Abstract
Several G-protein coupled receptors, such as the β1-adrenergic receptor (β1-AR), contain polyproline motifs within their intracellular domains. Such motifs in other proteins are known to mediate protein–protein interactions such as with Src homology (SH)3 domains. Accordingly, we used the proline-rich third intracellular loop of the β1-AR either as a glutathione S-transferase fusion protein in biochemical “pull-down” assays or as bait in the yeast two-hybrid system to search for interacting proteins. Both approaches identified SH3p4/p8/p13 (also referred to as endophilin 1/2/3), a SH3 domain-containing protein family, as binding partners for the β1-AR. In vitro and in human embryonic kidney (HEK) 293 cells, SH3p4 specifically binds to the third intracellular loop of the β1-AR but not to that of the β2-AR. Moreover, this interaction is mediated by the C-terminal SH3 domain of SH3p4. Functionally, overexpression of SH3p4 promotes agonist-induced internalization and modestly decreases the Gs coupling efficacy of β1-ARs in HEK293 cells while having no effect on β2-ARs. Thus, our studies demonstrate a role of the SH3p4/p8/p13 protein family in β1-AR signaling and suggest that interaction between proline-rich motifs and SH3-containing proteins may represent a previously underappreciated aspect of G-protein coupled receptor signaling.
β-adrenergic receptors (βARs) belong to a large family of G-protein-coupled receptors that contain seven transmembrane domains connected by three extracellular loops, three intracellular loops, and a carboxyl terminal tail. Three βAR subtypes, β1, β2, and β3, have been identified by molecular cloning (1). All three βAR subtypes couple to the stimulatory GTP-binding protein (Gs) to activate adenylyl cyclase. Notably, the βAR subtypes are distinguishable with respect to tissue distribution and their regulatory roles in specific physiological processes (2).
Our knowledge about the molecular mechanisms underlying βAR signaling has been greatly expanded in the past 10 years, largely as a result of studies using the β2-AR as a model system (3). Understanding the biology of both β1-AR and β3-AR signaling has significantly lagged behind, although it has long been recognized that the βAR subtypes might undergo distinct regulation (4–6) and signal through distinct mechanisms.
It has been previously shown by mutagenesis studies that the specificity of G protein coupling for βAR is dictated by the intracellular domains, particularly the third intracellular loop and the C-terminal tail (7, 8). Furthermore, these regions are also critical for binding of βAR regulatory proteins, primarily the G protein-coupled receptor kinases (GRKs) and the β-arrestins (9, 10). On agonist stimulation, GRKs are recruited to the plasma membrane and phosphorylate the activated β2 receptors (11). The β-arrestins then bind to the phosphorylated β2 receptors (12) to induce rapid desensitization and internalization of the receptors.
Close examination of the primary sequences of the β1- and β2-ARs (13) reveals that their third intracellular loops are highly conserved except for a 24-aa proline-rich segment in the middle of the β1 third intracellular loop. It has been suggested that this motif may be responsible for certain differences between β1-AR and β2-AR signaling (13). More intriguingly, this sequence feature also can be found in other G-protein coupled receptors (GPCRs), including β3-AR, α2A-AR, and dopamine D4 receptors (14). In the case of dopamine D4 receptors, these proline-rich motifs were demonstrated, in vitro, to serve as potential ligands for several SH3 domain-containing proteins, such as Nck and Grb2 (14).
In an attempt to better understand the biology of β1-AR signaling and the molecular determinants underlying the differences between β1-AR and β2-AR regulation, we set out to search for novel binding proteins for the β1-AR by using its third intracellular loop as bait in several protein interaction systems.
Materials and Methods
Antibodies and Plasmids.
Anti-Flag M2 affinity gel and biotinylated M2 anti-Flag antibody were obtained from Sigma. Rabbit polyclonal anti-hemagglutinin (HA) antibody from Babco (Richmond, CA) was used to detect HA-tagged proteins. Streptavidin horseradish peroxidase, anti-mouse horseradish peroxidase, and anti-rabbit horseradish peroxidase were obtained from Amersham Pharmacia.
The pcDNA3 Flag-β1-AR plasmid was constructed by PCR, incorporating the HA endoplasmic reticulum targeting peptide and the FLAG peptide at the amino terminus. The pcDNA3 Flag-β2-AR plasmid was constructed by subcloning the Flag-β2-AR sequence from pBK-CMV (15) into the BamHI and XbaI sites of pcDNA3. pcDNA3 Flag-(or HA)-SH3p4 and its mutants were used as indicated in the figure legends. Glutathione S-transferase (GST)–ALP1 (amphiphysin-like-protein 1), GST-Grb2 NSH3, and GST-Grb2 CSH3 constructs were generously provided by Ann M. Pendergast (Duke University Medical Center) and GST-Grb2 by A. R. Saltiel (Parke–Davis Pharmaceutical Research Institute). GST-Src Src homology (SH)3 construct was described previously (16).
Bovine Brain Tissue Extract Preparation and GST Pull-Down Assay.
GST fusion proteins encoding the β1- and β2-AR third intracellular loops were purified according to manufacturer’s instruction (Amersham Pharmacia). Frozen bovine brain (80 g) was thawed in 200 ml of buffer A (20 mM Tris⋅HCl, pH7.2/5 mM EDTA/10 mM PMSF/1 μg/ml leupeptin/1 μg/ml pepstatin/1 μg/ml aprotinin/10 μg/ml benzamidine⋅HCl) and was homogenized by using a polytron homogenizer. Brain extracts were fractionated by centrifugation at 150,000 × g for 30 min. GST-β1 (or β2) third intracellular loop fusion proteins (1–1.5 mg) conjugated on glutathione Sepharose 4B beads were subsequently added to 10-ml aliquots of the resulting supernatant. After incubation at 4°C with gentle rotation for 1 hr, the beads were recovered by centrifugation at 800 × g for 5 min and were washed extensively with PBS containing 0.1% Triton X-100. After washing, GST fusion proteins and putative interacting proteins were eluted with reduced 10 mM glutathione in 50 mM Tris⋅HCl (pH 8.0). Where indicated, the elute was concentrated by using a Centricon (Millipore). Protein concentration was determined with Bradford reagent (Bio-Rad). SDS sample buffer was added to the eluted samples, which were subjected to SDS/PAGE and stained with Coomassie blue. Proteins specifically interacting with GST-β1 third intracellular loop were subjected to in-gel trypsin digestion and peptide sequencing (Protein Chemistry Core Facility, Baylor College of Medicine, Houston, TX).
Yeast Two-Hybrid Screen.
The pAS2-1-β1 (third intracellular loop) plasmid was constructed by PCR using the pcDNA1 HA-β1AR construct as a template. The yeast PJ-69-4A strain was co-transformed with the pAS2-1-β1(third intracellular loop) plasmid and a rat brain cDNA library (CLONTECH) by using standard yeast transformation protocols (17, 18). Of ≈13 million independent clones, nine exhibited moderate to strong growth on either −His or −Ade selective plates. The library plasmids isolated from positive clones then were cotransformed into the PJ-69-4A strain with either the pAS2-1-β1(third intracellular loop) plasmid or pAS2-1, and the specificity of the interactions were confirmed by growth on −His and −Ade selective plates as well as by β-galactosidase activity (Yeast Protocols Handbook, CLONTECH).
ELISA Assay for Analyzing Protein Interaction in Vitro.
The pET 30a vector (Novagen) was used to express His-S-tagged β1-AR and β2-AR third intracellular loops in Escherichia coli, and expressed proteins were purified according to the manufacturer’s manual. His-S-tagged β1 or β2 third intracellular loop fusion protein (0.5 μg) diluted in 100 μl of PBS plus a mixture of protease inhibitors was coated per well in 96-well high affinity binding dishes (Fisher) overnight at 4°C. After the coating solution was removed, the wells were washed with PBS and then were blocked with 2% dry milk in PBS for 1 hr at room temperature. Subsequently, GST fusion proteins diluted in 2% milk in PBS plus protease inhibitors were added to the precoated wells at the indicated concentrations and were incubated for 1 hr at room temperature. After incubation, the wells were extensively washed with PBS. GST substrate (100 mM potassium phosphate buffer, pH 6.5/1 mM 1-chloro-2, 4-dinitrobenzene/1 mM reduced glutathione), 150 μl, was added to each well, and the absorbance at 340 nm was recorded to determine the reaction kinetics and endpoint readings. To ensure that equivalent amounts of His-S-tagged β1and β2 third intracellular loop proteins were bound to the 96-well dishes, some wells were developed by using anti-S alkaline phosphatase (Novagen) and alkaline phosphatase substrate (Bio-Rad). As an additional control, the GST activity per unit protein of the GST fusion proteins used in this study was determined.
Cell Culture and Transfection.
HEK293 cells were maintained in minimal essential medium plus 10% bovine serum and penicillin/streptomycin in a 5% C02 37°C incubator. For co-immunoprecipitation assays, 2 μg of pcDNA3 Flag-β1-AR or pcDNA3 Flag-β2-AR with either 4 μg of pcDNA3 HA-SH3p4, pcDNA3 HA-SH3P4 ΔSH3 or 4 μg of pcDNA3 empty vector was cotransfected into 50–80% confluent 10-cm tissue culture plates by using a calcium-phosphate transfection kit (GIBCO/BRL). To establish stable HEK293 cell lines, either 4 μg of pcDNA3 HA-SH3p4, pcDNA3 HA-SH3p4 ΔSH3 or 4 μg of pcDNA3 empty vector was transfected into HEK293 cells and cells selected with 400 μg/ml G418 (GIBCO/BRL) in medium. After selection for 10–15 days, the colonies were pooled together. Overexpression of transfected proteins was assessed by Western blot analysis. Stable cells were cultured in medium containing 200 μg/ml G418.
Coimmunoprecipitation.
Twenty-four hours after transfection, HEK293 cells were incubated in serum-free medium overnight before treatment with 10 μM isoproterenol. Treated plates were transferred to ice, and cells were lysed with 1 ml of ice-cold digitonin buffer (20 mM triethanolamine, pH 8.0/300 mM NaCl/2 mM EDTA/20% glycerol/1% digitoinin and protease inhibitors). The lysate was solubilized by incubation at 4°C for 30 min and was clarified by centrifugation at 21,000 × g for 20 min. The concentration of soluble protein was determined with the BCA protein assay kit (Pierce). Equal amounts of protein was used for all subsequent immunoprecipitations. A portion (25 μl) of anti-Flag M2 affinity gel (Sigma) was incubated overnight at 4°C with ≈1 ml of cell lysate to precipitate Flag-β1-AR or Flag-β2-AR. After extensive washing with digitonin buffer, immunoprecipitated proteins were eluted from the beads with 2× SDS sample buffer, were resolved by SDS/PAGE, and were subjected to Western blot analysis.
Sequestration Assay and Whole-Cell cAMP Assay.
For sequestration and whole-cell cAMP assays, 0.5–1.0 μg of pcDNA1 HA-β1-AR or pcDNA1 HA-β2-AR was transfected into HEK293 cells in 10-cm plates to obtain a receptor density of ≈500 fmol/mg protein (determined by 125I-cyanopindolol binding assay). After transfection, cells were split into one collagen-treated 6-well dish for sequestration assays or two 12-well dishes for cyclase assays.
The sequestration assays were performed essentially as described (12). Ten micromolar isoproterenol was used to stimulate the cells for 30 min. Sequestration was defined as the percentage of cell surface receptors lost after agonist treatment.
Whole-cell cAMP was determined as described (19). In brief, cells in 12-well dishes were labeled with [3H]adenine in growth medium overnight. Dobutamine was used as a β1-AR agonist, and endogenous β2-AR-induced adenylyl cyclase activity was measured by using isoproterenol. Both 125I-cyanopindolol binding (19) and immunofluorescence flow cytometry was used to determine total and cell surface HA-β1-AR expression.
Results
Identification of the SH3p4/p8/p13 Protein Family as Binding Partners for the Proline-Rich Third Intracellular Loop of the β1-Adrenergic Receptor.
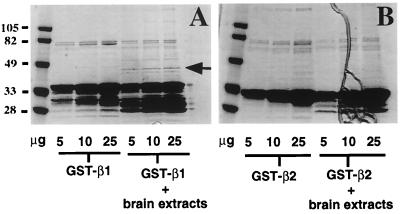
To search for potential binding partners for the third intracellular loop of the β1- and/or β2-AR, we used both a biochemical approach and the yeast two-hybrid system. In the biochemical approach, GST fusion proteins encompassing the third intracellular loop of the β1- and β2-AR were purified from bacteria, were conjugated to glutathione Sepharose 4B beads, and were incubated separately with bovine brain extracts. As shown in Fig. 1A, two proteins of Mr ≈40 and ≈50 were recovered from GST-β1-AR third intracellular loop fusion protein incubations with bovine brain extracts. The binding of these proteins to the β1-AR third intracellular loop appeared to be specific because the GST-β2-AR third intracellular loop fusion protein did not pull out these proteins under the same experimental conditions (Fig. 1B).
Figure 1.
Purification of GST-β1-AR third intracellular loop binding protein(s) from bovine brain extracts. A GST pull-down assay was performed, as described in Materials and Methods, by using the GST-β1-AR third intracellular loop (A) and GST-β2-AR third intracellular loop (B), respectively. For each incubation condition, 5, 10, and 25 μg of proteins was loaded on SDS/PAGE, stained with Coomassie blue, and dried. A 40-kDa protein, as indicated by an arrow, was specifically recovered after GST-β1-AR third intracellular loop incubation with bovine brain extracts.
We have initially focused on the 40-kDa protein. To identify this protein, the eluted material was concentrated, run on larger- scale SDS/PAGE, and stained with Coomassie blue. The 40-kDa peptide then was cut out and subjected to in-gel trypsin digestion and peptide sequencing. Three major peptide sequences were obtained (IPDEELRQALEK, QNFIDPLQNL, and DLREIQHHLK). blast searches of protein sequence databases revealed that all three fragments matched with sequences of the SH3p4/p8/p13 protein family. The first two peptide sequences are identical among all three members of this protein family. The R in the third peptide is specific to SH3p4. Thus, the 40-kDa protein we purified from bovine brain extract is SH3p4.
Concurrently, we also used the yeast two-hybrid system to search for potential binding partners for the β1-AR third intracellular loop. The β1-AR third intracellular loop was fused to the GAL4 DNA binding domain and was used as bait to screen a rat brain cDNA library by using the yeast strain PJ-69-4A. Approximately 13 million independent clones were analyzed, and nine exhibited moderate to strong growth on −HIS or −ADE media. Strikingly, eight of the nine clones encoded different versions of cDNAs for either SH3p4 or SH3p13. Activity of the reporter gene LacZ was measured to assess the relative affinities of the SH3p4 and SH3p13 clones with the β1-AR third intracellular loop in the yeast two-hybrid system. As shown in Table 1, constructs encoding SH3p4 induced the LacZ reporter gene between 3.09- and 6.24-fold over background whereas SH3p13 clones activated the reporter from 3.94- to 10.81-fold. The fact that no other SH3-containing protein was identified in the yeast two-hybrid system strongly suggests that the SH3p4/p8/p13 family of proteins may be specific binding partners for the β1-AR in vivo.
Table 1.
Description of clones identified by the yeast two-hybrid system
| Protein | Clone | Position | β-galactosidase activity, mean ± SE |
|---|---|---|---|
| SH3p4 | 2 | aa 24-end | 6.24 ± 0.49 |
| 100 | aa 104-end | 3.09 ± 0.31 | |
| SH3p13 | 6 | aa 57-end | 10.81 ± 1.23 |
| 1 | FL (+32 aa) | 4.26 ± 0.21 | |
| 3, 55 | FL (+36 aa) | 3.94 ± 0.2 | |
| 9 | FL (+39 aa) | 4.21 ± 1.47 | |
| 35 | FL (+37 aa) | ND |
The identity of the clone and the amino acids contained in each isolated clone are shown. FL indicates that the entire open reading frame including some of the 5′ untranslated region is contained in the clone. The β-galactosidase activity was determined by liquid assay using log phase cultures. The numbers represent fold stimulation of activity as determined by co-transformation of the indicated plasmid with the β1-AR third intracellular loop into the yeast strain PJ-69-4A in comparison with transformation of the indicated plasmid with empty vector. Values of β-galactosidase activity are mean ± SE of three independent experiments. ND, not determined.
In Vitro, SH3p4 Specifically Interacts with the Proline-Rich Third Intracellular Loop of the β1-AR.
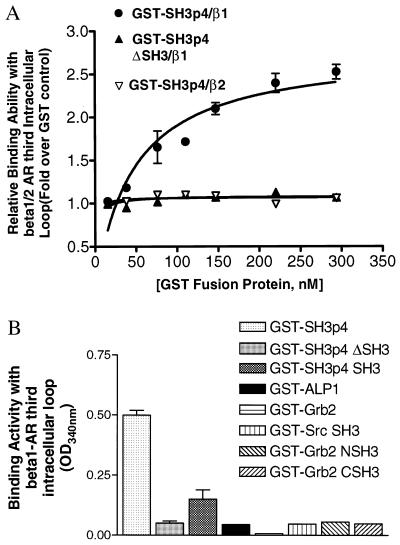
To further characterize the interaction of SH3p4 with the β1-AR third intracellular loop, we performed ELISA assays. His-S-tagged β1- and β2-AR third intracellular loop fusion proteins were purified from E. coli and were used to coat 96-well dishes. The coated wells then were overlaid with GST fusion proteins encoding GST alone as a negative control, or full length SH3p4. The amount of bound GST fusion protein was detected by measuring the enzymatic activity of the GST moiety by using a GST substrate. Absorbance at 340 nm then was recorded to obtain both the reaction kinetics and endpoint readings. As shown in Fig. 2A, the dose–response curve of GST-SH3p4 binding with His-S-tagged β1 third intracellular loop revealed a saturable, high-affinity interaction between these two polypeptides (Kd = 47 ± 9 nM). In contrast, there was no detectable interaction between His-S-tagged β2-AR third intracellular loop fusion protein and GST-SH3p4. SH3p4 contains a C-terminal SH3 domain that may represent the site of interaction with the proline-rich β1-AR third intracellular loop. Therefore, we also examined an SH3 domain-deleted SH3p4 mutant (i.e., SH3p4 ΔSH3) in the ELISA assay. As shown in Fig. 2A, deletion of the SH3 domain completely abolished the association of SH3p4 with the β1-AR third intracellular loop. Moreover, the SH3 domain of SH3p4 alone was also capable of binding to the β1-AR third intracellular loop, although with lower affinity than full length SH3p4 (Fig. 2B). The relatively weak binding of the SH3p4 SH3 domain alone to the β1-AR third intracellular loop is consistent with previous findings that isolated SH3 domains bind with lower affinity to their cognate proline-rich peptide ligands than do full-length proteins (20). These data suggest that regions other than SH3 domains are also important for the interactions between SH3 domains and proline-rich motif-containing proteins and dictate the specificity of such interactions.
Figure 2.
(A) GST-SH3p4 binds to His-S-β1-AR third intracellular loop in a saturable and SH3-dependent manner in vitro. An ELISA assay was carried out as described in detail in Materials and Methods. In brief, His-S-β1 or β2-AR third intracellular loop was coated on 96-well plates and was subsequently overlaid with GST (control), GST-SH3p4 (●,▿), or SH3p4 ΔSH3 (▴) for 1 hr at room temperature. After washing with PBS, GST substrate was added to each well, and the plates were read at 340 nm to record reaction kinetics (ΔOD/time). The binding activity of GST fusion proteins with His-S-β1 or β2-AR third intracellular loop is presented as the fold increase in enzymatic activity over a GST control. The results shown were obtained from three independent experiments performed in duplicate (mean ± SE). Note, for GST-SH3p4 ΔSH3 binding with His-β1-AR third intracellular loop and GST-SH3p4 binding with His-β2-AR third intracellular loop, the error bars are smaller than the symbols. (B) The β1-AR third intracellular loop specifically binds to SH3p4 but not to other SH3-containing proteins. The ELISA assay was performed essentially as described in A. The binding activity shown is the endpoint absorbance at 340 nm (subtracted by GST control absorbance) after incubation with GST substrate for 1 hr at room temperature. For GST-SH3p4, GST-SH3p4ΔSH3, and GST-SH3p4 SH3, the results shown were obtained from three independent experiments (mean ± SE). For all other GST fusion proteins, the results shown are the average of two independent experiments.
It has been well established that many cellular signaling events are driven by interaction between proline-rich motifs and SH3 domains. Given the great diversity of the signaling events occurring within cells, the specificity of such interactions must be intricately controlled. Therefore, we decided to test the specificity of the interaction between the SH3 domain of SH3p4 and the β1-AR third intracellular loop. We examined several SH3 domain-containing proteins for their ability to interact with the β1-AR third intracellular loop, including the well studied adapter protein Grb2, the tyrosine kinase Src, and the synaptic vesicle trafficking protein amphiphysin 2 (also referred to as amphiphysin-like-protein 1). As shown in Fig. 2B, neither GST-Grb2 nor GST- ALP1 fusion proteins show detectable binding to the β1-AR third intracellular loop. Similarly, GST fusion proteins containing the SH3 domains from Src and Grb2 exhibit no binding to the β1-AR third intracellular loop. Given the specific nature of the association of the β1-AR third intracellular loop with SH3p4, this interaction might have potential physiological relevance.
SH3p4 Associates with the β1-Adrenergic Receptors in HEK293 Cells.
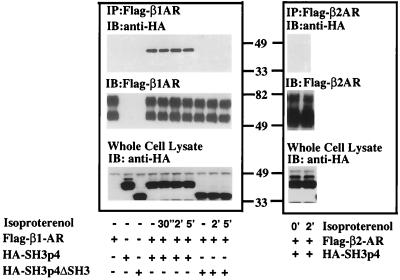
To verify that the in vitro interaction between SH3p4 and the β1-AR third intracellular loop reflects the ability of these proteins to interact in the cells, we overexpressed both proteins in HEK293 cells and examined their association by co-immunoprecipitation. HEK293 cells were transiently transfected with pcDNA3 Flag-β1-AR and pcDNA3 HA-SH3p4. After isoproterenol stimulation for various periods of time, cells were lysed, and Flag-β1-ARs from cell lysates were immunoprecipitated with anti-Flag affinity gel. The immunoprecipitates then were analyzed by SDS/PAGE and Western blotting. The blots were probed with anti-HA antibody to detect HA-SH3p4. Fig. 3 shows that SH3p4 interacts specifically with the β1-AR in intact cells. However, no obvious effect of receptor occupancy on the association of β1-ARs and SH3p4 was observed under our experimental conditions. Consistent with the in vitro data, the SH3 domain deletion mutant of SH3p4, although expressed in cells at similar levels to the wild-type protein, does not co-immunoprecipitate with the β1-AR. These data support the conclusion that the interaction between the β1-AR and SH3p4 is mediated by the SH3 domain of SH3p4 and, presumably, the proline-rich region of the β1 third intracellular loop. GST pull-down assays and in vitro ELISA assays demonstrate that the β2-AR third intracellular loop does not bind to SH3p4 (Figs. 1 and 2A). Consistent with these observations, no immunoreactive HA-SH3p4 was detected in the Flag-β2-AR immunoprecipitates (Fig. 3 Right). These data further support the contention that the SH3p4 protein may serve a specific role in β1-AR but not β2-AR signaling.
Figure 3.
Coimmunoprecipitation of HA-SH3p4 and Flag-β1-adrenergic receptor from HEK293 cells. pcDNA3 Flag-β1-AR (Left) or pcDNA3 Flag-β2-AR (Right) were transiently transfected into HEK293 cells in the absence or presence of pcDNA3 HA-SH3p4 or pcDNA3 HA-SH3p4 ΔSH3. Nonstimulated cells and cells stimulated with 10 μM isoproterenol for various periods of time were lysed in digitonin buffer. Flag-β1-AR was immunoprecipitated with M2 anti-Flag affinity gel from samples containing equal amounts of protein. Immunoprecipitates were subjected to SDS/PAGE and Western blot analysis. The blot was initially probed with anti-HA rabbit polyclonal antibody to detect HA-SH3p4 and HA-SH3p4 ΔSH3. The blot was subsequently stripped and reprobed with biotinylated-M2 anti-Flag antibody and streptavidin/horseradish peroxidase conjugates to detect Flag-βAR. Overexpression of HA-SH3p4 or HA-SH3p4ΔSH3 in whole cell lysate also was monitored. The experiment was repeated at least three times with similar results. The diffuse doublet of both Flag-β1-AR and Flag-β2-AR in the middle panel likely represents differentially glycosylated forms of these receptors.
Overexpression of SH3p4 Promotes Agonist-Induced Internalization and Decreases the Gs-Coupling Efficacy of the β1-Adrenergic Receptors in HEK293 Cells.
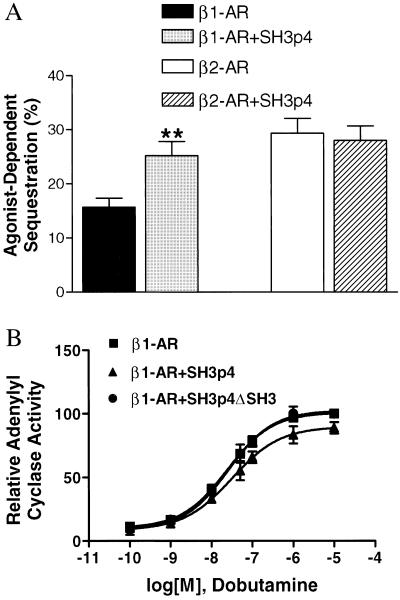
On agonist stimulation, β1- and β2-adrenergic receptors, like other GPCRs, internalize from the cell surface to cytoplasmic vesicles via a classical clathrin-mediated endocytosis pathway (9). Although the physiological significance of this receptor internalization is not fully understood, internalization of the β2-AR is critical for resensitization (21). Compared with the β2-AR, the β1-AR undergoes sequestration to a much lower extent. For example, it was previously reported that agonist stimulation led to >30% β2-AR internalization in Chinese hamster fibroblast cells whereas sequestration of the β1-AR was negligible (6). More detailed characterization revealed that the proline-rich region in the β1-AR third intracellular loop appears to be responsible for its distinctive pattern of sequestration (5). Therefore, we set out to determine whether SH3p4 plays a specific role in β1-AR internalization. HEK293 cells were transiently transfected with either pcDNA1 HA-β1-AR or pcDNA1 HA-β2-AR alone or together with pcDNA3 Flag-SH3p4. As shown in Fig. 4A, isoproterenol stimulation induced ≈15 and 30% sequestration for the β1-AR and the β2-AR, respectively. Notably, overexpression of SH3p4 increased agonist-induced β1-AR internalization to ≈25% but had no effect on the internalization of the β2-AR.
Figure 4.
(A) Internalization of HA-β1-AR in HEK293 cells is potentiated on overexpression of SH3p4. HEK293 cells were transiently transfected with 0.5–1 μg of pCDNA1 HA-β1-AR or HA-β2-AR to obtain a receptor expression level of 500 fmol/mg protein in the presence or absence of pcDNA3 Flag-SH3p4. Cells were stimulated with 10 μM isoproterenol for 30 min. Results are presented as the percentage loss of cell surface receptor. For HA-β1-AR, n = 9, P < 0.01(∗∗) for comparison of HA-β1-AR internalization in the presence versus absence of SH3p4 according to Student’s t test; for HA-β2-AR, n = 5. (B) Overexpression of SH3p4 in HEK293 cells attenuates HA-β1-AR stimulated adenylyl cyclase activity. Stable HEK293 cells, overexpressing pcDNA3 HA-SH3p4, pcDNA3 HA-SH3p4 ΔSH3, or pcDNA3 empty vector, were transfected with pCDNA1 HA-β1-AR to achieve a receptor concentration of ≈500 fmol/mg protein. HA-β1-AR was stimulated for 10 min with the indicated concentrations of dobutamine. Whole cell cAMP accumulation was measured as the percentage conversion of [3H]adenine to [3H]cAMP. cAMP accumulation on 50 μM forskolin stimulation was used to normalize dobutamine-induced cAMP production. The cyclase activity observed in the presence of HA-β1-AR overexpression in the pcDNA3-stable HEK293 cells at 10 μM dobutamine is set at 100. The other data points are shown as percentage of this maximal cyclase activity. The means ± SE from four independent experiments are shown.
In addition to differences in their ability to undergo agonist-induced receptor sequestration, the β1-AR and β2-AR also exhibit differential coupling to Gs. It has been demonstrated that the β1-AR couples to Gs less well than does the β2-AR (5, 13, 22). The EC50 of isoproterenol for activating adenylyl cyclase through the β1-AR is ≈5-fold higher than through the β2-AR (13). Once again, the proline-rich region of the β1-AR third intracellular loop has been suggested to contribute to this difference (13). To explore the possible role of SH3p4 in β1-AR signaling, we examined whole-cell cAMP accumulation mediated by the β1-AR in the presence or absence of SH3p4 overexpression. HEK293 cells stably expressing HA-SH3p4, HA-SH3p4ΔSH3 or pcDNA3 were established, and pcDNA1 HA-β1-AR transiently transfected into these cells to test their Gs coupling efficacy. The β1-AR-specific agonist dobutamine was used to stimulate the cells. Dose-response curves are presented in Fig. 4B. Analysis of the curves reveals that overexpression of SH3p4 causes the EC50 to shift from 22 ± 6 nM to 34 ± 6 nM (P < 0.01). In contrast, overexpression of SH3p4ΔSH3 did not affect the β1-AR-induced adenylyl cyclase activity. Dobutamine did not stimulate adenylyl cyclase through endogenous β2-ARs in HEK293 cells, indicating specificity of dobutamine for the β1-ARs (data not shown). Moreover, when isoproterenol was used to stimulate endogenous β2-AR in either pcDNA3 empty vector or pcDNA3 SH3p4 stably transfected cells, no difference in adenylyl cyclase activity was observed (data not shown). These results provide further support for the hypothesis that SH3p4 specifically affects β1-AR signaling.
Discussion
Here we report the identification of the members of SH3p4/p8/p13 protein family as binding partners for the β1-AR. The SH3p4/p8/p13 protein family, also referred to as endophilin 1/2/3, was first discovered as novel SH3-containing proteins by screening a mouse embryonic cDNA expression library using a proline-rich peptide ligand for the SH3 domain (20). With respect to tissue distribution, SH3p4 is restricted to the brain and SH3p8 is present in brain and testis whereas SH3p13 shows more ubiquitous expression (23, 24). Recently, the SH3p4/p8/p13 protein family has been shown to form complexes with several proteins, such as amphiphysin, synaptojanin, and dynamin, all of which are implicated in presynaptic vesicle trafficking in nerve terminals (24–26). Complex formation depends on the interaction between proline-rich motifs and SH3 domains. It also has been reported recently that human SH3p8 is associated with the Huntingtin protein in an SH3-dependent manner and that this association enhances the formation of polyglutamine-containing protein aggregates (27). Here we report yet another aspect of the potential physiological significance of this protein family by demonstrating a specific interaction between SH3p4 and a GPCR, the β1-AR. Furthermore, β1-AR/SH3p4 complex formation is shown to affect both β1-AR internalization and signaling.
β1-AR and β2-AR are closely related β-adrenergic receptor subtypes, sharing ≈54% overall sequence homology (1). However, these two βAR subtypes are biologically distinguishable in a number of respects. For instance, they show distinctive agonist and antagonist binding affinities (13). Although both subtypes couple to Gs and activate adenylyl cyclase on agonist stimulation, the Gs coupling efficacy of the β1-AR is lower than that of the β2-AR (5, 22). β2-ARs are also able to couple to Gi. Moreover, β1 and β2-AR display distinctive tissue distribution patterns and serve different physiological roles. The β1-AR is the predominant β adrenergic receptor in heart whereas the β2-AR predominates in liver, lung, and smooth muscle (2). Both transgenic (28–30) and gene “knock-out” mouse models (31, 32) have been established for the β1- and β2-ARs, resulting in different physiological and developmental outcomes.
It has been noted that the β1-AR third intracellular loop contains a unique 24-aa proline-rich segment, which has no counterpart in the β2-AR (13). Previous studies have suggested that this sequence disparity between the β1- and β2-ARs might function as one of the molecular determinants for their distinguishable signaling features (13). Our in vitro assay shows that SH3p4 interacts with the β1-AR third intracellular loop in a saturable and SH3-dependent manner. In intact cells, SH3p4 coimmunoprecipitates with the β1-AR, and the SH3 domain of SH3p4 is required for this association. Receptor occupancy does not appear to affect β1AR/SH3p4 complex formation. Agonist stimulation may, however, alter SH3p4 conformation or lead to the convalent modification of this protein, processes that may modulate downstream signaling events. There are precedents for such a possibility. For example, in the case of the AMPA receptor (α-amino-3-hydroxy-5-methyl-4-isoxazole propionate), a ligand-gated cation channel, the extent of physical association of the AMPA receptors with Lyn, an SH3-containing tyrosine kinase, is not altered after AMPA stimulation. Instead, agonist binding causes a significant increase in the tyrosine kinase activity of Lyn and subsequently leads to mitogen-activated protein kinase activation (33).
Interactions driven by SH3 domains and proline-rich motifs are among the most common events found in many cellular processes, such as cytoskeleton organization, subcellular localization of signaling molecules, and vesicle endocytosis (34). However, the role of SH3 domain-mediated interactions in β-adrenergic receptor and other GPCR signaling has not been extensively studied. Proline-rich regions have been found in intracellular domains of a number of GPCRs, including β1-AR, β3-AR, α2A-AR, and dopamine D4 receptor, and a few studies have been attempted to understand their potential roles in GPCR signaling. In vitro, the SH2-SH3 adapter proteins, Grb2 and Nck, have been shown to interact with the proline-rich third intracellular loop of dopamine D4 receptor (14). Furthermore, removal of the putative SH3 binding sites from the third intracellular loop resulted in constitutive internalization of the mutant D4 receptors and deficient activation of second messengers (14).
Our studies demonstrate that overexpression of SH3p4 increases agonist-induced sequestration and modestly decreases the Gs coupling efficacy of the β1-AR in HEK293 cells. What is the role of SH3p4 in β1-AR internalization and signaling? It has been well established, largely by using the β2-AR as a model system, that GRK-catalyzed phosphorylation followed by β-arrestin binding leads to endocytosis of the β2-AR(3). β-arrestin is thought to target the receptors to the clathrin-coated vesicles. It also has been shown that the β1-AR can be phosphorylated by GRKs (19). However, it was previously proposed that the β1-AR might be a poorer substrate for GRKs and β-arrestins than the β2-AR (35). It is possible that endocytosis of the β1-AR may use different protein components than the β2-AR. It has been reported recently that SH3-containing proteins, including SH3p4, function sequentially in endocytic clathrin-coated vesicle formation (36). Thus, SH3p4 may act as an adapter protein by directing the β1-AR to the endocytic machinery. In support of this hypothesis is the evidence that SH3p4 can form homodimers in vivo (N.R. and P.D., unpublished data). The dimerization of SH3p4 may facilitate simultaneous binding of SH3p4 with the β1-AR and its downstream binding partner(s). Although the downstream binding partner(s) for SH3p4 in β1-AR endocytosis remains to be identified, in vitro studies have indicated that SH3p4 is capable of interacting with the carboxy-terminal proline/arginine-rich domain (PRD) of the GTPase dynamin, which is recruited to the necks of the clathrin-coated pits and regulates the fission reaction (24, 26).
In addition to its effect on β1AR internalization, SH3p4 binding to the third intracellular loop alters the coupling of the β1-AR to the Gs protein. It was shown previously that the N- and C- terminal ends of the β-adrenergic receptor third intracellular loops dictate their G protein coupling efficacy and specificity (7, 8). It is conceivable that binding of SH3p4 to the middle segment of the β1-AR third intracellular loop alters stimulation of the Gs protein, either by physically interfering with receptor Gs protein coupling or by altering receptor conformation, thus lowering agonist binding affinity. In either case, rightward and downward shift of the dose-response curve on overexpression of SH3p4 is observed.
Previous studies have implicated the 24-aa proline-rich region of the β1-AR third intracellular loop as contributing to differences in the internalization and signaling properties of β1-ARs vs. β2-ARs in Chinese hamster fibroblast cells (13). Our studies further support the hypothesis that this proline-rich region may serve as a molecular determinant for β1-AR functionality by interacting with SH3 domain-containing protein(s). The molecular consequences of the β1-AR/SH3p4 interaction remain to be further elucidated. However, the findings reported here underscore the likelihood that interactions between proline-rich motifs in GPCRs and SH3-containing proteins are likely to play important roles in previously unappreciated aspects of GPCR signaling.
Acknowledgments
We thank Dr. Richard Cook for protein sequencing, Humphrey Kendall for DNA subcloning, Grace Irons for tissue culture assistance, Millie McAdams and Judy Phelps for DNA sequencing, and Donna Addison and Mary Holben for excellent secretarial assistance. This work was supported in part by a grant from the National Institutes of Health, Grant HL16037 (to R.J.L.). R.J.L. is an Investigator of the Howard Hughes Medical Institute.
Abbreviations
- AR
adrenergic receptor
- GPCR
G-protein coupled receptor
- GRK
G protein-coupled receptor kinase
- HA
hemagglutinin
- GST
glutathione S-transferase
- HEK
human embryonic kidney
- SH
Src homology
Note Added in Proof
Endophilin 1 (i.e., SH3p4) has recently been demonstrated to exhibit lysophosphatidic acid acyl transferase activity (37) and mediate synaptic vesicle invagination (38).
References
- 1.Frielle T, Kobilka B, Lefkowitz R J, Caron M G. Trends Neurosci. 1988;11:321–324. doi: 10.1016/0166-2236(88)90095-1. [DOI] [PubMed] [Google Scholar]
- 2.Bouvier M, Rousseau G. Adv Pharmacol. 1998;42:433–438. doi: 10.1016/s1054-3589(08)60781-4. [DOI] [PubMed] [Google Scholar]
- 3.Lefkowitz R J. J Biol Chem. 1998;273:18677–18680. doi: 10.1074/jbc.273.30.18677. [DOI] [PubMed] [Google Scholar]
- 4.Liggett S B, Freedman N J, Schwinn D A, Lefkowitz R J. Proc Natl Acad Sci USA. 1993;90:3665–3669. doi: 10.1073/pnas.90.8.3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Green S A, Holt B D, Liggett S B. Mol Pharmacol. 1992;41:889–893. [PubMed] [Google Scholar]
- 6.Suzuki T, Nguyen C T, Nantel F, Bonin H, Valiquette M, Frielle T, Bouvier M. Mol Pharmacol. 1992;41:542–548. [PubMed] [Google Scholar]
- 7.Cheung A H, Huang R R, Graziano M P, Strader C D. FEBS Lett. 1991;279:277–280. doi: 10.1016/0014-5793(91)80167-2. [DOI] [PubMed] [Google Scholar]
- 8.Rousseau G, Nantel F, Bouvier M. Mol Pharmacol. 1996;49:752–760. [PubMed] [Google Scholar]
- 9.Ferguson S S, Zhang J, Barak L S, Caron M G. Adv Pharmacol. 1998;42:420–424. doi: 10.1016/s1054-3589(08)60778-4. [DOI] [PubMed] [Google Scholar]
- 10.Pitcher J A, Freedman N J, Lefkowitz R J. Annu Rev Biochem. 1998;67:653–692. doi: 10.1146/annurev.biochem.67.1.653. [DOI] [PubMed] [Google Scholar]
- 11.Pitcher J A, Inglese J, Higgins J B, Arriza J L, Casey P J, Kim C, Benovic J L, Kwatra M M, Caron M G, Lefkowitz R J. Science. 1992;257:1264–1267. doi: 10.1126/science.1325672. [DOI] [PubMed] [Google Scholar]
- 12.Ferguson S S, Downey W E, III, Colapietro A M, Barak L S, Menard L, Caron M G. Science. 1996;271:363–366. doi: 10.1126/science.271.5247.363. [DOI] [PubMed] [Google Scholar]
- 13.Green S A, Liggett S B. J Biol Chem. 1994;269:26215–26219. [PubMed] [Google Scholar]
- 14.Oldenhof J, Vickery R, Anafi M, Oak J, Ray A, Schoots O, Pawson T, von Zastrow M, Van Tol H H. Biochemistry. 1998;37:15726–15736. doi: 10.1021/bi981634+. [DOI] [PubMed] [Google Scholar]
- 15.Hall R A, Premont R T, Chow C W, Blitzer J T, Pitcher J A, Claing A, Stoffel R H, Barak L S, Shenolikar S, Weinman E J, et al. Nature (London) 1998;392:626–630. doi: 10.1038/33458. [DOI] [PubMed] [Google Scholar]
- 16.Luttrell L M, Ferguson S S, Daaka Y, Miller W E, Maudsley S, Della Rocca G J, Lin F, Kawakatsu H, Owada K, Luttrell D K, et al. Science. 1999;283:655–661. doi: 10.1126/science.283.5402.655. [DOI] [PubMed] [Google Scholar]
- 17.Gietz R D, Schiestl R H. Methods Mol Cell Biol. 1995;5:255–269. [Google Scholar]
- 18.James P, Halladay J, Craig E A. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Freedman N J, Liggett S B, Drachman D E, Pei G, Caron M G, Lefkowitz R J. J Biol Chem. 1995;270:17953–17961. doi: 10.1074/jbc.270.30.17953. [DOI] [PubMed] [Google Scholar]
- 20.Sparks A B, Hoffman N G, McConnell S J, Fowlkes D M, Kay B K. Nat Biotechnol. 1996;14:741–744. doi: 10.1038/nbt0696-741. [DOI] [PubMed] [Google Scholar]
- 21.Krueger K M, Daaka Y, Pitcher J A, Lefkowitz R J. J Biol Chem. 1997;272:5–8. doi: 10.1074/jbc.272.1.5. [DOI] [PubMed] [Google Scholar]
- 22.Levy F O, Zhu X, Kaumann A J, Birnbaumer L. Proc Natl Acad Sci USA. 1993;90:10798–10802. doi: 10.1073/pnas.90.22.10798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Giachino C, Lantelme E, Lanzetti L, Saccone S, Bella Valle G, Migone N. Genomics. 1997;41:427–434. doi: 10.1006/geno.1997.4645. [DOI] [PubMed] [Google Scholar]
- 24.Ringstad N, Nemoto Y, De Camilli P. Proc Natl Acad Sci USA. 1997;94:8569–8574. doi: 10.1073/pnas.94.16.8569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Micheva K D, Kay B K, McPherson P S. J Biol Chem. 1997;272:27239–27245. doi: 10.1074/jbc.272.43.27239. [DOI] [PubMed] [Google Scholar]
- 26.de Heuvel E, Bell A W, Ramjaun A R, Wong K, Sossin W S, McPherson P S. J Biol Chem. 1997;272:8710–8716. doi: 10.1074/jbc.272.13.8710. [DOI] [PubMed] [Google Scholar]
- 27.Sittler A, Walter S, Wedemeyer N, Hasenbank R, Scherzinger E, Eickhoff H, Bates G P, Lehrach H, Wanker E E. Mol Cell. 1998;2:427–436. doi: 10.1016/s1097-2765(00)80142-2. [DOI] [PubMed] [Google Scholar]
- 28.Milano C A, Allen L F, Rockman H A, Dolber P C, McMinn T R, Chien K R, Johnson T D, Bond R A, Lefkowitz R J. Science. 1994;264:582–586. doi: 10.1126/science.8160017. [DOI] [PubMed] [Google Scholar]
- 29.Zolk O, Kilter H, Flesch M, Mansier P, Swynghedauw B, Schnabel P, Bohm M. Biochem Biophys Res Commun. 1998;248:801–805. doi: 10.1006/bbrc.1998.9030. [DOI] [PubMed] [Google Scholar]
- 30.Bertin B, Mansier P, Makeh I, Briand P, Rostene W, Swynghedauw B, Strosberg A D. Cardiovasc Res. 1993;27:1606–1612. doi: 10.1093/cvr/27.9.1606. [DOI] [PubMed] [Google Scholar]
- 31.Chruscinski A J, Rohrer D K, Schauble E, Desai K H, Bernstein D, Kobilka B K. J Biol Chem. 1999;274:16694–16700. doi: 10.1074/jbc.274.24.16694. [DOI] [PubMed] [Google Scholar]
- 32.Rohrer D K, Desai K H, Jasper J R, Stevens M E, Regula D P, Jr, Barsh G S, Bernstein D, Kobilka B K. Proc Natl Acad Sci USA. 1996;93:7375–7380. doi: 10.1073/pnas.93.14.7375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hayashi T, Umemori H, Mishina M, Yamamoto T. Nature (London) 1999;397:72–76. doi: 10.1038/16269. [DOI] [PubMed] [Google Scholar]
- 34.Pawson T. Nature (London) 1995;373:573–580. doi: 10.1038/373573a0. [DOI] [PubMed] [Google Scholar]
- 35.Zhou X M, Pak M, Wang Z, Fishman P H. Cell Signalling. 1995;7:207–217. doi: 10.1016/0898-6568(94)00091-o. [DOI] [PubMed] [Google Scholar]
- 36.Simpson F, Hussain N K, Qualmann B, Kelly R B, Kay B K, McPherson P S, Schmid S L. Nat Cell Biol. 1999;1:119–124. doi: 10.1038/10091. [DOI] [PubMed] [Google Scholar]
- 37.Schmidt A, Wolde M, Thiele C, Fest W, Kratzin H, Podtelejnikov A V, Witke W, Huttner W B, Söling H. Nature (London) 1999;401:133–141. doi: 10.1038/43613. [DOI] [PubMed] [Google Scholar]
- 38.Ringstad N, Gad H, Löw P, DiPaolo G, Brodin L, Shupliakov O, DeCamilli P. Neuron. 1999;24:143–154. doi: 10.1016/s0896-6273(00)80828-4. [DOI] [PubMed] [Google Scholar]