Abstract
Nm23 genes, which encode nucleoside diphosphate kinases, have been implicated in suppressing tumor metastasis. The motility of human breast carcinoma cells can be suppressed by transfection with wild-type nm23-H1, but not by transfections with two nm23-H1 mutants, nm23-H1S12OG and nm23-H1P96S. Here we report that nm23-H1 can transfer a phosphate from its catalytic histidine to aspartate or glutamate residues on 43-kDa membrane proteins. One of the 43-kDa membrane proteins was not phosphorylated by either nm23-H1P96S or nm23-H1S120G, and another was phosphorylated much more slowly by nm23-H1P96S and by nm23-H1S120G than by wild-type nm23-H1. Nm23-H1 also can transfer phosphate from its catalytic histidine to histidines on ATP-citrate lyase and succinic thiokinase. The rates of phosphorylation of ATP-citrate lyase by nm23-H1S120G and nm23-H1P96S were similar to that by wild-type nm23-H1. The rate of phosphorylation of succinic thiokinase by nm23-H1S120 was similar to that by wild-type nm23-H1, and the rate of phosphorylation of succinic thiokinase by nm23-H1P96S was about half that by wild-type nm23-H1. Thus, the transfer of phosphate from nm23-H1 to aspartates or glutamates on other proteins appears to correlate better with the suppression of motility than does the transfer to histidines.
Nm23 genes have been implicated in the control of tumor metastasis (1). For many, but not all, types of tumors, there is an inverse relationship between the level of nm23 expression and metastatic potential (reviewed in ref. 2). Overexpression of nm23 in highly metastatic murine melanoma cell lines (3–6), rat mammary adenocarcinoma cells (7), and human breast carcinoma cells (4, 8, 9) reduces their metastatic potentials, indicating that nm23 genes can suppress tumor metastasis. Increased levels of nm23 expression also have been associated with both cellular proliferation (10) and differentiation (11). In human tissues the two major forms of nm23 are nm23-H1 and nm23-H2. Expression of nm23-H1 has been correlated with suppression of metastasis (2).
The abnormal wing disc (awd) gene of Drosophila encodes a protein that is 78% identical to the human nm23 proteins (12). The awd gene is essential for normal Drosophila development (13). The killer of prune mutation in this gene (awdkpn) causes a dominant lethality but only in flies that do not have a functional prune gene. The prune gene has been cloned, but the biochemical function of the resulting protein is not known (14, 15).
Nm23 and awd genes encode nucleoside diphosphate kinases (NDPKs) (12). NDPKs catalyze the phosphorylation of nucleoside 5′-diphosphates to triphosphates (16). In the first step, a phosphate is transferred from a nucleoside 5′-triphosphate to a histidine at the catalytic site of the enzyme. This high-energy phosphate then is transferred to a phosphate group on a nucleoside 5′-diphosphate.
The synthesis of nucleoside triphosphates does not appear to explain the role of nm23 in suppressing metastasis (2). Similarly, the effect of the awdkpn mutation on Drosophila development does not appear to result from a change in nucleoside diphosphate kinase activity (17). However, the kinase activities of nm23/NDPK do appear to be necessary for their regulating these processes (18, 19). One possible explanation for this discrepancy is that in addition to phosphorylating nucleoside diphosphates NDPK can phosphorylate other substrates. Rat liver NDPK can phosphorylate the histidine at the catalytic site of ATP-citrate lyase (20), nm23-H1 can phosphorylate a histidine on succinic thiokinase (21), and NDPKs transfer a phosphate to serines or threonines on several proteins in a heat-inactivated cell extract in the presence of 1 M urea (22).
Transfection with wild-type nm23-H1 suppresses the motility of human breast carcinoma cells (23), but transfections with nm23-H1P96S and by nm23-H1S120G do not suppress the motility of these cells. The serine 120 to glycine mutation of nm23-H1S120G was found in 6 of 28 aggressive childhood neuroblastomas (24). The nm23-H1P96S contains a proline to serine substitution at residue 96 and is homologous to the Drosophila awdkpn mutation (12, 23).
Here we report that the rates of phosphorylation of ATP-citrate lyase by nm23-H1S120G and nm23-H1P96S were similar to that by wild-type nm23-H1. We also describe the phosphorylation of a 43-kDa protein or proteins from bovine brain membranes by rat liver NDPK and nm23-H1. This phosphorylation occurred on either an aspartate or glutamate residue. The phosphorylation of aspartate or glutamate residues by NDPK is analogous to phosphorylation reactions carried out by prokaryotic histidine kinases (25). The 43-kDa protein(s) either were not phosphorylated or phosphorylated much more slowly by nm23-H1P96S and nm23-H1S120G than by wild-type nm23-H1.
EXPERIMENTAL PROCEDURES
Rat Liver NDPK, PC12 Cell ATP-Citrate, and Porcine Succinic Thiokinase.
Rat liver NDPK and PC12 cell ATP-citrate lyase were purified as described previously (20). Porcine heart succinic thiokinase (succinyl CoA synthetase) was purchased from Sigma.
Purification of Recombinant nm23-H1, nm23-H1P96S, and nm23-H1S120G.
Escherichia coli strain BL21(DE3) containing expression plasmids for nm23-H1, nm23-H1P96S, and nm23-H1S120G (21, 26) were grown overnight in Luria–Bertani (LB) media containing 100 μg/ml ampicillin at 37°C. Aliquots were diluted 1/100 in LB media and grown until A600 was 0.8. Expression of nm23 cDNAs was induced by the addition of 1 mM isopropyl-1-thio β-d-galactopyranoside. After 2 hr, the cells were pelleted and washed twice with 20 mM Tris and 1 mM EDTA, pH 8.4 at 4°C.
Cells (5 g) were sonicated at 4°C in 25 ml of 20 mM Tris, pH 8.4/1 mM EDTA/2 mM MgCl2/1 mM DTT/0.2 mM phenylmethylsulfonyl fluoride (PMSF)/1 μg/ml leupeptin/2 mM benzamidine/0.1 mg/ml lysozyme. Cell debris was removed by centrifugation at 20,000 × g for 20 min at 4°C. The resulting supernatant was fractionated with (NH4)2SO4. The 40–75% (NH4)2SO4 pellet was dialyzed overnight at 4°C against buffer A (20 mM Tris, pH 7.5/2 mM MgCl2/1 mM EDTA/1 mM DTT/0.2 mM PMSF/2 mM benzamidine). The dialysate was made to 0.3 M NaCl, clarified, and loaded onto a Pharmacia S200 column (1.6 × 80 cm) equilibrated in buffer A containing 0.3 M NaCl. Column fractions were assayed for NDPK activity (16). Peak fractions were pooled, concentrated, and dialyzed overnight against 20 mM Tris, pH 8.0/2 mM MgCl2/1 mM EDTA/100 mM NaCl/1 mM DTT/2 mM benzamidine. The dialysate was clarified and applied to a Pharmacia Mono Q HR 5/5 column equilibrated in this buffer. Nm23-H1 was eluted with a linear gradient of 0.1–0.4 M NaCl. Peak fractions containing nm23-H1 were pooled, diluted, and applied to a Mono Q HR 5/5 column equilibrated in 20 mM Tris, pH 7.5/2 mM MgCl2/1 mM EDTA/100 mM NaCl/1 mM DTT/2 mM benzamidine. Nm23-H1 was eluted using linear gradient of 0.1–0.2 M NaCl. Nm23-H1P96S and nm23-H1S120G were purified using the same isolation procedure. Nm23-H1, nm23-H1P96S, and nm23-H1S120G were greater than 99% pure.
Phosphorylation of Rat Liver NDPK and nm23 Proteins.
Rat liver NDPK (20 μM) was incubated with 100 μM [γ-32P]ATP (4 × 1015 dpm/mol) for 10 min in 100 mM NaCl/5 mM MgCl2/1 mM DTT/20 mM Tris, pH 7.5. Phosphorylated NDPK, [32P]NDPK, was separated from [γ-32P]ATP by gel filtration on a PD10 column (Pharmacia) equilibrated in 100 mM NaCl/1 mM DTT/20 mM Tris, pH 7.5 (20). The same procedure was used to make phosphorylated nm23 proteins.
Phosphorylation of ATP-Citrate Lyase, Succinic Thiokinase, and Bovine Brain Membrane Extract by [32P]NDPK and [32P]nm23-H1.
Protein samples (ATP-citrate lyase, succinic thiokinase, or column fractions from a bovine brain membrane extract) were incubated with either rat liver [32P]NDPK or [32P]nm23-H1 at room temperature in 100 mM NaCl/5 mM MgCl2/1 mM DTT/20 mM Tris, pH 7.5 (membrane fractions also contained 0.25–0.5% sodium cholate and either 1 mM EDTA or 0.5 mM EGTA). After 15 min or the time indicated, the phosphorylation was stopped by the addition of 0.5 volume of SDS sample buffer (5% SDS/25% glycerol/180 mM Tris, pH 8.8/0.01% bromophenol blue/50 mM DTT/50 μg/ml leupeptin/4 mM PMSF). Unless stated otherwise, the samples were frozen on dry ice, thawed, and loaded without boiling onto a SDS/polyacrylamide gel. After electrophoresis, the gels were dried without fixing. Phosphorylated proteins were detected by autoradiography.
Partial Purification of a Protein from Bovine Brain Membranes That Can Be Phosphorylated by NDPK.
Membranes (100 g) from the gray matter of bovine brains (27) in 50 mM Tris, pH 7.5/5 mM EDTA/10% sucrose were extracted with 150 ml of 3% sodium cholate/50 mM NaCl/2 mM DTT/0.4 mM PMSF/10 μg/ml leupeptin/10 μg/ml soybean trypsin inhibitor by stirring for 30 min at 4°C. The extract was clarified by centrifugation at 150,000 × g for 90 min and loaded onto a DEAE Sephacel column (2.6 × 40 cm) equilibrated in 1% sodium cholate/25 mM NaCl/1 mM EGTA/1 mM DTT/20 mM Tris, pH 7.5 at 4°C. The column was washed with 100 ml of equilibration buffer and then eluted with a 600-ml linear gradient of 25–425 mM NaCl. Fractions (10 ml) were collected, and 50-μl aliquots incubated with 0.4–0.8 μM [32P]NDPK and analyzed by SDS/PAGE and autoradiography.
Fractions 16–24 (flow through) from the DEAE Sephacel column were pooled and loaded onto a 1.6 × 30-cm Blue Sepharose column (Pharmacia) equilibrated in 1% sodium cholate/25 mM NaCl/2 mM EDTA/1 mM DTT/0.2 mM PMSF/20 mM Tris, pH 7.5 at 4°C. The column was washed with 100 ml of equilibration buffer and then eluted with a 600-ml linear gradient of 0–400 mM NaCl. Fractions (10 ml) were collected, and 50-μl aliquots incubated with 0.4–0.8 μM [32P]NDPK and analyzed by SDS/PAGE and autoradiography. Fractions from this column enriched in a 43-kDa protein, which was phosphorylated when incubated with [32P]NDPK, were pooled, concentrated by ultrafiltration using an Amicon PM10 membrane, and loaded onto an 1.6 × 90-cm Sephacryl S200 column equilibrated in 0.5% sodium cholate/100 mM NaCl/2 mM EGTA/2 mM MgCl2/1 mM DTT/0.1 mM PMSF/20 mM Tris, pH 7.5 at 4°C. Fractions (4 ml) were collected, and 50-μl aliquots incubated with 0.4–0.8 μM [32P]NDPK and analyzed by SDS/PAGE and autoradiography. Fractions from this column enriched in a 43-kDa protein, which was phosphorylated when incubated with [32P]NDPK, were pooled and concentrated to 2.5 ml by ultrafiltration. This fraction is referred to as the Blue Sepharose fraction.
Fractions 31–42 from the DEAE Sephacel column were pooled, and proteins precipitating in 25–45% saturated (NH4)2SO4 dialyzed overnight against 0.5% sodium cholate/100 mM NaCl/2 mM EGTA/2 mM MgCl2/1 mM DTT/0.1 mM PMSF/20 mM Tris, pH 7.5 at 4°C and fractionated on 1.6 × 90-cm Sephacryl S200 column as described above. Fractions from the S200 column enriched in a 43-kDa protein, which was phosphorylated when incubated with [32P]NDPK, were pooled and concentrated to 2.5 ml by ultrafiltration. This sample is referred to as S200-B.
RESULTS
Phosphorylation of a Membrane 43-kDa Protein by NDPK.
Rat liver NDPK was phosphorylated on its active site histidine by incubation with [γ-32P]ATP, and phosphorylated NDPK, [32P]NDPK, was separated from [γ-32P]ATP by gel filtration (20). After gel filtration, NDPK contained 0.4–0.5 mol of 32P/mol of NDPK 18-kDa subunit. The concentrations of [32P]NDPK given in this paper refer to the concentration of 32P bound to the NDPK. Thin-layer chromatography showed that 1 μM [32P]NDPK contained less than 10 nM [γ-32P]ATP.
Incubation of PC12 cell cytosolic extract with [32P]NDPK results in primarily the phosphorylation of ATP-citrate lyase (20). A variety of other extracts also have been examined for potential substrates for [32P]NDPK. A persistent problem in the identification of protein substrates for [32P]NDPK has been the formation of [γ-32P]ATP when [32P]NDPK reacts with ADP present in cell or tissue extracts. The formation of [γ-32P]ATP was a problem even after several days of dialysis and/or column fractionations, and it was frequently difficult to distinguish between direct transfer of phosphate from [32P]NDPK to a protein and indirect transfer through the formation of [γ-32P]ATP. For this reason, we chose to try to identify proteins that were phosphorylated by [32P]NDPK but not by either [γ-32P]ATP or [γ-32]GTP. Below we describe the partial purification of one, and possibly two, proteins that appear to be directly phosphorylated by [32P]NDPK.
Washed bovine brain membranes were extracted with sodium cholate and fractionated on a DEAE Sephacel column. A large number of proteins in the fractions from this column were phosphorylated when incubated with [32P]NDPK, but additional fractionation indicated that most of these phosphorylations resulted from the formation of [γ-32P]ATP. On the basis of the phosphorylation pattern, fractions from this column were pooled and subjected to further fractionation.
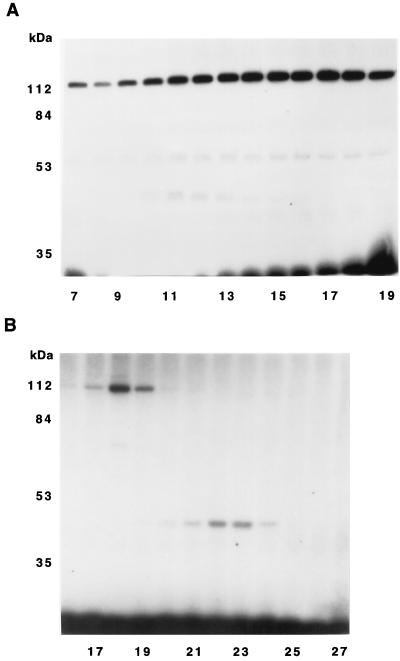
Proteins that eluted from the DEAE column with equilibration buffer were fractionated on a Blue Sepharose column. Fractions from the Blue Sepharose column were incubated with [32P]NDPK, and protein phosphorylation examined by SDS/PAGE and autoradiography (Fig. 1A). (After drying, the SDS gels were cut to remove the [32P]NDPK, which was much more radioactive than the other protein bands.) Three major phosphorylated polypeptides (120 kDa, 55 kDa, and 43 kDa) were observed. Fractions 10–13 were pooled, concentrated, and fractionated on a S200 column. Fractions from this column were incubated with [32P]NDPK and protein phosphorylation examined by SDS/PAGE and autoradiography (Fig. 1B). Fractions containing a 43-kDa protein that was phosphorylated when incubated with [32P]NDPK were pooled and concentrated. This sample is referred to as the Blue Sepharose fraction.
Figure 1.
An autoradiogram showing the partial purification of a 43-kDa protein phosphorylated by [32P]NDPK. (A) Fractions 16–24 from the DEAE Sephacel column were pooled and fractionated on a Blue Sepharose column. (B) Fractions 10–13 from the Blue Sepharose column were pooled, concentrated, and fractionated on a S200 column. Aliquots (50 μl) of the column fractions were incubated with 0.4 μM [32P]NDPK for 15 min at room temperature. The samples were analyzed by SDS/PAGE and autoradiography.
Fractions 25–30 (pool A) and fractions 31–42 (pool B) from the DEAE column were pooled separately, and proteins precipitating in 25–45% saturated (NH4)2SO4 dialyzed and fractionated on S200 columns. Fractions from these columns were incubated with [32P]NDPK, and protein phosphorylation examined by SDS/PAGE and autoradiography. Fractions from the S200 column of pool B containing a 43-kDa protein that was phosphorylated when incubated with [32P]NDPK (fractions 23–25) were pooled and concentrated. This sample is referred to as S200-B fraction. None of the fractions from the S200 column of pool A contained significant amounts of proteins that could be phosphorylated by [32P]NDPK.
Transfer of 32P From a Histidine on NDPK to a Aspartate or Glutamate on the 43-kDa Protein(s).
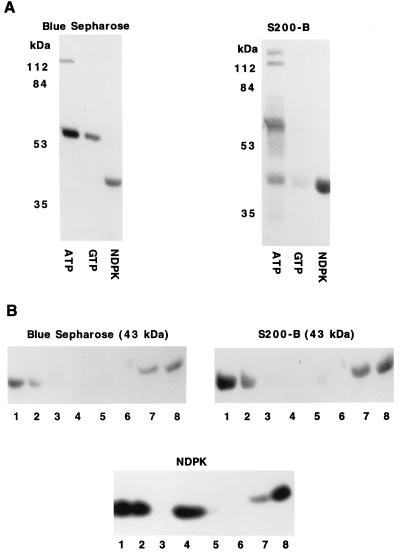
The 43-kDa protein in the Blue Sepharose fraction was not phosphorylated when this fraction was incubated with [γ-32P]ATP or [γ-32P]GTP, but it was phosphorylated by [32P]NDPK (Fig. 2A). A 43-kDa protein in the S200-B fractions was phosphorylated when this fraction was incubated with either [γ-32P]ATP or with [32P]NDPK, but the extent of phosphorylation by [32P]NDPK was more than five times that by [γ-32P]ATP and more than 10 times that by [γ-32P]GTP (Fig. 2A). Several other proteins in the Blue Sepharose and S200-B fractions were phosphorylated when these fractions were incubated [γ-32P]ATP, but they were not phosphorylated by [32P]NDPK. The S200-B fraction contained some endogenous NDPK (data not shown), which may have contributed to the phosphorylation of the 43-kDa protein when this fraction was incubated with [γ-32P]ATP.
Figure 2.
Phosphorylation of 43-kDa proteins in the Blue Sepharose and S200-B fraction by [32P]NDPK. (A) Blue Sepharose and S200-B fractions were incubated for 15 min at room temperature with 1 μM [γ-32P]ATP, 1 μM [γ-32P]GTP, or 1 μM [32P]NDPK. The samples were analyzed by SDS/PAGE and autoradiography. (B) Blue Sepharose and S200-B fractions were incubated for 15 min at room temperature with 1 μM [32P]NDPK. After these incubations, the samples were treated as follows: lanes 1, frozen immediately after adding SDS sample buffer; lanes 2, incubated for 10 min at 37°C in SDS sample buffer; lanes 3, incubated for 10 min at 37°C in SDS sample buffer containing 0.5 m HCl; lanes 4, incubated for 10 min at 37°C in SDS sample buffer containing 0.5 M NaOH; lanes 5, incubated in SDS sample buffer in boiling water for 2 min; lanes 6, incubated for 10 min at 37°C in 0.8 M hydroxylamine at pH 5.4; lanes 7, incubated for 10 min at 37°C in 0.8 M NaCl at pH 5.4; and lanes 8, incubated for 10 min at 37°C at pH 7.5. After these incubations, the samples were neutralized or SDS sample buffer added, and they were analyzed by SDS/PAGE and autoradiography.
The phosphates incorporated into the 43-kDa protein(s) in both the Blue Sepharose and S200-B fractions by incubation with [32P]NDPK were acid labile (Fig. 2B, lanes 3), base labile (Fig. 2B, lanes 4), lost upon boiling (Fig. 2B, lanes 5), and removed by hydroxylamine (Fig. 2B, lanes 6). These stability measurements indicate that the phosphorylated residues were either aspartates or glutamates as acylphosphates are unstable at both high and low pH (28–30). The phosphohistidine in NDPK was base stable (Fig. 2B, lane 4). Simply incubating the phosphorylated 43-kDa protein(s) in SDS sample buffer for 10 min at 37°C resulted in a substantial loss of 32P (Fig. 2B, lanes 2). The phosphate incorporated into the 43-kDa protein in the S200-B fraction by incubation with [γ-32P]ATP was acid labile, base labile, and lost upon boiling, indicating that it was either a phosphoaspartate or phosphoglutamate.
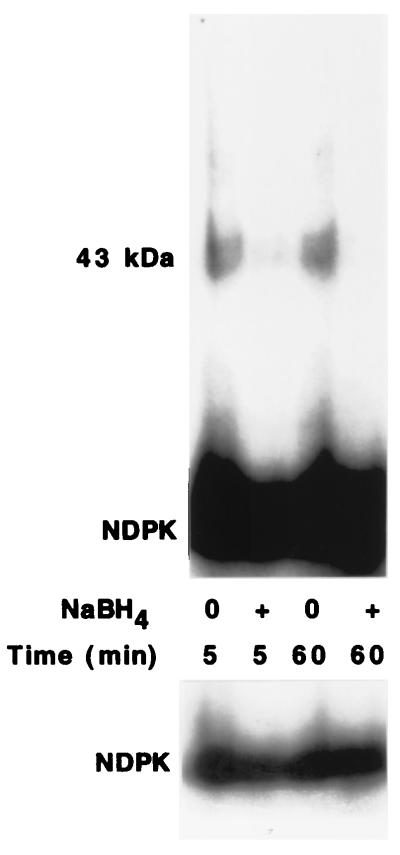
Phosphoaspartate and phosphoglutamate can be reduced with NaBH4 (29, 30) to give free phosphate and either δ-hydroxy-α-amino-valeric acid or homoserine. Phosphohistidines are not reduced by NaBH4. The phosphates incorporated in the 43-kDa protein(s) by incubation with [32P]NDPK were lost upon treatment with NaBH4 (Fig. 3), but the phosphate incorporated into NDPK was not removed by NaBH4.
Figure 3.
Removal of 32P from the 43-kDa protein by NaBH4. The 43-kDa protein in the Blue Sepharose fraction was phosphorylated by incubation with 0.6 μM [32P]NDPK for 10 min at room temperature. The proteins were precipitated on ice in 16% trichloroacetic acid. The protein pellets were rinsed with 1 ml of cold 10 mM HCl and dried under vacuum. The pellets were dissolved in dimethyl sulfoxide and incubated at room temperature in the presence and absence of 25 mM NaBH4 (30). After 5 or 60 min, SDS sample buffer was added, and the samples analyzed by SDS/PAGE and autoradiography. The lower panel shows a shorter exposure of the autoradiogram of the part of the gel containing [32P]NDPK.
The phosphorylation of the 43-kDa protein(s) in both the Blue Sepharose and S200-B fractions by 0.4 μM [32P]NDPK was rapid with maximum incorporation occurring within 1 min at room temperature. Incubation of these fractions in boiling water for 1 min greatly reduced or eliminated the subsequent phosphorylation of the 43-kDa protein(s) by [32P]NDPK, and EDTA blocked the phosphorylation of the 43-kDa protein(s) by [32P]NDPK.
Purity of the 43-kDa Proteins.
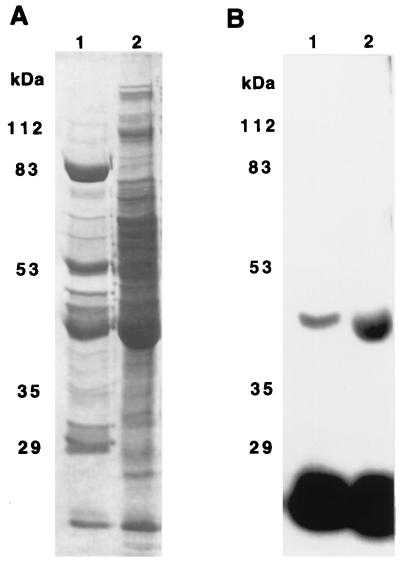
Coomassie blue-stained gels of the Blue Sepharose and S200-B fractions showed that these fractions contained many different proteins (Fig. 4A). The 43-kDa polypeptides phosphorylated by [32P]NDPK (Fig. 4B) aligned with one of the minor bands just above the major bands at 42 kDa seen in Coomassie blue-stained gels. The 43-kDa protein in the Blue Sepharose fraction moved slightly slower on SDS/PAGE than did the 43-kDa protein in the S200-B fraction (Fig. 4B). The 43-kDa protein(s) in both the Blue Sepharose and S200-B fractions may be oligomeric or bound to other proteins as they eluted from the S200 column with an apparent molecular mass greater than 120 kDa. At this time, it is not known whether the 43-kDa proteins in these two fractions are the same protein.
Figure 4.
Comparison of 43-kDa protein in Blue Sepharose and S200-B fractions by SDS/PAGE. Blue Sepharose (lanes 1) and S200-B (lanes 2) fractions were incubated for 15 min at room temperature with 250 nM [32P]NDPK. After these incubations the samples were separated on SDS/PAGE. The gel was stained with Coomassie blue in 10% acetic acid and 25% isopropanol for 1.5 hr, destained in 10% acetic acid for 2 hr, and dried. A is the Coomassie brilliant blue-stained gel, and B is the autoradiogram of this gel.
Based on 32P incorporation and assuming stoichiometric phosphorylation, the Blue Sepharose and S200-B fractions were estimated to contain 40–100 nM 43-kDa proteins.
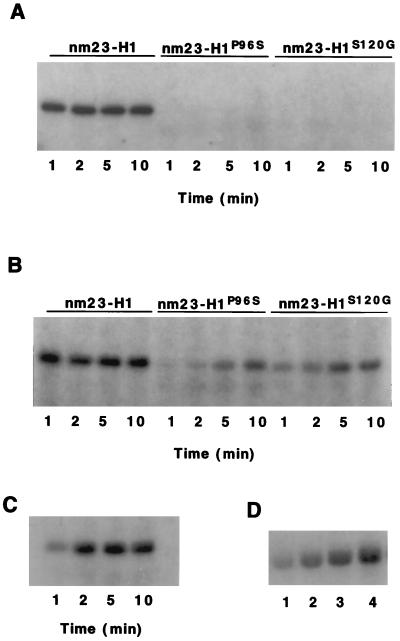
The 43-kDa Protein in the Blue Sepharose Fraction Was Not Phosphorylated by Either nm23-H1P96S or nm23-H1S120G.
The phosphorylation of the 43-kDa proteins in both the Blue Sepharose and S200-B fractions by [32P]nm23-H1 and two mutant forms of nm23-H1, [32P]nm23-H1P96S and [32P]nm23-H1S120G, were examined (Fig. 5). The rates phosphorylation of the 43-kDa proteins by 0.4 μM [32P]nm23-H1 were rapid with maximum phosphorylations occurring within 1 min. In contrast, there was no detectable phosphorylation of the 43-kDa protein in the Blue Sepharose fraction by either [32P]nm23-H1P96S or [32P]nm23-H1S120G (Fig. 5A). The rate of phosphorylation of the 43-kDa protein in the S200-B fraction (Fig. 5B) by [32P]nm23-H1P96S was less than 10% of that by [32P]nm23-H1, and the rate phosphorylation by [32P]nm23-H1S120G was about 20% that by [32P]nm23-H1 (Table 1). Under similar conditions, the nucleoside diphosphate kinase activities of nm23-H1P96S and nm23-H1S120G were about 90% those of nm23-H1.
Figure 5.
Phosphorylation of 43-kDa proteins in Blue Sepharose and S200-B fractions by nm23-H1, nm23-H1P96S, and nm23-H1S120G. (A) The Blue Sepharose fraction was incubated with 370 nM [32P]nm23-H1, [32P]nm23-H1P96S, or [32P]nm23-H1S120G for 1, 2, 5, and 10 min. (B) The S200-B fraction was incubated with 500 nM [32P]nm23-H1, [32P]nm23-H1P96S, or [32P]nm23-H1S120G for 1, 2, 5, and 10 min. (C) Time course of phosphorylation. The Blue Sepharose fraction was incubated with 170 nM [32P]nm23-H1 for 1, 2, 5, and 10 min. (D) Phosphorylation by varying concentrations of nm23-H1. The Blue Sepharose fraction was incubated for 2 min with (lane 1) 40 nM, (lane 2) 90 nM, (lane 3) 180 nM, and (lane 4) 280 nM [32P]nm23-H1. The samples were analyzed by SDS/PAGE and autoradiography.
Table 1.
Comparison of the protein phosphotransferase activities of nm23-H1, nm23-H1P96S, and nm23-H1S120G
| Substrate | Relative rates of phosphorylation by [32P]nm23-H1
|
||
|---|---|---|---|
| Wild type | P96S | S120G | |
| ATP-citrate lyase | 100 | 80-100 | 90-100 |
| Succinic thiokinase | 100 | 40-70 | 70-100 |
| 43-kDa protein in Blue Sepharose fraction | 100 | 0 | 0 |
| 43-kDa protein in S200-B fraction | 100 | 5-10 | 15-20 |
The rates of phosphorylation of the different substrates are expressed relative to the rate of phosphorylation of that substrate by wild-type nm23-H1. The range of values reflect results obtained with different preparations of [32P]nm23 proteins, and reactions carried out on different substrate preparations.
The time course of the phosphorylation of the 43-kDa protein in the Blue Sepharose fraction by 170 nM [32P]nm23-H1 is shown in Fig. 5C. At this concentration maximum phosphorylation occurred within 2 min. The dependence of phosphorylation of the 43-kDa protein in the Blue Sepharose fraction on [32P]nm23-H1 concentration is shown in Fig. 5D.
Phosphorylation of ATP-Citrate Lyase and Succinic Thiokinase by nm23-H1, nm23-H1P96S, and nm23-H1S120G.
When ATP-citrate lyase is incubated with [32P]NDPK, phosphate is transferred from the catalytic histidine on NDPK to the catalytic histidine on ATP-citrate lyase (20). The rate of phosphorylation of ATP-citrate lyase by [32P]nm23-H1P96S and [32P]nm23-H1S120G were similar to that by wild-type [32P]nm23-H1 (Table 1).
It recently has been reported that nm23-H1 can phosphorylate succinic thiokinase on a histidine residue (21). The rates phosphorylation of succinic thiokinase by [32P]nm23-H1P96S and [32P]nm23-H1S120G were compared with that by [32P]nm23-H1. Whereas the rate of phosphorylation of succinic thiokinase by [32P]nm23-H1S120G was comparable to that by [32P]nm23-H1, the rate of phosphorylation of succinic thiokinase by [32P]nm23-H1P96S was less than by [32P]nm23-H1 (Table 1). This lower rate of succinic thiokinase phosphorylation was observed using several different preparations of nm23-H1P96S. Freije et al. (21) reported that the phosphorylation of succinic thiokinase by [32P]nm23-H1P96S is about 15% that by [32P]nm23-H1.
DISCUSSION
Although a number of studies have demonstrated that expression of nm23 cDNAs in highly metastatic cells lines reduces their metastatic potentials, the mechanisms by which it accomplishes this is unknown (2). The nucleoside diphosphate kinase activity of nm23 doesn’t appear to account for its ability to inhibit metastasis (2). A number of alternate activities for nm23 have been suggested. Several papers have reported that NDPK can function as a protein kinase or protein phosphotransferase (20–22).
The present study provides additional data on the phosphorylation of ATP-citrate lyase by rat liver NDPK and recombinant nm23-H1 and confirms the phosphorylation of succinic thiokinase by nm23-H1. It also describes the transfer of a phosphate from a histidine on NDPK to aspartate or glutamate residues on bovine brain membrane proteins and compares the protein kinase activities of two mutant forms of nm23-H1 with those of wild-type nm23-H1.
The phosphorylations of ATP-citrate lyase and succinic thiokinase by NDPK appear to be useful model systems to characterize the protein kinase activity of NDPK. However, the biological significance of these phosphorylations is unclear. NDPK phosphorylates histidines at the catalytic sites of ATP-citrate lyase and succinic thiokinase (20). These histidines are phosphorylated much more rapidly by ATP and GTP. NDPK and nm23-H1 had no obvious effects on the enzymatic activities of ATP-citrate lyase or succinic thiokinase (unpublished observations).
The phosphorylation of a bovine brain membrane 43-kDa protein or proteins by rat liver NDPK or nm23-H1 is analogous to phosphorylation reactions carried out by prokaryotic histidine kinases (25). These histidine kinases autophosphorylate on a histidine residue and then transfer the phosphate to an acyl group, usually an aspartate, on the same or different proteins. These bacterial “two-component signaling” systems regulate many different activities, including chemotaxis, osomoregulation, sporulation, and gene regulation (25). Two-component histidine kinases have been identified in Saccharomyces cerevisiae (31–33) and Arabidopsis thaliana (34). Yeast SLN1 histidine kinase is involved in sensing changes in osmomolarity, and Arabidopsis ETR1 protein is involved in sensing changes in ethylene.
The 43-kDa protein in the Blue Sepharose fraction from extracts of bovine brain was phosphorylated by rat liver NDPK and nm23-H1. It did not autophosphorylate, and there was no phosphorylation when NDPK was not added. The 43-kDa protein in the S200-B fraction from extracts of bovine brain also was phosphorylated by rat liver NDPK and nm23-H1. However, there was a low level of phosphorylation of this protein in the absence of added NDPK. The S200-B fraction contained a low amount of NDPK, and the phosphorylation of the 43-kDa protein observed when only ATP was added may have resulted from its phosphorylation by endogenous NDPK.
Human MDA-MB-435 breast carcinoma cells have been transfected with wild-type and mutant forms of nm23-H1 (23). Transfection with wild-type nm23-H1 suppresses the motility of these cells to either fetal calf serum or to autotaxin. Transfection with nm23-H1P96S or nm23-H1S12OG does not suppress the motility of these cells. The inability of these mutants to suppress motility has been compared with their reduced histidine kinase activities. Nm23-H1P96S has normal autophosphorylation and nucleoside diphosphate kinase activities, but it has lower histidine kinase activities when succinic thiokinase and other nm23 proteins were used as substrates (21). However, the rate of phosphorylation of ATP-citrate lyase by nm23-H1P96S was about the same by wild-type nm23-H1 (Table 1). Nm23-H1S120G has only slightly less histidine activity than does wild-type nm23-H1 when either succinic thiokinase (21) or ATP-citrate lyase were used as substrates (Table 1).
The phosphorylation of the 43-kDa protein(s) on an aspartate or glutamate residue by nm23-H1S120G and nm23-H1P96S was much slower than by wild-type nm23-H1. When the Blue Sepharose fraction was used, the 43-kDa protein was maximally phosphorylated by nm23-H1 in 1 min, but there was no detectable phosphorylation of the 43-kDa protein after 10-min incubation with either nm23-H1P96S or nm23-H1S120G. The rate of phosphorylation of the 43-kDa protein in the S200-B fraction by nm23-H1 was about five times faster than by nm23-H1S120G and about 10 times faster than by nm23-H1P96S. As the S200-B fraction contained a low level of endogenous NDPK, it is possible that the phosphorylation of the 43-kDa protein might result from the transfer of phosphate from the added [32P]nm23 to the endogenous NDPK and then to the 43-kDa protein. Both nm23-H1P96S or nm23-H1S120G can phosphorylate other nm23 proteins (21).
Whereas there is currently no direct link between the inability of m23-H1P96S and nm23-H1S120G to suppress motility in breast carcinoma cells and their low kinase activity toward the 43-kDa protein(s), these observations do suggest that this transfer of phosphate from the histidine on nm23 to aspartate or glutamate residues on other proteins may be biologically important. Two-component histidine kinases are involved in regulating a number of cell signaling processes in both prokaryotes and eukaryotes (25). For example, the yeast SLN1 histidine kinase regulates the HOG1 MAP kinase cascade (32). Several papers have suggested that nm23 has a role in signal transduction (3, 4, 35, 36). The results presented here demonstrate that NDPK has a two-component histidine kinase-like activity. It seems possible that NDPK phosphorylates proteins that are part of two-component signaling systems. The suppression of metastasis by nm23 could result from its phosphorylating proteins that are part of a two-component signaling pathway that regulates metastasis.
ABBREVIATIONS
- NDPK
nucleoside diphosphate kinase
- PMSF
phenylmethylsulfonyl fluoride
References
- 1.Steeg P S, Bevilacqua G, Kopper L, Thorgeirsson U P, Talmadge J E, Liotta L A, Sobel M E. J Natl Cancer Inst. 1988;80:200–204. doi: 10.1093/jnci/80.3.200. [DOI] [PubMed] [Google Scholar]
- 2.DeLaRosa A, Williams R L, Steeg P S. BioEssays. 1995;17:53–62. doi: 10.1002/bies.950170111. [DOI] [PubMed] [Google Scholar]
- 3.Leone A, Flatow U, King C R, Sandeen M, A, Margulies, Liotta L A, Steeg P S. Cell. 1991;65:25–35. doi: 10.1016/0092-8674(91)90404-m. [DOI] [PubMed] [Google Scholar]
- 4.Kantor J D, McCormick B, Steeg P S, Zetter B R. Cancer Res. 1993;53:1971–1973. [PubMed] [Google Scholar]
- 5.Parhar R S, Shi Y, Zou M, Farid N R, Ernst P, Al-Sedairy S. Int J Cancer. 1995;60:204–210. doi: 10.1002/ijc.2910600213. [DOI] [PubMed] [Google Scholar]
- 6.Baba H, Urano T, Okada K, Furukawa K, Nakayama E, Tanaka H, Iwasaki K, Shiku H. Cancer Res. 1995;55:1977–1981. [PubMed] [Google Scholar]
- 7.Fukuda M, Ishii A, Yasutomo Y, Shimada N, Ishikawa N, Hanai N, Nagata N, Irimura T, Nicolson G L, Kimura N. Int J Cancer. 1996;65:531–537. doi: 10.1002/(SICI)1097-0215(19960208)65:4<531::AID-IJC23>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 8.Leone A, Flatow U, VanHoutte K, Steeg P S. Oncogene. 1993;8:2325–2333. [PubMed] [Google Scholar]
- 9.Howlett A R, Petersen O W, Steeg P S, Bissell M J. J Natl Cancer Inst. 1994;86:1838–1844. doi: 10.1093/jnci/86.24.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Keim D, Hailat N, Melhem R, Zhu X X, Lascu I, Veron M, Strahler J, Hanash S M. J Clin Invest. 1992;89:919–924. doi: 10.1172/JCI115672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lakso M, Steeg P S, Westphal H. Cell Growth Differ. 1992;3:873–879. [PubMed] [Google Scholar]
- 12.Biggs J, Hersperger E, Steeg P S, Liotta L A, Shearn A. Cell. 1990;63:933–940. doi: 10.1016/0092-8674(90)90496-2. [DOI] [PubMed] [Google Scholar]
- 13.Dearolf C, Tripoulas N, Biggs J, Shearn A. Dev Biol. 1988;129:169–178. doi: 10.1016/0012-1606(88)90171-6. [DOI] [PubMed] [Google Scholar]
- 14.Teng D H, Engele C M, Venkatesh T R. Nature (London) 1991;353:437–440. doi: 10.1038/353437a0. [DOI] [PubMed] [Google Scholar]
- 15.Frolov M V, Zverlov V V, Alatortsev V E. Mol Gen Genet. 1994;242:478–483. doi: 10.1007/BF00281800. [DOI] [PubMed] [Google Scholar]
- 16.Parks R E, Agarwal R P. In: The Enzymes. 3rd Ed. Boyer P D, editor. Vol. 8. New York: Academic; 1973. pp. 307–334. [Google Scholar]
- 17.Lascu I, Chaffotte A, Limbourg-Bouchon B, Veron M. J Biol Chem. 1992;267:12775–12781. [PubMed] [Google Scholar]
- 18.Timmons L, Xu J, Hersperger G, Deng X-F, Shearn A. J Biol Chem. 1995;270:23021–23030. doi: 10.1074/jbc.270.39.23021. [DOI] [PubMed] [Google Scholar]
- 19.Xu J, Liu L Z, Deng X F, Timmons L, Hersperger E, Steeg P S, Veron M, Shearn A. Dev Biol. 1996;177:544–557. [PubMed] [Google Scholar]
- 20.Wagner P D, Vu N-D. J Biol Chem. 1995;270:21758–21764. doi: 10.1074/jbc.270.37.21758. [DOI] [PubMed] [Google Scholar]
- 21.Freije J M P, Blay P, MacDonald N J, Manrow R E, Steeg P S. J Biol Chem. 1997;272:5525–5532. doi: 10.1074/jbc.272.9.5525. [DOI] [PubMed] [Google Scholar]
- 22.Engel M, Veron M, Theisinger B, Lacombe M-L, Seib T, Dooley S, Welter C. Eur J Biochem. 1995;234:200–207. doi: 10.1111/j.1432-1033.1995.200_c.x. [DOI] [PubMed] [Google Scholar]
- 23.MacDonald N J, Freije J M P, Stracke M L, Manrow R E, Steeg P S. J Biol Chem. 1996;271:25107–25116. doi: 10.1074/jbc.271.41.25107. [DOI] [PubMed] [Google Scholar]
- 24.Chang C, Zhu X, Thoraval D, Ungar D, Rawwas J, Hora N, Strahler J, Hanash S, Radany E. Nature (London) 1994;370:335–336. doi: 10.1038/370335a0. [DOI] [PubMed] [Google Scholar]
- 25.Alex L A, Simon M I. Trends Genet. 1994;10:133–138. doi: 10.1016/0168-9525(94)90215-1. [DOI] [PubMed] [Google Scholar]
- 26.MacDonald N J, De La Rosa A, Benedict M A, Freije J M P, Krutsch H, Steeg P S. J Biol Chem. 1993;268:25780–25789. [PubMed] [Google Scholar]
- 27.Sternweis P C, Robishaw J D. J Biol Chem. 1984;259:13806–13813. [PubMed] [Google Scholar]
- 28.Fujitaki J M, Smith R A. Methods Enzymol. 1984;107:23–36. doi: 10.1016/0076-6879(84)07004-x. [DOI] [PubMed] [Google Scholar]
- 29.Weiss V, Magasanik B. Proc Natl Acad Sci USA. 1988;85:8919–8923. doi: 10.1073/pnas.85.23.8919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sanders D A, Gillece-Castro B L, Stock A M, Burlingame A L, Koshland D E. J Biol Chem. 1989;264:21770–21778. [PubMed] [Google Scholar]
- 31.Ota I M, Varshavsky A. Science. 1993;262:566–569. doi: 10.1126/science.8211183. [DOI] [PubMed] [Google Scholar]
- 32.Maeda T, Wurgler-Murphy S M, Saito H. Nature (London) 1994;369:242–245. doi: 10.1038/369242a0. [DOI] [PubMed] [Google Scholar]
- 33.Posas F, Wurgler-Murphy S M, Maeda T, Witten E A, Thai T C, Saito H. Cell. 1996;86:865–875. doi: 10.1016/s0092-8674(00)80162-2. [DOI] [PubMed] [Google Scholar]
- 34.Chang C, Kwok S F, Bleecker A B, Meyerowitz E M. Science. 1993;262:539–544. doi: 10.1126/science.8211181. [DOI] [PubMed] [Google Scholar]
- 35.Zinyk D L, McGonnigal B G, Dearolf C R. Nat Genet. 1993;4:195–201. doi: 10.1038/ng0693-195. [DOI] [PubMed] [Google Scholar]
- 36.Hsu S, Huang F, Wang L, Banerjee S, Winawer S, Friedman E. Cell Growth Differ. 1994;5:909–917. [PubMed] [Google Scholar]