, Kloetzel P M. J Biol Chem. 1995;270:23808–23815. doi: 10.1074/jbc.270.40.23808. [DOI] [PubMed] [Google Scholar]
- 21.Groettrup M, Baron A, Griffiths G, Palacios R, von Boehmer H. EMBO J. 1992;11:2735–2746. doi: 10.1002/j.1460-2075.1992.tb05339.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frentzel S, Kuhn H I, Gernold M, Gött P, Seelig A, Kloetzel P M. Eur J Biochem. 1993;216:119–126. doi: 10.1111/j.1432-1033.1993.tb18123.x. [DOI] [PubMed] [Google Scholar]
- 23.Frentzel S, Pesold-Hurt B, Seelig A, Kloetzel P. J Mol Biol. 1994;236:975–981. doi: 10.1016/0022-2836(94)90003-5. [DOI] [PubMed] [Google Scholar]
- 24.Schmidtke G, Kraft R, Kostka S, Henklein P, Frömmel C, Löwe J, Huber R, Kloetzel P M, Schmidt M. EMBO J. 1996;15:6887–6898. [PMC free article] [PubMed] [Google Scholar]
- 25.Chen P, Hochstrasser M. Cell. 1996;86:961–972. doi: 10.1016/s0092-8674(00)80171-3. [DOI] [PubMed] [Google Scholar]
- 26.Arnold D, Driscoll J, Androlewicz M, Hughes E, Cresswell P, Spies T. Nature (London) 1993;360:171–174. doi: 10.1038/360171a0. [DOI] [PubMed] [Google Scholar]
- 27.Groll M, Ditzel L, Löwe J, Satock D, Bochtler M, Bartunik H D, Huber R. Nature (London) 1997;386:463–471. doi: 10.1038/386463a0. [DOI] [PubMed] [Google Scholar]
- 28.Löwe J, Stock D, Jap B, Zwickl P, Baumeister W, Huber R. Science. 1995;268:533–539. doi: 10.1126/science.7725097. [DOI] [PubMed] [Google Scholar]
- 29.Seemüller E, Lupas A, Stock D, Löwe J, Huber R, Baumeister W. Science. 1995;268:579–582. doi: 10.1126/science.7725107. [DOI] [PubMed] [Google Scholar]
- 30.Enenkel C, Lehmann H, Kipper J, Gueckel R, Hilt W, Wolf D H. FEBS Lett. 1994;341:193–196. doi: 10.1016/0014-5793(94)80455-9. [DOI] [PubMed] [Google Scholar]