Abstract
The repair of chromosomal double-strand breaks (DSBs) is necessary for genomic integrity in all organisms. Genetic consequences of misrepair include chromosomal loss, deletion, and duplication resulting in loss of heterozygosity (LOH), a common finding in human solid tumors. Although work with radiation-sensitive cell lines suggests that mammalian cells primarily rejoin DSBs by nonhomologous mechanisms, alternative mechanisms that are implicated in chromosomal LOH, such as allelic recombination, may also occur. We have examined chromosomal DSB repair between homologs in a gene targeted mammalian cell line at the retinoblastoma (Rb) locus. We have found that allelic recombinational repair occurs in mammalian cells and is increased at least two orders of magnitude by the induction of a chromosomal DSB. One consequence of allelic recombination is LOH at the Rb locus. Some of the repair events also resulted in other types of genetic instability, including deletions and duplications. We speculate that mammalian cells may have developed efficient nonhomologous DSB repair processes to bypass allelic recombination and the potential for reduction to homozygosity.
Multiple genetic alterations such as point mutations, chromosomal translocations, and loss of heterozygosity (LOH) contribute to cellular carcinogenesis. LOH leads to tumorigenesis when defective tumor suppressor loci become homozygous (1). Retinoblastoma is the prototype tumor for which mechanisms leading to LOH have been examined (1). These mechanisms include chromosome loss, chromosome loss followed by duplication, and somatic recombination between alleles. Although the mechanism responsible for a particular LOH event may be difficult to determine due to the lack of appropriate markers, somatic recombination may contribute to as many as 75% of the LOH events at the retinoblastoma (Rb) locus (1). Spontaneous allelic recombination in mammalian cell lines has been shown to be an extremely rare event (2–4). However, LOH may be an expected outcome of recombinational repair of chromosomal double-strand breaks (DSBs) by gene conversion mechanisms, with a reduction to homozygosity initiated from the site of the DSB. Cells having undergone LOH by allelic recombination may then have a proliferative advantage when growth control genes are affected.
DSBs can be caused by ionizing radiation or other DNA damaging agents or by normal DNA metabolic processes. In mitotically growing yeast, DSBs have been demonstrated to induce recombination between alleles (5). However, repair of DSBs in mammalian cells has been thought to be fundamentally different from DSB repair in yeast in terms of the types of repair mechanisms that predominate (6). Ionizing-radiation-sensitive yeast mutants exhibit defects in homologous recombination, demonstrating a key role for recombinational repair of DSBs in yeast (7). By contrast, radiosensitive mammalian cell mutants that are defective in DSB repair demonstrate impaired nonhomologous rejoining of broken ends (6). Impairment of nonhomologous recombination in mammalian cells has been measured either by V(D)J recombination assays (8), because this site-specific recombination process involves rejoining of DSBs (9), or by endonuclease-induced chromosomal DSB repair assays (10). Where examined, these cell mutants are proficient in the repair of chromosomal DSBs by homologous recombination (10).
Recently, a system to experimentally introduce defined DSBs into the genome of mammalian cells has been developed, allowing an investigation into the role that homology plays in DSB repair events. In this system, a rare-cutting site-specific endonuclease expressed in mammalian cells is used to cleave chromosomal DNA in vivo at its recognition site (11). With this system, DSBs in the genome of mammalian cells lines have been found to stimulate homologous recombination as much as three orders of magnitude when homology is provided from exogenous DNA in gene targeting experiments or as a tandemly repeated chromosomal sequence (10, 12–14). These results with exogenous and intrachromosomal DNA suggest that recombination between homologous chromosomes, though rarely observed spontaneously, may also be induced by a DSB in one of the homologs. By using marked Rb alleles in embryonic stem (ES) cells, we now test whether allelic recombination is an outcome of chromosomal DSB repair and whether allelic recombinational repair results in a reduction to homozygosity at the site of repair.
MATERIALS AND METHODS
DNA Constructions.
Rb alleles were gene targeted by modifying previously described gene targeting vectors (15). The mutant neo genes, Sneo and Pneo, were incorporated into the Rb pgkhyg and pgkhprt targeting vectors, respectively. In these targeting vectors, the mouse pgk1 (phosphoglycerate kinase) promoter drives expression of the hygromycin-resistance gene (pgkhyg) and the hypoxanthine phosphoribosyltransferase gene (pgkhprt). Sneo has an insertion of an oligonucleotide, which is composed of the 18-bp I-SceI site and the 4-bp NcoI overhangs, into the NcoI site in the 3′ end of the pMC1neo gene, as described (12). Pneo has an insertion of an oligonucleotide containing the PacI site in the 5′ end of the pMC1neo gene. Pneo was constructed by blunting the overhangs of an EagI site and ligating the linker 5′-CCTTAATTAAGG-3′. The EagI and NcoI sites are 526 bp apart. To construct the Rb+ K and L alleles, the mutant neo genes and selectable markers were cloned into the NheI site of intron 18 of the Rb locus by the addition of NheI linkers to the marker genes. To construct the Rb− M and N alleles, the mutant neo genes and selectable markers were cloned into the BglII/EcoRV sites of the Rb locus, deleting exon 19 and a portion of intron 19. The targeting constructs contain approximately 10 kb of Rb homology and were cleaved away from the plasmid backbone sequences prior to electroporation. As a mock DNA control in subsequent transfections, a plasmid containing pgklacZ cloned into pim-1 locus DNA (unpublished results) was used.
Cell Transfections and Southern Blot Analysis.
Hprt− ES cell line E14TG2a (16) was grown in medium supplemented with LIF at 105 units/ml (GIBCO/Life Technologies). For gene targeting experiments, 0.8 ml of cells at 2 × 107 cells per ml was electroporated at 800 V and 3 μF with 50 μg of the linearized gel-purified Rb targeting fragment. Selection medium was added 24 h after transfection, containing either hygromycin (150 μg/ml) or HAT (0.1 mM hypoxanthine/0.8 μM aminopterin/20 μM thymidine), as appropriate. Targeted clones were identified by screening Southern blots of genomic DNA cleaved with either EcoRI or PstI and hybridizing with probe A (15).
Spontaneous recombinants were scored by plating ES cells on multiple plates in nonselective medium for 24 h and then selecting cells on all but one of the plates in G418 at 200 μg/ml (GIBCO) for 10 days. Cells on the remaining plate were trypsinized and counted to determine the number of cells at the time selective medium was added. Recombination frequencies were calculated by dividing the number of G418-resistant (G418R) colonies obtained at 10 days by the total cell number present at the time selective medium was added.
To obtain colonies with DSB-induced events, cell lines with marked Rb alleles (i.e., K, L, KL, M, and MN) were electroporated at 250 V and 960 μF with 50 μg of the I-SceI expression vector, pPGK3xnlsI-SceI (G. Donoho, M.J., and P. Berg, unpublished results) or mock DNA. One day after electroporation, cells on one plate were trypsinized and counted to determine the number of cells surviving electroporation. G418 was added to the remaining plates at 200 μg/ml. After 10 days of selection, G418R colonies were counted and individually expanded. Southern blots of genomic DNA from the neo+ clones were probed with the EagI–NcoI neo gene probe.
RESULTS
Design of Allelic Recombination Substrates.
To examine recombination between homologs, gene targeting constructs were designed to differentially mark the two retinoblastoma (Rb) alleles in mouse ES cells (Fig. 1). The targeting constructs contained either of two defective neomycin phosphotransferase (neo) genes that would serve as selectable recombination substrates. The defective neo genes, termed Sneo and Pneo, differ in the type and position of the mutation. The Sneo gene is mutated near its 3′ end by the insertion of the 18-bp I-SceI cleavage site. The I-SceI site was inserted into the 4-bp overhangs of an NcoI site, creating a 4-bp duplication flanking the I-SceI site. The site can be cleaved in vivo on expression of the I-SceI endonuclease (17). Pneo, the second neo gene, is mutated near its 5′ end by the insertion of a 12-bp oligonucleotide containing a PacI restriction site.
Figure 1.
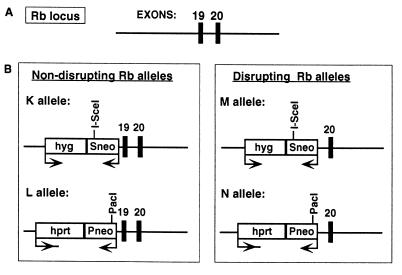
Gene-targeted region of the Rb locus in ES cells containing recombination substrates. (A) The Rb locus at exons 19 and 20. (B) Gene-targeted Rb alleles containing substrates to detect DSB-induced allelic recombination. Alleles K and L are nondisrupting Rb alleles that have the recombination substrates gene targeted to intron 18. Alleles M and N are disrupting Rb alleles that have the recombination substrates targeted to exon 19. Each Rb allele contains a defective neo gene. The K and M alleles contain the Sneo gene that is mutated by the presence of an I-SceI site at the 3′ end of the neo gene. The 18-bp I-SceI site can be cleaved in vivo by expression of I-SceI. The L and N alleles contain the Pneo gene that is mutated at its 5′ end by the insertion of a PacI linker. Transcription of the neo genes is opposite to that of the Rb gene. Drug selection markers hyg+ and hprt+ are transcribed in the same orientation as the Rb gene and were used to select for gene targeting in the hprt− ES cell line. Single rounds of gene targeting were used to construct cell lines with each of the K, L, M, and N alleles. Consecutive rounds of gene targeting were used to construct cell lines with KL and MN alleles. The KL cell line is effectively Rb+/+, whereas the MN cell line is Rb−/−.
To assess the effect of Rb mutation on recombination, two different Rb alleles were created with each neo gene, one that disrupts the Rb coding sequence and one that does not (Fig. 1). In the nondisrupting Rb alleles, termed K and L, the defective neo genes were inserted at intron 18 of the Rb gene. The K allele contains the Sneo gene, whereas the L allele contains the Pneo gene. In the disrupting alleles, termed M and N, the defective neo genes were inserted at exon 19 of the Rb gene. In this case, the M allele contains the Sneo gene and the N allele contains the Pneo gene.
In cell lines containing the Sneo gene, I-SceI cleavage will create a DSB at the Rb allele containing the recognition site. Repair of the I-SceI-generated DSB is predicted to give rise to a neo+ gene by either of two mechanisms, nonhomologous DNA end-joining or homologous recombination. Nonhomologous end-joining can give rise to a neo+ gene if the DNA ends are rejoined at the 4-bp microrepeat flanking the I-SceI site. This type of repair event has previously been observed in mouse and hamster cells (10, 12). Homologous recombination with a Pneo gene at the other Rb allele also has the potential to restore a neo+ gene. These two types of events can be distinguished by restriction fragment length polymorphisms, if recombination involves gene conversion that extends beyond the end of the neo gene or if crossing-over occurs.
Gene Targeting and Spontaneous Allelic Recombination.
The gene targeting constructs were introduced into the hprt− ES cell line E14TG2a. One or both Rb alleles in this cell line were targeted to contain either the Sneo or Pneo gene, selecting for the linked hyg+ or hprt+ marker, respectively. Southern blot analysis was used to identify gene-targeted clones. With each targeting construct, 10–20% of the selected clones were correctly targeted to the Rb allele (data not shown). Singly targeted cell lines K, L, M, and N contain one gene-targeted Rb allele and one wild-type Rb allele. Doubly targeted KL and MN cell lines contain two gene-targeted alleles. The KL cell line containing the K and L alleles is effectively Rb+/+, whereas the MN cell line with the M and N alleles is Rb−/−. MN ES cells grew similarly to nontargeted and to intron-targeted KL ES cells under normal cell culture conditions, a finding consistent with the normal development of Rb−/− mice until embryonic days 10.5–12.5 (18–20).
To measure reversion of the singly targeted alleles and spontaneous allelic recombination of the doubly targeted alleles, cell lines were grown in G418. Neo+ revertants were not detected for the control K, L, M, and N cell lines (<10−9). Rare neo+ recombinants were detected for the KL and MN cell lines, arising at a frequency of approximately 10−8. This is in the range of previous estimates of spontaneous allelic recombination in mammalian cell lines (2–4).
DSBs Induce Allelic Recombination and Loss of Heterozygosity in Mammalian Cells.
To determine whether a DSB would stimulate recombination between alleles, cells were electroporated with the expression vector for the I-SceI endonuclease, pPGK3xnlsI-SceI. Cleavage of the genome in cells transfected with the I-SceI expression vector is highly restricted to the Sneo gene at the Rb locus, due to the long recognition site of the I-SceI endonuclease. Neo+ colonies were selected in G418. G418R colonies were obtained from Sneo-containing K, M, KL, and MN cell lines electroporated with the I-SceI expression vector at a frequency of 1.1–4.8 × 10−6 (Table 1). When the much lower reversion and spontaneous recombination frequencies are considered, neo+ colonies would be expected to result from repair events initiated by an I-SceI-generated DSB at the Sneo gene. This expectation was confirmed when the Sneo gene in each of the G418R colonies was found to have lost the I-SceI site (see below). In contrast, no colonies were obtained from the Pneo-containing L cell line electroporated with the I-SceI expression vector or from any of the cell lines electroporated with a mock DNA.
Table 1.
DSB-induced allelic recombination and end-joining in mouse ES cells
| Transfected DNA | No. G418R | No. analyzed | No. EJ or STGC | No. LTGC (+NH) | No. duplications |
|---|---|---|---|---|---|
| K cell line | |||||
| pPGK3xnlsI-SceI | 13 | 12 | 12 (EJ) | 0 | 1 |
| pgklacZ | 0 | ||||
| L cell line | |||||
| pPGK3xnlsI-SceI | 0 | ||||
| pgklacZ | 0 | ||||
| KL cell line | |||||
| pPGK3xnlsI-SceI | 25 | 21 | 16 (EJ/STGC) | 5 (3) | 1 |
| pgklacZ | 0 | ||||
| M cell line | |||||
| pPGK3xnlsI-SceI | 9 | 8 | 8 (EJ) | 0 | 0 |
| pgklacZ | 0 | ||||
| MN cell line | |||||
| pPGK3xnlsI-SceI | 40 | 38 | 26 (EJ/STGC) | 12 (1) | 13 |
| pgklacZ | 0 |
Approximately 1.6 × 107 cells for each cell line were electroporated with the I-SceI endonuclease expression vector pPGK3xnlsI-SceI or, as a mock DNA control, pgklacZ and selected in G418. Correcting for 50% cell killing, the frequency of neo+ colonies was 1.1–1.6 × 10−6 (K and M cell lines) or 3.0–4.8 × 10−6 (KL and MN cell lines). Allelic recombination with LTGCs in the KL and MN cell lines was identified by Southern blot analysis (Fig. 2). Some LTGC clones displayed altered bands consistent with coincident nonhomologous (+NH) events, the number of such clones being indicated in parentheses. K and M alleles undergo nonhomologous DNA end-joining (EJ) at the Sneo gene to give rise to G418R clones. In addition, in the KL and MN cell lines, the K and M alleles can also undergo allelic recombination with STGCs, due to the presence of the L and N alleles, respectively. EJ and STGC cannot be distinguished. Duplications of the Rb locus were also detected in some of the G418R clones, as identified by additional bands and/or increased hybridization intensity (Figs. 2 and 3).
Neo+ colonies from the K and M cell lines were obtained at a frequency of 1.1–1.6 × 10−6 (Table 1). These colonies were derived by nonhomologous DNA end-joining at the 4-bp duplication flanking the I-SceI cleavage site in the Sneo gene. Southern blot analysis of each of the neo+ K and M clones was consistent with this type of event. Although the I-SceI site was lost from the Sneo gene in each of the neo+ clones, no other size alteration to the locus was found for any of the clones. HindIII/PacI and PstI digestion was used to demonstrate this (Fig. 2). Each of the G418R subclones from the K and M cell lines (e.g., K clones A12, B1, and A11; M clones B7 and B8) contained the parental bands of 15 kb and 1.6 kb with HindIII/PacI and PstI digestion, respectively (Fig. 2; data not shown).
Figure 2.
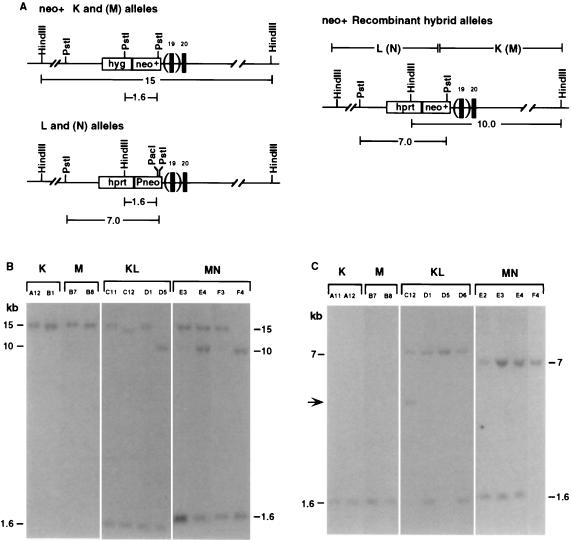
Southern blot analysis of G418R subclones obtained after transfection of cell lines with the I-SceI expression vector. (A) Structure of Rb alleles with relevant restriction sites. Parentheses indicate absence of exon 19 in the M and N alleles. HindIII–PacI and PstI restriction fragment sizes in kb are indicated below each allele. Neo+ clones derived from DNA end-joining or allelic recombination with gene conversion tracts of less than 300 bp (i.e., STGC) give indistinguishable products in which the only detectable alteration of the K and M alleles is loss of the I-SceI site from the neo gene, as indicated. Recombinant hybrid alleles are derived from allelic recombination with gene conversions tracts that are greater than 300 bp (i.e., LTGC). LTGCs that include the entire hprt gene are at least 2.8 kb. In addition to the neo+ hybrid allele, each recombinant cell line has an intact L or N allele, as shown. (B) Southern blot analysis of genomic DNA cleaved with HindIII/PacI. (C) Southern blot analysis of genomic DNA cleaved with PstI. Genomic DNA of expanded K, M, KL, and MN G418R clones was digested with restriction enzymes as noted and hybridized to a 32P-labeled neo EagI–NcoI fragment. Rb alleles in these clones have fragment sizes as shown in A, except for KL neo+ clone C12. The hybrid Rb allele in clone C12 has an approximate 2.5-kb deletion. This is detected by the faster mobility of the upper band in B and is indicated by the arrow in C.
The KL and MN cell lines, which each contain both the Sneo and Pneo alleles, gave rise to neo+ clones at an increased frequency relative to the K and M cell lines, at 3.0–4.8 × 10−6 (Table 1). We considered that the increase in the number of neo+ clones may result from allelic recombination. To test this possibility, Southern blot analysis was performed on genomic DNA from the clones. Allelic recombination with gene conversion extending beyond the 3′ end of the cleaved Sneo gene (i.e., long tract gene conversion or LTGC) or allelic recombination involving crossing-over will result in hybrid L–K or N–M alleles that are distinguishable from the parental alleles by restriction digestion. These hybrid alleles will contain the 5′ end of the Sneo gene and the 3′ end of the Pneo gene, restoring a wild-type neo gene, as well as the hprt gene of Pneo (Fig. 2A).
Digestion with HindIII/PacI was used to identify these hybrid alleles. The parental K and M alleles that contain the Sneo gene gave rise to an approximately 15-kb HindIII fragment, whereas the L and N alleles that contain the Pneo gene gave rise to a 1.6-kb HindIII–PacI fragment (Fig. 2A). This parental pattern for the HindIII/PacI digest was seen in some of the KL and MN neo+ clones (e.g., KL clones C11 and D1; MN clones E3 and F3; Fig. 2B). However, in KL clone D5 and MN clone F4, the 15-kb HindIII fragment of the K and M alleles was lost and replaced with the 10-kb fragment expected from recombinant L–K and N–M alleles, respectively. These two clones maintained the 1.6-kb HindIII–PacI band derived from an intact L and N allele (clones D5 and F4, respectively).
Other restriction digestions also gave rise to band patterns expected from allelic recombination events. For example, PstI digestion of the parental K and M alleles gave rise to a 1.6-kb band, whereas PstI digestion of the L and N alleles gave rise to an approximately 7.0-kb band (Fig. 2 A and C). PstI digestion of recombinant alleles would also be expected to give rise to the 7.0-kb band (Fig. 2A), with loss of the 1.6-kb band. In clones containing recombinant alleles, the 1.6-kb parental K or M allele band was lost, whereas the 7.0-kb band was retained. Because clones containing recombinant alleles demonstrate both the recombinant allele and the parental Pneo allele by HindIII/PacI digestion, the single PstI band must be derived from both the recombinant and the parental alleles.
A consequence of allelic recombination is that heterozygosity was lost at the targeted Rb locus in clones that repaired the DSB by LTGC. The LOH was evident downstream of the I-SceI cleavage site, at the position of the selectable marker genes. Although parental clones contained a hyg+ gene at one Rb allele and a hprt+ gene at the other, recombinant clones having undergone LOH contained an hprt+ gene at both alleles. Clones resulting from LTGC arose at a frequency of approximately 10−6 after electroporation of the I-SceI expression vector, a 100-fold induction over spontaneous events. No clones arising from allelic recombination with an associated cross-over were observed. In this case, both parental bands would have been lost and replaced with two recombinant alleles. However, in all 17 clones in which one recombinant allele was observed, the parental allele used to repair the chromosomal DSB (i.e., the L or N allele) was intact and the reciprocal recombinant allele was not observed.
Approximately 24–32% of the neo+ clones derived from the KL and MN cell lines contained recombinant hybrid alleles with observed LOH. The remaining clones, 76% of the KL and 68% of the MN neo+ clones, showed no fragment size alteration from the parental pattern at the Rb locus (e.g., KL clones C11, D1, and D6; MN clones E3, F3, and E2; Fig. 2). These clones may be the result of nonhomologous DNA end-joining at the Sneo gene, as described above for the G418R K and M clones. Alternatively, a portion of these clones may be derived from allelic recombination in which gene conversion tracts are short (i.e., short tract gene conversion or STGC), so as not to extend beyond the end of the Sneo gene. The increase in the number of clones relative to those derived from the singly marked K and M clones supports this possibility, especially regarding the MN clone (Table 1). Further alterations to the locus will allow us to conclusively determine the range of gene conversion tract lengths in allelic recombination events in mammalian cells.
DSB-Induced Allelic Recombination with Associated Nonhomologous Events.
Some of the G418R clones derived from the KL and MN cell lines were found to have undergone a nonhomologous event in conjunction with homologous allelic recombination. In these clones, homologous recombination between the cleaved Sneo gene and the Pneo gene restored a wild-type neo gene. However, new bands of unexpected sizes were found to have arisen from nonhomologous events downstream of the neo gene. For example, the recombinant alleles in KL neo+ clones C6 (Fig. 3; L–K-deleted allele) and C12 (Fig. 2) had deletions of 1.6 kb (clone C6) and 2.5 kb (clone C12) downstream of the neo+ gene. These deletions were indicative of nonhomologous junctions between the K and L alleles. One other KL clone and one of the MN clones had nonhomologous components to their structure (data not shown). The MN clone appeared to contain an insertion downstream of the neo gene. Because several novel genotypes are found in uniquely obtained clones, it is likely that other recombinant colonies are also independently derived.
Figure 3.
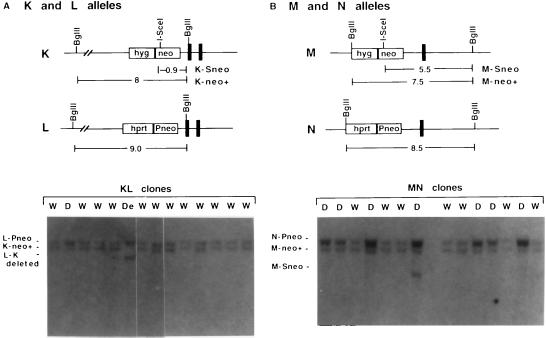
Southern blot analysis demonstrating loss of the I-SceI site and duplication of the Rb locus. Lanes are marked with W (for wild-type) if alleles are unaltered in copy number and D if an allele is duplicated. The neo gene probe is as in Fig. 2. (A) Analysis of genomic DNA cleaved with BglII/I-SceI from KL neo+ clones. Restriction fragment sizes are indicated for each of the alleles, with both the parental and neo+ alleles shown for K (K-Sneo and K-neo+, respectively). The KL clone in lane De has a deletion in the recombinant L–K allele (clone C6; see text). (B) Analysis of genomic DNA cleaved with BglII/I-SceI from MN neo+ clones. Restriction fragment sizes are indicated for each of the alleles, with both the parental and neo+ allele shown for M (M-Sneo and M-neo+, respectively). All MN clones in lanes labeled D have duplicated N alleles, except one clone that has an additional parental M-Sneo allele, as indicated.
Genomic Instability at the Rb Locus.
Neo+ clones derived from the Rb−/− MN cell line were frequently found to contain duplications of one of the Rb alleles (Table 1). Nearly 35% of the MN clones were found to contain duplications, whereas duplications were rarely observed for the K, KL, and M clones (1 of 12 clones, 1 of 21 clones, and 0 of 8 clones, respectively). One example of a cell line containing an Rb locus duplication is MN clone E4. It has a recombinant N–M allele, as well as intact N and M alleles (Fig. 2). Another example, MN clone E3, has an M allele that is intact except for having lost the I-SceI site (Fig. 2). It also has two copies of the N allele, as detected by hybridization intensity relative to the M allele (Fig. 2; data not shown).
Duplications were especially apparent by Southern blot analysis after BglII/I-SceI restriction analysis (Fig. 3). In many clones, two bands of equal intensity were detected, the two bands being derived from the K or M (neo+) and L or N (parental) alleles. However, in the clones with duplications, the intensity of one of the bands was enhanced (Fig. 3, D lanes). Typically, the band derived from the uncleaved donor allele was the one that was duplicated (Fig. 3, upper band), suggesting that duplications may arise during DSB repair between alleles. Of the 38 MN clones examined, 13 had duplications, 10 of which were duplications of the N allele. This contrasts with the 21 KL clones in which only 1 clone had a duplication. The appearance of duplications of the uncleaved N allele in MN clones that do not contain recombinant N–M alleles suggests that these clones may be derived from STGC rather than DNA end-joining. Further analysis will be necessary to distinguish whether the increased frequency of duplications arising in the Rb−/− cells is a consequence of DSB repair involving allelic recombination or is a general feature of increased genetic instability in Rb-deficient cells.
DISCUSSION
We have marked both Rb alleles in mouse ES cells with defective neo genes, one of which contains an I-SceI endonuclease cleavage site. We find that a DSB introduced into one Rb allele will induce allelic recombination at least 100-fold. One outcome of allelic recombination is LOH of sequences downstream of the reconstructed neo+ gene. The LOH results from a gene conversion tract length of at least 2.8 kb. In some cases, resolution of the allelic recombination event is accompanied by an associated nonhomologous event.
Although DSBs in mammalian cells have been thought to be repaired primarily by nonhomologous mechanisms (6), the ability to use a homolog for repair emphasizes the role that homology can have in mammalian DSB repair. Previously, it had been shown that DSBs in the genome of mammalian cells will profoundly stimulate both gene targeting and intrachromosomal recombination (10, 12–14, 21). Intrachromosomal recombination, i.e., recombination between tandem chromosomal repeats, may be expected to occur from both sister-chromatid and intrachromatid interactions, although this has not been demonstrated. Whereas intrachromatid recombinational repair is specific to gene repeats, sister-chromatid recombinational repair is possible for any chromosomal sequence after DNA replication. The finding that homologs can also be used in the repair of chromosomal DSBs suggests that homologs, as well as sister chromatids, provide a second global homology partner for recombinational repair of DSBs in mammalian cells. In budding yeast, sister chromatids are the preferred recombination partner, although homolog recombination readily occurs (22). Comparison of results from two separate studies suggests that sister chromatids may also be the preferred homology partner in mammalian cells, because DSB-promoted intrachromosomal recombination may occur in up to 1% of mammalian cells (10), whereas allelic events occur only at a frequency of 10−6.
The ability to manipulate chromosomal breakage events at the molecular level may be expected to yield important insights into the propensity for allelic repair processes to involve LOH. Allelic recombination in cell lines has been shown in several cases to be an extremely rare event (2–4), and initial studies to detect allelic recombination induced by chromosomal breakage were unsuccessful (4). The I-SceI system has the advantage over earlier studies of chromosomal break repair by introducing one DSB (or very few DSBs) into chromosomal DNA. The 100-fold induction of allelic recombination upon I-SceI expression is likely an underestimate of the total effect of DSBs on recombination between homologs, because only a fraction of the cells have demonstrated cleavage of the chromosomal I-SceI site during transient I-SceI expression (F. Liang and M.J., unpublished results). We chose the Rb locus for analysis of allelic recombination and associated LOH due to its propensity to have undergone LOH in human tumors. The observation that imprinted chromosomal domains of homologs may reside in close proximity to each other in the nucleus (23) and that LOH typically involves specific chromosomal regions (1) raises the possibility that certain alleles may undergo recombination at elevated rates.
Our assay for DSB repair also detects nonhomologous repair. Nonhomologous repair with the restoration of a neo+ gene involves end-joining at a 4-bp microrepeat flanking the I-SceI cleavage site. This type of repair event is typical of nonhomologous repair in mammalian cells (12, 24–26), but neo+ events constitute only a portion of repair events that have been observed at the I-SceI cleavage site (12). In this study, the frequency of end-joining restoring a neo+ gene in the singly targeted cell lines is similar to the frequency of allelic recombination with long gene conversion tracts in the doubly targeted cell lines. Because only one of the possible end-joining events is detected in the neo+ selections, it is likely that nonhomologous end-joining events occur more frequently than allelic recombination for the repair of DSBs in mammalian cells. Although the factors responsible for the use of a homologous or nonhomologous DSB repair pathway are unknown, it may be that mammalian cells have efficient nonhomologous DSB repair processes to bypass allelic recombination and the potential for reduction to homozygosity of loci.
Interestingly, we have found that some allelic recombination events are resolved by nonhomologous recombination, suggesting a possible mutagenic component to allelic repair. The association of a homologous recombination event with a nonhomologous one is reminiscent of “one-sided” DSB-promoted gene targeting events in mammalian cells (12), as well as some plasmid recombination events (27). Heterology between our two Rb alleles may promote such events or associated nonhomologous events may be a general feature of allelic recombination in mammalian cells, reflecting the overall propensity of mammalian cells to undergo nonhomologous recombination.
The observation of chromosomal duplications in a significant fraction of clones from the Rb−/− MN cell line suggests that the Rb−/− phenotype may contribute to genomic instability. Duplications may involve only the chromosomal region around the Rb locus, potentially arising from the induction of a chromosomal DSB at the locus, or they may involve entire chromosomes. Of note, karyotypes of the parental MN cell line demonstrated a normal number of chromosomes (unpublished results). These results raise the possibility that the Rb−/− cells have a propensity toward increased genetic instability, which may be related to one of the functional roles of the Rb protein in maintaining the G1-phase restriction point (28, 29). Oversimplified, it has been assumed that inactivation of the Rb pathway promotes cancer by uncontrolled cellular growth, whereas loss of DNA-damage checkpoints by p53 mutation promotes cancer by increasing genomic instability. Experimental evidence involving the Rb pathway in genomic instability was first noted in human fibroblasts expressing the E7 oncoprotein of type 16 human papillomavirus (30). Cells expressing the E7 protein displayed increased aneuploidy while under selection for CAD gene amplification. Because the E7 protein binds other proteins in addition to Rb, alternative mechanisms rather than Rb inactivation may have been involved in the generation of aneuploidy. The frequent duplications of the Rb locus in the Rb−/− cells, as well as a small (2-fold) increase in LOH events, supports a role for the Rb pathway in maintaining genetic stability.
ES cells provide a particularly intriguing model for studying LOH. Although they are genetically intact, they have growth characteristics that are unlike normal somatic cells (31). They continuously cycle in an undifferentiated state and have low levels of Rb protein and undetectable levels of D-type cyclins (32, 33). If injected into syngeneic mice, they have the ability to form tumors (31). These features of ES cells are analogous to cancer cells and to other self-renewing cells at risk for becoming cancerous over the lifetime of an organism, making them a particularly interesting model for genetic instability studies.
A prediction from the current study is that cells with DNA metabolism defects that cause increased chromosome breakage will have increased rates of allelic recombination and LOH and that people with such cellular defects would be cancer prone. Cells from patients with Bloom syndrome and ataxia telangiectasia are characterized by increased chromosome breakage, and patients with these syndromes develop cancers at high frequency (34–36). The elevated occurrence of symmetrical four-armed (quadriradial) chromosome configurations in Bloom syndrome cells is likely a cytological manifestation of molecular homologous exchange events (see also ref. 37). Although other types of genomic alterations may participate in tumorigenesis in patients with these syndromes, our experiments suggest that increased LOH is also likely to play a major role. In normal cells, spontaneous chromosomal DSBs occur much less frequently yet have the potential of uncovering mutations in tumor suppressor loci by allelic recombinational repair.
Acknowledgments
We thank Hein te Riele (Amsterdam) for materials and Christine Richardson for comments on the manuscript. M.E.M. was funded by an American Society of Clinical Oncology Young Investigator Award. This work was supported by a grant from the National Science Foundation (MCB-9419507) to M.J.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: DSB, DNA double-strand break; LOH, loss of heterozygosity; Rb, retinoblastoma gene; ES, embryonic stem; LTGC, long tract gene conversion; STGC, short tract gene conversion; G418R, G418 resistant.
References
- 1.Lasko D, Cavenee W, Nordenskjöld M. Annu Rev Gen. 1991;25:281–314. doi: 10.1146/annurev.ge.25.120191.001433. [DOI] [PubMed] [Google Scholar]
- 2.Subramani S, Seaton B L. In: Homologous Recombination in Mitotically Dividing Mammalian Cells. Kucherlapati R, Smith G R, editors. Washington, DC: Am. Soc. Microbiol.; 1988. pp. 549–573. [Google Scholar]
- 3.Benjamin M B, Potter H, Yandell D W, Little J B. Proc Natl Acad Sci USA. 1991;88:6652–6656. doi: 10.1073/pnas.88.15.6652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Godwin A R, Bollag R J, Christie D-M, Liskay R M. Proc Natl Acad Sci USA. 1994;91:12554–12558. doi: 10.1073/pnas.91.26.12554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Petes T D, Malone R E, Symington L S. In: The Molecular and Cellular Biology of the Yeast Saccharomyces. Broach J R, Pringle J R, Jones E W, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1991. pp. 407–521. [Google Scholar]
- 6.Jackson S P, Jeggo P A. Trends Biochem Sci. 1995;20:412–415. doi: 10.1016/s0968-0004(00)89090-8. [DOI] [PubMed] [Google Scholar]
- 7.Haber J E. BioEssays. 1995;17:609–620. doi: 10.1002/bies.950170707. [DOI] [PubMed] [Google Scholar]
- 8.Taccioli G E, Rathbun G, Oltz E, Stamato T, Jeggo P A, Alt F W. Science. 1993;260:207–210. doi: 10.1126/science.8469973. [DOI] [PubMed] [Google Scholar]
- 9.Roth D B, Menetski J P, Nakajima P B, Bosma M J, Gellert M. Cell. 1992;70:983–991. doi: 10.1016/0092-8674(92)90248-b. [DOI] [PubMed] [Google Scholar]
- 10.Liang F, Romanienko P J, Weaver D T, Jeggo P A, Jasin M. Proc Natl Acad Sci USA. 1996;93:8929–8933. doi: 10.1073/pnas.93.17.8929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jasin M. Trends Genet. 1996;12:224–228. doi: 10.1016/0168-9525(96)10019-6. [DOI] [PubMed] [Google Scholar]
- 12.Rouet P, Smih F, Jasin M. Mol Cell Biol. 1994;14:8096–8106. doi: 10.1128/mcb.14.12.8096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smih F, Rouet P, Romanienko P J, Jasin M. Nucleic Acids Res. 1995;23:5012–5019. doi: 10.1093/nar/23.24.5012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Choulika A, Perrin A, Dujon B, Nicolas J-F. Mol Cell Biol. 1995;15:1963–1973. doi: 10.1128/mcb.15.4.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.te Riele H, Maandag E R, Berns A. Proc Natl Acad Sci USA. 1992;89:5128–5132. doi: 10.1073/pnas.89.11.5128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hooper M, Hardy K, Handyside A, Hunter S, Monk M. Nature (London) 1987;326:292–295. doi: 10.1038/326292a0. [DOI] [PubMed] [Google Scholar]
- 17.Rouet P, Smih F, Jasin M. Proc Natl Acad Sci USA. 1994;91:6064–6068. doi: 10.1073/pnas.91.13.6064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clarke A R, Maandag E R, van Roon M, van der Lugt N M T, van der Valk M, Hooper M L, Berns A, te Riele H. Nature (London) 1992;359:328–330. doi: 10.1038/359328a0. [DOI] [PubMed] [Google Scholar]
- 19.Jacks T, Fazeli A, Schmitt E M, Bronson R T, Goodell M A, Weinberg R A. Nature (London) 1992;359:295–300. doi: 10.1038/359295a0. [DOI] [PubMed] [Google Scholar]
- 20.Lee E Y-H P, Chang C-Y, Hu N, Wang Y-C, Lai C-C, Herrup K, Lee W-H, Bradley A. Nature (London) 1992;359:288–294. doi: 10.1038/359288a0. [DOI] [PubMed] [Google Scholar]
- 21.Brennemen M, Gimble F S, Wilson J H. Proc Natl Acad Sci USA. 1996;93:3608–3612. doi: 10.1073/pnas.93.8.3608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kadyk L C, Hartwell L H. Genetics. 1992;132:387–402. doi: 10.1093/genetics/132.2.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.LaSalle J M, Lalande M. Science. 1996;272:725–728. doi: 10.1126/science.272.5262.725. [DOI] [PubMed] [Google Scholar]
- 24.Phillips J W, Morgan W F. Mol Cell Biol. 1994;14:5794–5803. doi: 10.1128/mcb.14.9.5794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lukacsovich T, Yang D, Waldman A S. Nucleic Acids Res. 1994;22:5649–5657. doi: 10.1093/nar/22.25.5649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thacker J. Int J Radiat Biol. 1994;66:591–596. doi: 10.1080/09553009414551671. [DOI] [PubMed] [Google Scholar]
- 27.Sakagami K, Tokinaga Y, Yoshikura H, Kobayashi I. Proc Natl Acad Sci USA. 1994;91:8527–8531. doi: 10.1073/pnas.91.18.8527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Elledge S J. Science. 1996;274:1664–1672. doi: 10.1126/science.274.5293.1664. [DOI] [PubMed] [Google Scholar]
- 29.Sherr C J. Science. 1996;274:1672–1677. doi: 10.1126/science.274.5293.1672. [DOI] [PubMed] [Google Scholar]
- 30.White A E, Livanos E J, Tlsty T D. Genes Dev. 1994;8:666–677. doi: 10.1101/gad.8.6.666. [DOI] [PubMed] [Google Scholar]
- 31.Robertson E J. In: Embryo-Derived Stem Cell Lines. Robertson E J, editor. Washington, DC: IRL; 1987. pp. 71–112. [Google Scholar]
- 32.Savatier P, Huang S, Szekely L, Wiman K G, Samarut J. Oncogene. 1994;9:809–818. [PubMed] [Google Scholar]
- 33.Savatier P, Lapillonne H, van Grunsven L A, Rudkin B B, Samarut J. Oncogene. 1996;12:309–322. [PubMed] [Google Scholar]
- 34.German J. Medicine. 1993;72:393–406. [PubMed] [Google Scholar]
- 35.Blocher D, Sigut D, Hannan M A. Int J Radiat Biol. 1991;60:791–802. doi: 10.1080/09553009114552601. [DOI] [PubMed] [Google Scholar]
- 36.Meyn M S. Cancer Res. 1995;55:5591–6001. [PubMed] [Google Scholar]
- 37.Groden J, Nakamura Y, German J. Proc Natl Acad Sci USA. 1990;87:4315–4319. doi: 10.1073/pnas.87.11.4315. [DOI] [PMC free article] [PubMed] [Google Scholar]