Abstract
Some group I introns self-splice in vitro, but almost all are thought to be assisted by proteins in vivo. Mutational analysis has shown that the splicing of certain group I introns depends upon a maturase protein encoded by the intron itself. However the effect of a protein on splicing can be indirect. We now provide evidence that a mitochondrial intron-encoded protein from Aspergillus nidulans directly facilitates splicing in vitro. This demonstrates that a maturase is an RNA splicing protein. The protein-assisted reaction is as fast as that of any other known group I intron. Interestingly the protein is also a DNA endonuclease, an activity required for intron mobilization. Mobile elements frequently encode proteins that promote their propagation. Intron-encoded proteins that also assist RNA splicing would facilitate both the transposition and horizontal transmission of introns.
Keywords: homing endonuclease/maturase, mitochondria, mobile element, RNA splicing protein
Many biological processes are catalyzed by complexes of RNA and protein. In some instances the catalytic activity is provided by the RNA component known as a ribozyme. Evidence is accumulating that ribozymes are active in central cellular processes such as pre-mRNA splicing and protein synthesis (1, 2). Since these are very complicated, research has also focused on simpler examples such as ribonuclease P (3) and the splicing of group I introns (4). We now provide evidence for a particularly interesting ribozyme-protein interaction involving the AnCOB group I intron from the Aspergillus nidulans apocytochrome b gene and a protein encoded by the intron itself.
In 1980 mutational analysis suggested that excision of a group I intron from a yeast mitochondrial gene required expression of an intron-encoded protein (5–9). The protein was termed a maturase. Shortly afterwards two somewhat contradictory developments took place: (i) It was shown that a group I intron was a ribozyme in that it could self-splice in vitro in the absence of proteins (10). (ii) It was shown that the excision of many group I introns required or utilized the assistance of proteins (reviewed in ref. 4). The contradiction is likely to be more apparent than real because there is no evidence to suggest that the proteins have usurped the catalytic role of the intron RNA; those that have been studied in vitro stabilize RNA tertiary structure (11–13).
The role of the maturase has proved difficult to determine because the purified protein has never been observed to directly stimulate splicing. Biochemical analysis verified that mutations expected to prevent correct translation of the intron ORF led to accumulation of unspliced RNA precursor in vivo (14, 15); however, this did not exclude the possibility that the maturase’s mode of action was indirect as is the case for the yeast Suv3p protein (16). We now present evidence that a maturase directly stimulates RNA splicing.
Several group I intron-encoded maturases have been identified in Saccharomyces cerevisiae and other species of yeast (reviewed in refs. 17–19). They show sequence similarity with an unusual class of intron-encoded DNA endonucleases (reviewed in ref. 20) that can mobilize the intron to an equivalent site lacking the intron, a process known as homing. Similar endonuclease sequences are also found in inteins (21). Sequence similarity is most striking in two related LAGLIDADG dodecapeptide sequences know as P1 and P2 (22–24). Despite the fact that group I intron ORFs in S. cerevisiae only encode either a maturase or a DNA endonuclease, there is strong evidence that the two activities are very closely associated: in vivo analysis has shown that an intron ORF in Saccharomyces capensis encodes both activities (19); a homing intron (presumed to encode endonuclease activity) encodes a maturase in Schizosaccharomyces pombe (18) and a point mutation in the S. cerevisiae cox1-I4α intron-encoded endonuclease activates latent maturase activity (25). In this work we show that the Aspergillus intron-encoded protein is not only a maturase but also a DNA endonuclease.
A considerable amount is known about the DNA endonucleases in yeast and other organisms as they can be assayed biochemically in vitro; in contrast, however, very little is known about the maturases. This situation should improve rapidly now that we have a direct in vitro assay for maturase activity.
METHODS
Expression and Purification of the Maturase.
The maturase coding sequence was obtained from pSPAnCOB (26), which had been constructed by inserting a PCR fragment spanning the AnCOB intron into the EcoRI/HindIII sites of the transcription vector pSP65 (Promega). Intron sequence from position 202 to 1082 (22) (Fig. 1A) was inserted as a BglII/HindIII fragment into the BamHI/HindIII sites of the Escherichia coli expression vector pET28b (Novagen) after two mitochondrial UGA tryptophan codons had been mutated to UGG. The resulting plasmid, pEC, was shuttled into BL21DE3 and the AnCOB protein expressed as recommended (27). It was purified by Ni++—chelated affinity chromatography using the vector-encoded histidine tag (Fig. 1B) (Novagen) as recommended except as follows. The column was normally washed with 20 mM Tris (pH 7.9), 500 mM NaCl (TN buffer), and 30 mM imidazole. In one preparation the cleared lysate loading buffer included 6 M urea. The column in this preparation was first washed in TN buffer and 5 mM imidazole; and sequentially 6, 4, 2, 1, and 0 M urea. The protein was eluted with 3 × 1 ml TN buffer + 60 mM imidazole. The first ml was discarded along with several minor bacterial contaminants. Elution was completed with 3 × 0.5 ml TN buffer and 100 mM imidazole. Purity was about 90%, as determined by densitometry analysis of a Coomassie blue-stained SDS polyacrylamide gel. A second affinity chromatography step, using an antibody (Novagen) to the 11-amino acid, T7 gene 10, leader sequence (Fig. 1B), was performed as recommended, with 1 M urea in the loading buffer to improve epitope recognition. The protein was concentrated using a 10-K MW cutoff Centricon microconcentrator (Amicon) and stored in 20 mM Tris (pH 7.9) 50 mM NaCl, 1 mM DTT, and 50% glycerol. The purity was now >96% (as determined above) with the major impurity likely to have been a proteolytic form of the AnCOB protein (see Results and Fig. 3). The size of the burst in the multiple turnover experiments implied that most of the protein molecules were active. Cloning of the control intron-encoded protein from AnOX2 [in pET28a (Novagen)] will be described elsewhere (unpubished data).
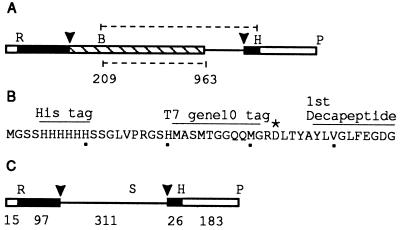
Figure 1.
Plasmid constructs. (A) Map of a region of pSPAnCOB. □, vector sequence; ▪, A. nidulans cobA gene exon; ▧, intron ORF; —, intron sequence. R = EcoRI, H = HindIII, P = PvuII, B = BglII Arrowheads, splice-sites. The upper dotted line denotes the region cloned into the expression vector pET28b. The lower dotted line denotes the region replaced by a SalI site to yield pCOBsal. (B) Amino-terminal amino acid sequence of the 32-kDa maturase protein expressed from pET28b. ∗, start of the Aspergillus sequence. (C) Map of a region of pCOBsal. The region shown corresponds precisely to the precursor used for all splicing assays. The number of bases in each segment are shown (the intron ORF remaining from pSPAnCOB is not shown as ▧). S = SalI.
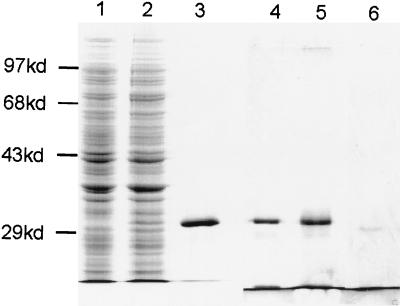
Figure 3.
Purification of the AnCOB maturase. 10% SDS/PAGE, stained with Coomassie blue (lanes 1–3). Lanes 1 and 2: lysate of induced BL21DE3 with pET28b (the expression vector) or pEC (pET28b + the AnCOB intron), respectively. Lane 3: Ni++ chromatography purification of AnCOB protein (2.4 μg). A second gel was silver stained (lanes 4–6). Lane 4: Ni++ purification of AnCOB (1.2 μg). Lane 5: immunoaffinity chromatography purification of material from lane 4 (0.66 μg). Lane 6: carbonic anhydrase control (0.02 μg). It was not possible to load more than 0.66 μg of the immunoaffinity column purified material.
RNA Precursor Synthesis.
RNA precursors were transcribed from pCOBsal (26) (Fig. 1C) as described (28). The AnCOB precursor generated from pCOBsal linearized with PvuII contains 112 and 209 nucleotides of 5′ and 3′ exon respectively of which 97 and 26 nucleotides are from the cobA gene (Fig. 1C). The intron is only 311 nucleotides because 753 nucleotides were deleted from the P8/L8 stem-loop (for the secondary structure of AnCOB, see ref. 29) and replaced with the sequence TGTCGACATA containing a SalI site—the deletion has little effect at 25 mM MgCl2 and approximately doubles the rate of self-splicing at 50 mM (26). Precursor RNA was purified by denaturing PAGE (28).
Splicing Reactions.
Precursor RNA was heated to 90°C in 10 mM Tris (pH 7.6), 0.1 mM EDTA for 1 min, prefolded in splicing buffer (50 mM Tris, pH 7.6/0.1 M KCl/5 or 25 mM MgCl2, as shown) for 5 min at 37°C. Single turnover reactions (RNA at 1 nM) were started by adding 1 mM GTP with 10 nM maturase (all concentrations are final). For multiple turnover, 20 nM RNA (prefolded as above) was preincubated for 5 min with 2 or 4 nM maturase in splicing buffer (5 mM MgCl2) and the reaction started with GTP. Reactions were stopped with phenol and EDTA (100 mM final), organically extracted, and analyzed on a 6% denaturing polyacrylamide gel.
Kinetic Analysis.
The rate of the first splicing step was estimated from the data in Fig. 2A, which was performed under single turnover conditions. An autoradiogram was quantitated by laser densitometry (Molecular Dynamics). Using the Marquardt–Levenberg algorithm and nonlinear regression analysis (PSI-plot), the maturase assisted data were fitted to both a single and double exponential: Fpre = Ae−kt + (1 − A) and Fpre = A1e−k1t + A2e−k2t + (1 − A1 − A2); where Fpre is the fraction of precursor remaining and (1 − A) and (1 − A1 − A2) represent the fraction of unreacted RNA for the single and double exponential equations, respectively. Extra time points (0.33, 0.83, 1.0, and 10 min) not shown in Fig. 2A were included.
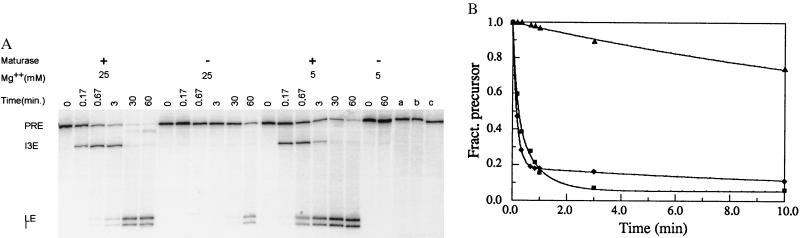
Figure 2.
The AnCOB intron encodes a maturase that directly assists RNA splicing. (A) Group I introns react by two sequential transesterification reactions: First, a guanosine attacks the 5′ splice site and is transferred to the first base of the intron. The exposed 3′OH end of the upstream exon (5′ exon) then attacks the 3′ splice site, releasing the intron and leaving the exons ligated (58). PRE, precursor; I3E, intron + 3′ exon splicing intermediate; I, excised intron; LE, ligated exons. The 5′ exon is short and is not shown. Its presence exactly mirrors that of the I3E molecule, being generated during 5′splice-site cleavage and disappearing as RNA molecules complete the second step. (Lane a) Prior to mixing with RNA, the protein extract was treated with 2 μg/μl of proteinase K for 30 min at 37°C. (lanes b and c) The pre-RNA was incubated as above (5 mM MgCl2) with extracts purified by Ni++ affinity chromatography (as for the AnCOB protein) from BL21DE3 either containing just the pET28b vector (lane b) or expressing the endonuclease encoded by a different intron, AnOX2 (37) (lane c). (B) The rate of the first splicing step was determined as described in Materials and Methods. ▴, 25 mM Mg2+, no maturase; ⧫, 5 mM Mg2+ + maturase; ▪, 25 mM Mg2+ + maturase.
The 5 mM Mg2+ data showed a better fit to a double exponential (the SE was 10× smaller); the curve in Fig. 2B has the form: Fpre = 0.82e−6.21t + 0.13e−0.08t + 0.06. This gave a kobs for the first step of splicing of 6 min−1. The 25 mM Mg2+ reaction did not show a significantly better fit to a double exponential (the SE only being 3× smaller). Unpublished data suggests that a biphasic curve is more likely; the curve in Fig. 2B has the form: Fpre = 0.51e−6.22t + 0.44e−1.28t + 0.05. kobs ranged from 6 to 3 min−1 derived from a double and single exponential fit respectively. Repeats of the 5 and 25 mM Mg2+ reactions gave a range of 4–10 min−1 for kobs. The self-splicing reaction (in 25 mM Mg2+ without maturase) was fitted to a single exponential (see above), using time points out to 60 min and the curve in Fig. 2B has the form: Fpre = 0.64e−0.053t + 0.36.
In the slow phase of the multiple turnover reaction, the protein was close to saturation: doubling the amount of RNA substrate did not change the rate (data not shown). This was consistent with the size of the burst in Fig. 5. We therefore estimated the turnover rate as: nmol of RNA precursor reacted per min per nmol protein present.
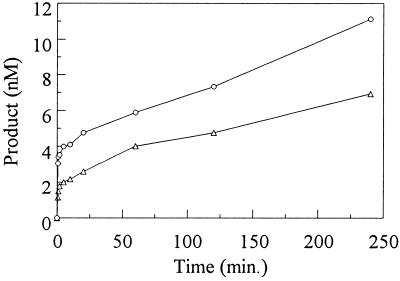
Figure 5.
The AnCOB maturase shows a stoichiometric burst under multiple turnover conditions. ▵, 2 nM maturase; ○, 4 nM maturase. Product was determined as follows: ([IVS] + [I3E])/([IVS] + [I3E] + [PRE]) where IVS, intron; I3E, intron attached to 3′ exon; PRE, precursor (Fig. 2).
Binding Assays.
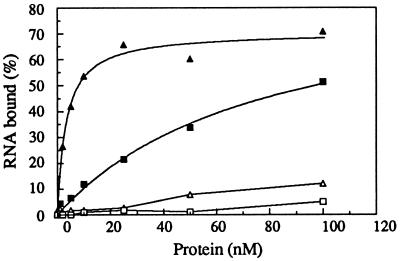
AnCOB pre-RNA and AnOX2 pre-RNA (transcribed from pSPAnOX2 (26) were prepared as for splicing except that the 3′ exon was only 30 nucleotides. RNAs were heated at 90°C as for splicing and then prefolded in binding buffer (50 mM Tris, pH 7.5/0.1 M KCl/25 mM MgCl2/10% glycerol/50 μg/ml BSA/50 μg/ml tRNA). After pre-equilibration at 28°C, RNA (0.05 nM) was mixed with protein for 20 min, then filtered through nitrocellulose (prewetted with binding buffer), and washed three times with 0.3 ml binding buffer. The filters were dried and counted in Tru-Count scintillation fluid (Tru Lab Supply). Assuming a simple bimolecular interaction, the data were fitted to: % total RNA bound (measured) = A × P/(P + Kd), where A = maximum % RNA bound and P = total protein (nM). The apparent Kd for the AnCOB specific binding was 3 nM (a range of 2–10 nM has been obtained under varying conditions). The AnOX2 protein/AnOX2 RNA binding data fitted the equation above but the Kd value was very dependent on the fitted value of A − saturation binding has not yet been attempted.
Determination of DNA Endonuclease Cleavage Site.
A cDNA sequence spanning nucleotide −97 to −1 and 1056 to 1082 (22) was amplified by reverse transcriptase-PCR of A. nidulans total RNA using the following primers: cobA5, 5′-AGGAATTCGTCAAATGAGTTTATGAGGTGCTAC; and cobA3b, 5′-TGGAAGCTTATTAAATGCATTAATGCTAAAGCAG. The EcoRI and HindIII sites in the PCR primers were digested and the fragment cloned into pIBI24 to yield pCOBLE. This was used as a template (30) for sequencing reactions using either primer cobA5 or cobA3b, Sequenase 2.0, 5 μCi (1 Ci = 37 GBq) [35S]dATP, and the protocol recommended by Amersham. The reactions were phenolized and ethanol precipitated. A portion was digested in 50 mM Tris (pH 8.0), 50 mM NaCl, 10 mM MnCl2, 500 ng AnCOB protein for 30 min at 37°C and analyzed as for a sequencing gel (Fig. 6).
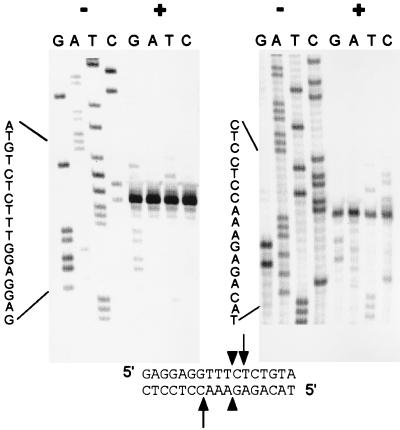
Figure 6.
The AnCOB intron-encoded protein is also a DNA endonuclease. Two DNA chain termination sequencing reactions were performed, one on either strand of a cDNA clone that spans the intron insertion site (Materials and Methods). Half of each reaction remained uncut (−) and the other half was digested (+) with the AnCOB protein preparation. Newly synthesized DNA molecules that terminate before reaching or completing the recognition site will remain uncut (including those that extend beyond the cleavage site but fail to complete the entire recognition site; see ref. 30). Strands that extend beyond the recognition site will all be cleaved and give the same-sized, primer-proximal, labeled fragment and a distal unlabeled fragment that will vary in size (the radioactive label is primarily incorporated very close to the primer; see Materials and Methods). Arrows indicate the cleavage sites and the arrowheads the site at which the intron would be inserted.
RESULTS
The AnCOB Intron Encodes a Maturase that Facilitates Splicing in Vitro.
Previous unsuccessful attempts to demonstrate maturase-dependent splicing in vitro were made with group I introns that do not self-splice. The yeast ScCOBi5 intron self-splices in a high concentration of Mg2+ but requires a nuclear encoded protein in low Mg2+ (31, 32). We suspected that it might be easier to identify a maturase that simply reduced a self-splicing intron’s requirement for Mg2+ rather than one that rescued a nonself-splicing intron. The group I intron (AnCOB) from the A. nidulans mitochondrial cytochrome b gene (cobA) (22) self-splices providing the concentration of Mg2+ is 25 mM or above (26). The intron contains an ORF that shows (in the carboxyl two-thirds) high homology (66%) to a maturase in the yeast ScCOBi3 intron (33).
Maturase/endonuclease precursors are usually expressed as read-through products in frame with the upstream exon and then processed. It is known that a maturase can function with only a few amino acids upstream of the first dodecapeptide repeat sequence, P1 (34). Furthermore, the first 200 nucleotides of the AnCOB and ScCOBi3 introns are similar but read in a different reading frame and so their encoded amino acids are unlikely to be required for protein function. For these reasons, the AnCOB ORF was expressed by inserting into the E. coli expression vector, pET28b, the sequence from just upstream of the first dodecapeptide sequence to a position in the 3′ exon (Fig. 1A). The 32-kDa AnCOB protein was expressed and purified as described in Materials and Methods.
Initial experiments indicated that the rate of splicing of the AnCOB RNA precursor at high Mg2+ was dramatically increased by addition of the AnCOB protein preparation and that splicing was activated at low Mg2+ (Fig. 2). The protein preparation led to very rapid accumulation of the splicing intermediates that are the intron attached to the 3′ exon (I3E) and the 5′ exon (not shown). Upon further incubation these reacted to give ligated exon RNA and the excised intron.
The splicing activity was proteinaceous in character since it was sensitive to proteinase K (Fig. 2A, lane a), heat (65°C, 30 min), and phenol (data not shown). Proteinase K itself was not an inhibitor; a control reaction showed that it did not inhibit self-splicing in 25 mM Mg2+ (data not shown).
A Bacterial Protein Is Not Responsible for the Splicing Activity.
It has been reported that bacterial proteins can nonspecifically facilitate RNA splicing by acting as RNA chaperones (35, 36). We therefore verified that the AnCOB protein was responsible for the splicing activity and not a contaminating bacterial protein. The protein is fused at its amino end to a histidine tag and an 11-amino acid leader of the T7 phage gene 10 product (Fig. 1). It had initially been purified by Ni++ affinity chromatography using the histidine tag. Two further steps were taken. (i) The protein was further purified using an antibody affinity column against the T7 leader (Fig. 3). There was no decrease in the specific activity of the protein preparation, contrary to what would be expected if a bacterial protein were responsible. (ii) To reduce the possibility of a bacterial contaminant binding to AnCOB and copurifying with it, the Ni++ column was loaded in the presence of 6 M urea; this preparation was as active as previous ones.
Densitometer analysis of a silver stained gel indicated that the Ni++ affinity chromatography had already achieved a significant degree of purity: a loading of 1.2 μg of protein revealed two minor contaminants of ≈26 and 19 kDa representing about 4 and 2% of the preparation (Fig. 3). The 26-kDa contaminant was probably a proteolytic form of the AnCOB protein because it was still present at ≈4% after the immunoaffinity column, was rather diffuse, and increased slightly in MW in a variant of AnCOB that had 12 extra amino acids at its amino end. This larger variant did not show a band at 32 kDa indicating that a 32-kDa bacterial contaminant does not copurify and comigrate with the standard AnCOB protein (data not shown). Analysis of 660 ng of the immunoaffinity purified preparation by silver staining revealed no other contaminating bands under conditions where 20 ng of carbonic anhydrase (29 kDa) was clearly visible (Fig. 3). A highly overdeveloped gel with 660 ng protein did reveal two other bands of about 2–4 ng based on visible carbonic anhydrase standards of 1 and 10 ng.
Any bacterial proteins contaminating the AnCOB preparation would also be expected in preparations obtained when purifying other plasmid encoded proteins. The Ni2+ affinity column procedure used to purify the AnCOB protein was repeated in an identical fashion with lysates from an induced bacterial strain carrying one of two plasmids: either the expression vector alone or a construct designed to express the protein encoded by the second intron (AnOX2) of the cytochrome oxidase subunit I gene (37). However neither protein preparation supported splicing of the AnCOB precursor (Fig. 2A, lanes b and c). We note that the AnOX2 encoded protein preparation itself was active as a DNA endonuclease and had no inhibitory effect on self-splicing in 25 mM Mg2+ (data not shown). The fact that the AnCOB protein preparation was extremely active and the AnOX2 preparation showed no activity on the AnCOB precursor in one hour is strong evidence that a bacterial contaminant is not responsible for stimulating RNA splicing.
The E. coli ribosomal S12 protein and the stpA gene product assist splicing of the T4 phage td group I intron (35, 36). We had previously obtained the S12 protein from the Belfort laboratory and shown that it did not promote AnCOB RNA splicing (26). We also note these proteins were used (35, 36) at 20–100 times the concentration used for the AnCOB protein.
Binding and Kinetic Analysis.
A filter binding assay (Fig. 4) showed that the AnCOB protein extract bound tightly to the AnCOB precursor (Kd = 3 nM) and very poorly to a precursor containing the AnOX2 intron. In contrast, the AnOX2 intron-encoded protein bound very poorly to the AnCOB precursor but significantly to the AnOX2 precursor. (The AnOX2 protein does not facilitate splicing of the AnOX2 precursor in vitro). The binding specificities of the protein preparations for their respective introns cannot easily be attributed to a bacterial protein given that both proteins were purified in the same manner from the same strain except for the DNA sequence in the vector’s expression site.
Figure 4.
Specific binding of the AnCOB and AnOX2 intron-encoded proteins. ▴ and ▵: AnCOB protein with AnCOB and AnOX2 pre-RNA, respectively. ▪ and □: AnOX2 protein with AnOX2 and AnCOB pre-RNA, respectively.
Under single turnover reaction conditions in 25 mM Mg2+ with 10 nM maturase, kobs for the first splicing step was 3–6 min−1 (see Materials and Methods). This is 60–120× greater than the value for the self-splicing reaction (0.05 min−1) (Fig. 2B). The maturase converted the rate limiting step from the first to the second splicing reaction and thus increased the overall rate of splicing (Fig. 2). In 5 mM Mg2+ with 10 nM maturase, kobs was 6 min−1, a rate increase of at least 4 × 104, assuming that 1% self-splicing would have been detected in 1 hr. In 5 and 25 mM Mg2+, 80 and 90% of the reactive precursor completed the first splicing step in < 1 min. By fitting the disappearance of the intermediates to a simple exponential, the rate constants for the second step were estimated to be ≥1.5 and ≥0.5 min−1, respectively.
Multiple turn-over experiments were also performed in 5 mM MgCl2, conditions in which self-splicing does not occur: 20 nM RNA was preincubated with 2 or 4 nM AnCOB protein and the reaction started with GTP. A rapid burst was observed whose size was almost stoichiometric (0.9 ± 0.1:1) to the amount of protein added (Fig. 5). This was followed by a much slower phase with turnover rate constant = 0.01–0.008 min−1 (it was assumed that the protein was saturated; see Materials and Methods). With a different lot of protein the sizes of the bursts were similar but the rate constants of the slow phase were 0.004–0.003 min−1. We interpret the experiment as follows: The burst resulted from the rapid reaction of RNA that had prebound to the maturase during preincubation. The ensuing slow phase was due to tight binding of the reaction products leading to slow turnover of the protein.
The AnCOB Maturase Is Also a DNA Endonuclease.
Group I introns frequently encode DNA endonucleases that mediate insertion of the intron into the equivalent DNA sequence lacking the intron, a process called homing (20). A plasmid was constructed containing a short region of cDNA spanning the intron insertion site but lacking the intron itself (see Materials and Methods). Restriction mapping indicated that the AnCOB protein cut close to the site of intron insertion (data not shown). The precise site of endonucleolytic activity was identified by digesting a sequence reaction spanning this region (Fig. 6). Digestion occurs at +1/−3 with respect to the intron insertion site. By convention (38), the endonuclease should be called I-AniI.
DISCUSSION
We have isolated a splicing factor from a bacterial strain expressing a protein encoded by the AnCOB intron. A series of experiments provided strong evidence that the AnCOB protein is responsible for the splicing activity rather than a contaminating bacterial protein. (i) We have purified the protein to a high level of purity by performing immunoaffinity chromatography on a preparation that was already fairly pure using Ni++ column chromatography. This did not reduce the specific activity of the preparation as would be expected if a bacterial protein were involved.
(ii) If a bacterial protein in the AnCOB protein preparation were responsible for stimulating cleavage of half the precursor in < 0.17 min (single turnover reaction, Fig. 2 A and B) it is very unlikely that the AnOX2 intron-encoded protein preparation would then have had insufficient bacterial protein to detect at least some activity in 1 hr (Fig. 2A, lane c) since the proteins were purified in an identical manner. (iii) The AnCOB and AnOX2 protein preparations bound specifically to their respective introns (Fig. 4)—this is not readily explained by a bacterial protein. It is possible that a bacterial protein copurified by being bound to the AnCOB protein. However it would have to show no binding to the AnOX2 protein and remain tightly bound to the AnCOB protein in 6 M urea present on one occasion in the Ni++ affinity column loading buffer. This explanation does not explain the fourth point below or readily explain the binding data above.
(iv) The characteristics of the multiple turnover experiments, especially the stoichiometric size of the bursts were consistent with each molecule of AnCOB protein promoting cleavage of one RNA molecule and then turning over slowly (this was seen with two different lots of protein). These data were also compatible with the filter binding assay: assuming formation of a simple bimolecular complex, a Kd of 3 nM predicts that about 85% of the protein should have prebound to the RNA and thus should have contributed immediately to the burst upon addition of GTP; the slow phase (Fig. 5) can be attributed to slow product release due to relatively tight binding of the intron to the protein as might be expected from the binding assay (Fig. 4). By contrast, given the purity of the AnCOB protein (Fig. 3), a contaminating bacterial protein would have had to have turned over >50 times for about a minute and then suddenly reacted about a thousand times slower thereafter. This is much less plausible.
The definition of a maturase is vague at present because little is known about their function. They have three primary characteristics (4): (i) A maturase is an intron-encoded protein, usually expressed in frame with the preceding exon. (ii) If not expressed correctly, the intron encoding it is not excised. (iii) Maturases from group I introns share sequence homology with a class of DNA endonucleases that contain conserved dodecapeptide sequences. The AnCOB protein has all these features: it is a splicing factor, a DNA endonuclease and it shares 66% homology with the ScCOBi3 maturase, comparable to the flanking apocytochrome b sequence. We therefore conclude that the AnCOB intron encodes a maturase with activity in vitro.
The protein splicing factors of several group I introns have been identified (4, 39): examples include the product of the nuclear gene CBP2, which acts on the yeast mitochondrial ScCOBi5 intron (32), and the product of the cyt-18 gene, a mitochondrial tyrosyl tRNA synthetase, whose targets include the Neurospora crassa large rRNA intron (40). The CYT-18 protein acts on several subclasses of intron and is believed to act somewhat nonspecifically by stabilizing several RNA helices that form the catalytic core of the intron (41–43). The Cbp2 protein is thought to be specific for the ScCOBi5 intron (12, 44) and to trap a transiently folded RNA tertiary structure (13).
Little is known about the action of maturases. The maturase encoded by the yeast ScCOBi4 intron assists splicing of both its own intron and the ScOXIi4 intron located in the cytochrome oxidase subunit I gene (reviewed in ref. 4). In general however, maturases are believed to be specific for their own introns. The AnCOB maturase was not able to activate splicing of introns ScCOBi3 and AnOX2 (data not shown). However, both of these introns are not self-splicing (26) and may require more than one protein. Data in Fig. 2 indicates that the rate of the in vitro maturase-assisted reaction is comparable to that of the fastest characterized group I introns for both splicing steps (44).
The mobility or homing activity of the S. cerevisiae group I introns has been analyzed in considerably more detail than that of the Aspergillus introns (17, 45). However evidence that the AnCOB intron is mobile has been discussed previously (46). The data in Fig. 6 indicate that the recognition sequence of the maturase’s endonuclease is about 14–16 bp. This and the generation of a 3′ overhang of four bases is observed with many other homing endonucleases (17).
Mechanistically what is the function of the AnCOB maturase and what is the relation between the endonuclease and maturase activity? It would seem most likely that the maturase activity consists primarily of an RNA binding site but not a catalytic site, since the latter resides in the RNA. Maturases and endonucleases show the strongest sequence homology in two related dodecapeptide regions (22–24) know as P1 and P2 or LAGLIDADG sequences. In the yeast coxI-I4 α protein (with its latent maturase function activated by mutation), mutations in P2 primarily affect maturase activity, whereas mutations in P1 affect endonuclease activity (47).
The Aspergillus AnCOB and yeast ScCOBi3 introns are similar in their RNA core structure sequences, are inserted in equivalent sites and encode similar maturases (22, 33). It will therefore be of considerable interest to know if the maturase from the yeast intron ScCOBi3 also supports splicing in vitro and if the Aspergillus maturase can rescue ScCOBi3 maturase-deficient mutants in vivo. Splicing of the ScCOBi3 intron in vivo also requires the gene product of the MRS1 gene (48) but this may play an indirect role as is the case for the Suv3p protein (16).
Group II introns also encode proteins with multiple activities, including reverse transcriptase, DNA endonuclease and maturase activity in a single protein (49–51). Group I and II intron-encoded proteins do not share sequence similarity. To our knowledge, in vitro experiments have not shown that a group II maturase directly facilitates RNA splicing. It has also been observed that some ribosomal proteins have DNA binding domains (52).
It has been proposed that intron-encoded proteins of both group I and II introns initially promoted mobilization and that they later acquired an ancillary role in RNA splicing (4, 17, 50). The demonstration here that the AnCOB endonuclease directly facilitates RNA splicing makes this model more plausible. Group I introns may transpose to new sites and possibly transfer horizontally between organisms (17, 53–55). Intron-encoded proteins that directly assist RNA splicing would facilitate such a process, especially in compact genomes with a high density of coding sequence and particularly if splicing were suboptimal at the new site (56, 57).
Acknowledgments
We thank the Belfort laboratory for the S12 protein sample, H. Rappaport for reading the manuscript; E. Gruberg and J. Sheffield for their encouragement; and Ray Colliton for technical assistance. This work was supported by a National Science Foundation Research Experiences for Undergraduates site grant and a Research Incentive Fund from Temple University.
Note Added in Proof
The Cbp2 protein also stimulates splicing of the omega intron of yeast mitochondria (59).
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Noller H F, Hoffarth V, Zimniak L. Science. 1992;256:1416–1419. doi: 10.1126/science.1604315. [DOI] [PubMed] [Google Scholar]
- 2.Madhani H D, Guthrie C. Annu Rev Genet. 1994;28:1–26. doi: 10.1146/annurev.ge.28.120194.000245. [DOI] [PubMed] [Google Scholar]
- 3.Brown J W, Nolan J M, Haas E S, Rubio M A T, Major F, Pace N R. Proc Natl Acad Sci USA. 1996;93:3001–3006. doi: 10.1073/pnas.93.7.3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lambowitz A M, Perlman P S. Trends Biochem Sci. 1990;15:440–444. doi: 10.1016/0968-0004(90)90283-h. [DOI] [PubMed] [Google Scholar]
- 5.Lazowska J, Jacq C, Slonimski P P. Cell. 1980;22:333–348. doi: 10.1016/0092-8674(80)90344-x. [DOI] [PubMed] [Google Scholar]
- 6.Perlman P S, Mahler H R, Dhawale S, Hanson D, Alexander N J. In: Organization and Expression of the Mitochondrial Genome. Saccone C, Kroon A, editors. New York: North–Holland; 1980. pp. 161–172. [Google Scholar]
- 7.van Ommen G J, Boer P H, Groot G S, de Haan M, Roosendaal E, Grivell L A, Haid A, Schweyen R J. Cell. 1980;20:173–183. doi: 10.1016/0092-8674(80)90245-7. [DOI] [PubMed] [Google Scholar]
- 8.Schmelzer C, Haid A, Grosch G, Schweyen R J, Kaudewitz F. J Biol Chem. 1981;256:7610–7619. [PubMed] [Google Scholar]
- 9.Burke J M. Gene. 1988;73:273–294. doi: 10.1016/0378-1119(88)90493-3. [DOI] [PubMed] [Google Scholar]
- 10.Cech T R, Zaug A J, Grabowski P J. Cell. 1981;27:487–496. doi: 10.1016/0092-8674(81)90390-1. [DOI] [PubMed] [Google Scholar]
- 11.Guo Q, Lambowitz A M. Genes Dev. 1992;6:1357–1372. doi: 10.1101/gad.6.8.1357. [DOI] [PubMed] [Google Scholar]
- 12.Shaw L C, Lewin A S. J Biol Chem. 1995;270:21552–21562. doi: 10.1074/jbc.270.37.21552. [DOI] [PubMed] [Google Scholar]
- 13.Weeks K M, Cech T R. Science. 1996;271:345–348. doi: 10.1126/science.271.5247.345. [DOI] [PubMed] [Google Scholar]
- 14.Labouesse M, Netter P, Schroeder R. Eur J Biochem. 1984;144:85–93. doi: 10.1111/j.1432-1033.1984.tb08434.x. [DOI] [PubMed] [Google Scholar]
- 15.Lamb M R, Anziano P Q, Glaus K R, Hanson D K, Klapper H J, Perlman P S, Mahler H R. J Biol Chem. 1983;258:1991–1999. [PubMed] [Google Scholar]
- 16.Margossian S P, Li H L, Zassenhaus H P, Butow R A. Cell. 1996;84:199–209. doi: 10.1016/s0092-8674(00)80975-7. [DOI] [PubMed] [Google Scholar]
- 17.Lambowitz A M, Belfort M. Annu Rev Biochem. 1993;62:587–622. doi: 10.1146/annurev.bi.62.070193.003103. [DOI] [PubMed] [Google Scholar]
- 18.Schafer B, Wilde B, Massardo D R, Manna F, Del Giudice L, Wolf K. Curr Genet. 1994;25:336–341. doi: 10.1007/BF00351487. [DOI] [PubMed] [Google Scholar]
- 19.Szczepanek T, Lazowska J. EMBO J. 1996;15:3758–3767. [PMC free article] [PubMed] [Google Scholar]
- 20.Dujon B. Gene. 1989;82:91–114. doi: 10.1016/0378-1119(89)90034-6. [DOI] [PubMed] [Google Scholar]
- 21.Cooper A A, Stevens T H. Trends Biochem Sci. 1995;20:351–356. doi: 10.1016/s0968-0004(00)89075-1. [DOI] [PubMed] [Google Scholar]
- 22.Waring R B, Davies R W, Scazzocchio C, Brown T A. Proc Natl Acad Sci USA. 1982;79:6332–6336. doi: 10.1073/pnas.79.20.6332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Michel F, Jacquier A, Dujon B. Biochemistry. 1982;64:867–881. doi: 10.1016/s0300-9084(82)80349-0. [DOI] [PubMed] [Google Scholar]
- 24.Hensgens L A, Bonen L, de Haan M, van der Horst G, Grivell L A. Cell. 1983;32:379–389. doi: 10.1016/0092-8674(83)90457-9. [DOI] [PubMed] [Google Scholar]
- 25.Dujardin G, Jacq C, Slonimski P P. Nature. 1982;298:628–632. doi: 10.1038/298628a0. [DOI] [PubMed] [Google Scholar]
- 26.Hur M. Ph.D. thesis. Philadelphia: Temple University; 1995. [Google Scholar]
- 27.Studier F W, Rosenberg A H, Dunn J J, Dubendorff J W. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 28.Suh E R, Waring R B. Mol Cell Biol. 1990;10:2960–2965. doi: 10.1128/mcb.10.6.2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Waring R B, Davies R W. Gene. 1994;28:277–291. doi: 10.1016/0378-1119(84)90145-8. [DOI] [PubMed] [Google Scholar]
- 30.Wenzlau J M, Saldanha R J, Butow R A, Perlman P S. Cell. 1989;56:421–430. doi: 10.1016/0092-8674(89)90245-6. [DOI] [PubMed] [Google Scholar]
- 31.Partono S, Lewin A S. Mol Cell Biol. 1988;8:2562–2571. doi: 10.1128/mcb.8.6.2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gampel A, Nishikimi M, Tzagoloff A. Mol Cell Biol. 1989;9:5424–5433. doi: 10.1128/mcb.9.12.5424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lazowska J, Claisse M, Gargouri A, Kotylak Z, Spyridakis A, Slonimski P P. J Mol Biol. 1989;205:275–289. doi: 10.1016/0022-2836(89)90341-0. [DOI] [PubMed] [Google Scholar]
- 34.Banroques J, Perea J, Jacq C. EMBO J. 1987;6:1085–1091. doi: 10.1002/j.1460-2075.1987.tb04862.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Coetzee T, Herschlag D, Belfort M. Genes Dev. 1994;8:1575–1588. doi: 10.1101/gad.8.13.1575. [DOI] [PubMed] [Google Scholar]
- 36.Zhang A X, Derbyshire V, Galloway Salvo J L, Belfort M. RNA. 1995;1:783–793. [PMC free article] [PubMed] [Google Scholar]
- 37.Waring R B, Brown T A, Ray J A, Scazzocchio C, Davies R W. EMBO J. 1984;9:2121–2128. doi: 10.1002/j.1460-2075.1984.tb02100.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dujon B, Belfort M, Butow R A, Jacq C, Lemieux C, Perlman P S, Vogt V M. Gene. 1989;82:115–118. doi: 10.1016/0378-1119(89)90035-8. [DOI] [PubMed] [Google Scholar]
- 39.Saldanha R, Mohr G, Belfort M, Lambowitz A M. FASEB J. 1993;7:15–24. doi: 10.1096/fasebj.7.1.8422962. [DOI] [PubMed] [Google Scholar]
- 40.Akins R A, Lambowitz A M. Cell. 1987;50:331–345. doi: 10.1016/0092-8674(87)90488-0. [DOI] [PubMed] [Google Scholar]
- 41.Mohr G, Zhang A, Gianelos J A, Belfort M, Lambowitz A M. Cell. 1992;69:483–494. doi: 10.1016/0092-8674(92)90449-m. [DOI] [PubMed] [Google Scholar]
- 42.Mohr G, Caprara M G, Guo Q, Lambowitz A M. Nature (London) 1994;370:147–150. doi: 10.1038/370147a0. [DOI] [PubMed] [Google Scholar]
- 43.Caprara M G, Mohr G, Lambowitz A M. J Mol Biol. 1996;257:512–531. doi: 10.1006/jmbi.1996.0182. [DOI] [PubMed] [Google Scholar]
- 44.Weeks K M, Cech T R. Biochemistry. 1995;34:7728–7738. doi: 10.1021/bi00023a020. [DOI] [PubMed] [Google Scholar]
- 45.Belfort M, Perlman P S. J Biol Chem. 1995;270:30237–30240. doi: 10.1074/jbc.270.51.30237. [DOI] [PubMed] [Google Scholar]
- 46.Scazzocchio C. Trends Genet. 1989;5:168–172. doi: 10.1016/0168-9525(89)90068-1. [DOI] [PubMed] [Google Scholar]
- 47.Henke R M, Butow R A, Perlman P S. EMBO J. 1995;14:5094–5099. doi: 10.1002/j.1460-2075.1995.tb00191.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kreike J, Schulze M, Ahne F, Lang B F. EMBO J. 1987;6:2123–2129. doi: 10.1002/j.1460-2075.1987.tb02479.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Michel F, Lang B F. Nature (London) 1985;316:641–643. doi: 10.1038/316641a0. [DOI] [PubMed] [Google Scholar]
- 50.Kennell J C, Moran J V, Perlman P S, Butow R A, Lambowitz A M. Cell. 1993;73:133–146. doi: 10.1016/0092-8674(93)90166-n. [DOI] [PubMed] [Google Scholar]
- 51.Moran J V, Mecklenburg K L, Sass P, Belcher S M, Mahnke D, Lewin A S, Perlman P S. Nucleic Acids Res. 1994;22:2057–2064. doi: 10.1093/nar/22.11.2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wool I G. Trends Biochem Sci. 1996;21:164–165. [PubMed] [Google Scholar]
- 53.Lang B F. EMBO J. 1984;3:2129–2136. doi: 10.1002/j.1460-2075.1984.tb02102.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Biniszkiewicz D, Cesnaviciene E, Shub D A. EMBO J. 1994;13:4629–4635. doi: 10.1002/j.1460-2075.1994.tb06785.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Beagley C T, Okada N A, Wolstenholme D R. Proc Natl Acad Sci USA. 1996;93:5619–5623. doi: 10.1073/pnas.93.11.5619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Belfort M. Annu Rev Genet. 1990;24:363–385. doi: 10.1146/annurev.ge.24.120190.002051. [DOI] [PubMed] [Google Scholar]
- 57.Hur M, Waring R B. Nucleic Acids Res. 1995;23:4466–4470. doi: 10.1093/nar/23.21.4466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cech T R. Annu Rev Biochem. 1990;59:543–568. doi: 10.1146/annurev.bi.59.070190.002551. [DOI] [PubMed] [Google Scholar]
- 59.Shaw L C, Lewin A S. Nucleic Acids Res. 1997;25:1597–1604. doi: 10.1093/nar/25.8.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]