Abstract
TFIIH is a multifunctional RNA polymerase II transcription factor that possesses DNA-dependent ATPase, DNA helicase, and protein kinase activities. Previous studies have established that TFIIH enters the preinitiation complex and fulfills a critical role in initiation by catalyzing ATP-dependent formation of the open complex prior to synthesis of the first phosphodiester bond of nascent transcripts. In this report, we present direct evidence that TFIIH also controls RNA polymerase II activity at a postinitiation stage of transcription, by preventing premature arrest by very early elongation complexes just prior to their transition to stably elongating complexes. Unexpectedly, we observe that TFIIH is capable of entering the transcription cycle not only during assembly of the preinitiation complex but also after initiation and synthesis of as many as four to six phosphodiester bonds. These findings shed new light on the role of TFIIH in initiation and promoter escape and reveal an unanticipated flexibility in the ability of TFIIH to interact with RNA polymerase II transcription intermediates prior to, during, and immediately after initiation.
TFIIH is a multifunctional protein (1) that was originally identified by its requirement in transcription initiation by RNA polymerase II. In addition to its role in initiation, TFIIH has been shown to play a key role in nucleotide excision repair (NER) of damaged DNA and to possesses DNA-dependent ATPase and DNA helicase activities, as well as a protein kinase activity capable of phosphorylating the carboxyl-terminal domain (CTD) of the largest RNA polymerase II subunit. The two largest subunits of TFIIH are ATP-dependent DNA helicases encoded by the NER XP-B and XP-D genes, and the TFIIH-associated CTD kinase is composed of the kinase/cyclin pair cdk7/cyclin H.
Substantial evidence argues that TFIIH enters the RNA polymerase II preinitiation complex (2, 3) and activates it just prior to initiation, in a reversible reaction that is catalyzed by the TFIIH DNA helicase and involves ATP-dependent formation of an open complex that decays to a closed complex with a half-life of ≈40 s (4–11). In subsequent studies, our laboratory identified (12) a distinct postinitiation role for ATP in preventing premature arrest by very early RNA polymerase II elongation complexes prior to their transition to stably elongating complexes. We observed that, in the absence of a hydrolyzable ATP cofactor, a significant fraction of very early RNA polymerase II elongation complexes suffers arrest at promoter-proximal sites 10–14 bp downstream of the transcriptional start site. Further investigation revealed (i) that addition of ATP to transcription reactions prior to arrest by polymerase at these sites is sufficient to suppress arrest and (ii) that a fraction of arrested elongation complexes can be reactivated by addition of ATP.
In this report, we present direct evidence that TFIIH mediates ATP-dependent suppression of arrest by very early RNA polymerase II elongation complexes. Characterization of the mechanism of action of TFIIH in this process led to the unexpected discovery that TFIIH is capable of entering the transcription cycle not only during assembly of the preinitiation complex but also after initiation and synthesis of as many as four to six phosphodiester bonds. These findings bring to light a novel role for TFIIH in controlling the activity of early RNA polymerase II elongation complexes.
MATERIALS AND METHODS.
Preparation of DNA Templates for Transcription.
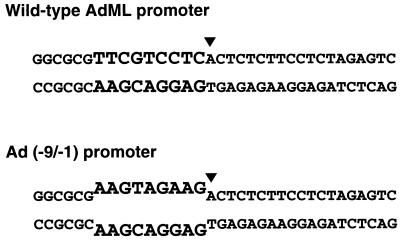
To prepare the double-stranded DNA fragments containing the adenovirus 2 major late (AdML) promoter and the premelted Ad(−9/−1) promoter (Fig. 1), the EcoRI-HindIII fragment from pDN-AdML (5) was introduced into the polylinker of double-stranded M13 mp19 linearized with the same restriction enzymes to generate M13 mp19-AdML. After transformation of Escherichia coli XL-1 Blue with M13 mp19-AdML, phage were grown in XL-1 Blue bacterial cultures, and single-stranded phage DNA, which corresponds to the bottom (coding) strand of the AdML promoter, was prepared as described (13). Oligonucleotides, one with the sequence of the top (noncoding) strand of an AdML promoter region encompassing nucleotides −59 to +17 from pDN-AdML and the other with the same sequence except for a 9-bp mismatch sequence AAGTAGAAG at the transcriptional start site (Fig. 1), were used as primers to direct synthesis of double-stranded DNA fragments. Approximately 150 fmol of each oligonucleotide was mixed with ≈50 fmol of single-stranded M13 mp19-AdML DNA in a hybridization solution containing 25 mM Tris⋅HCl (pH 7.6), 50 mM KCl, and 8 mM MgCl2. The mixtures were heated to 94°C for 5 min and then cooled to room temperature over 2 hr. To extend the oligonucleotide primers, the annealed templates were incubated with 10 units of the Klenow fragment of DNA polymerase I (Promega) and all four ultrapure deoxyribonucleoside triphosphates (Pharmacia; each at 100 μM) at 37°C for 3 hr. The DNA was then digested with KpnI, which cleaves the DNA ≈50 bp upstream of the transcriptional start site, extracted once with phenol/chloroform, and ethanol-precipitated. The DNA was then digested with AvaII to generate ≈410-bp fragments and purified by agarose gel electrophoresis.
Figure 1.
Structures of the transcriptional start sites of the wild-type AdML and Ad(−9/−1) promoters. The DNA strands of the wild-type AdML promoter (Upper) are fully complementarty, whereas the Ad(−9/−1) promoter (Lower) contains a 9-bp mismatch just upstream of the in vivo transcriptional start site. The bottom (coding) strand of the Ad(−9/−1) promoter is identical to the wild-type AdML promoter sequence. ▾, Position of the in vivo transcriptional start site of the AdML promoter.
Preparation of RNA Polymerase II and Transcription Factors.
RNA polymerase II (14) and TFIIH [rat δ, TSK SP-5-PW fraction (15)] were purified as described from rat liver nuclear extracts. Recombinant yeast TATA box-binding protein [TBP; AcA 44 fraction (16, 17)], and TFIIB [rat α (18)] were expressed in E. coli and purified as described. Recombinant TFIIE was prepared as described (19), except that the 56-kDa subunit was expressed in E. coli BL21(DE3)-pLysS. Recombinant TFIIF was purified as described (20) from E. coli JM109(DE3) coinfected with M13 mpET-RAP30 and M13 mpET-RAP74.
Assay of Transcription.
Transcription reactions were performed essentially as described (12). Except as indicated, preinitiation complexes were assembled at the AdML promoter at 28°C by a 45-min preincubation of 35-μl reaction mixtures containing 20 mM Hepes⋅NaOH (pH 7.9), 20 mM Tris⋅HCl (pH 7.9), 60 mM KCl, 4 mM MgCl2, 0.1 mM EDTA, 1 mM dithiothreitol, bovine serum albumin at 0.5 mg/ml, 2% (wt/vol) polyvinyl alcohol, 7% (vol/vol) glycerol, 6 units of RNasin, ≈10 ng of the M13mp19-AdML-derived KpnI–AvaII fragment containing either the wild-type AdML promoter or the Ad(−9/−1) promoter, ≈50 ng of recombinant yeast TBP, ≈10 ng of recombinant TFIIB, ≈20 ng of recombinant TFIIF, ≈20 ng of recombinant TFIIE, ≈10 ng of TFIIH, and ≈0.01 unit of RNA polymerase II. As indicated, transcription was initiated by addition of various combinations of ribonucleoside triphosphates.
Isolation of Early RNA Polymerase II Elongation Complexes by AcA 34 Gel Filtration.
A 1 cm × 12 cm AcA 34 column was packed and equilibrated at room temperature with buffer containing 20 mM Hepes⋅NaOH (pH 7.9), 20 mM Tris⋅HCl (pH 7.9), 60 mM KCl, 4 mM MgCl2, 0.1 mM EDTA, 1 mM dithiothreitol, bovine serum albumin at 0.5 mg/ml, and 7% glycerol. Five transcription reactions were performed as described in Fig. 4 and applied to the column, and material was eluted at room temperature with the same buffer. Fractions of 100 μl were collected.
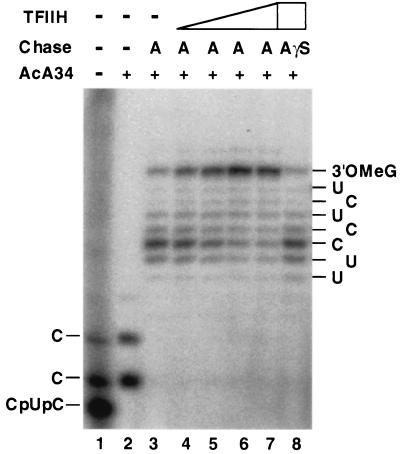
Figure 4.
TFIIH can interact functionally with early RNA polymerase II elongation complexes to suppress promoter-proximal arrest. Transcription reactions were performed. Preinitiation complexes were assembled at the Ad(−9/−1) promoter in the absence of TFIIH. Transcription was initiated with 200 μM CpU, 5 μM ATP, 0.01 μM UTP, and 0.5 μM [α-32P]CTP). After a 15-min incubation at 28°C, elongation complexes containing short (5–7 nt) transcripts were separated from initiating nucleotides by AcA 34 gel filtration. Short transcripts were then chased into longer transcripts in the presence of 100 μM ATP (A) or 100 μM ATP[γS] (AγS) and various amounts of TFIIH. Transcripts were analyzed as described (12) by electrophoresis through a 25% acrylamide/3% N,N′-methylenebisacrylamide/7.0 M urea gel. Lanes: 1, transcripts before AcA 34 gel filtration; 2, transcripts associated with isolated elongation complexes before the chase; 3–8, transcripts resulting from chase of isolated elongation complexes. The transcription reactions in lanes 4–8 contained ≈1.5 ng (lane 4), ≈6 ng (lane 5), ≈15 ng (lane 6), or ≈30 ng (lanes 7 and 8) of TFIIH (fraction 18 from TSK SP-5-PW fractionation shown in Fig. 5D). TFIIH was added to reaction mixtures immediately before addition of chase nucleotides. The labels on either side of the figure indicate the nucleotides at the 3′ end of transcripts. 3′OMeG, 3′-O-MeG-terminated transcripts.
RESULTS
As described above, we recently discovered a postinitiation role for ATP in suppression of premature arrest by RNA polymerase II at promoter-proximal sites, in experiments carried out with a transcription system reconstituted with recombinant TBP, TFIIB, TFIIE, TFIIF, purified RNA polymerase II, and TFIIH from rat liver (12). Because TFIIH is the only one of these proteins known to possess associated ATPase activity, we sought to determine whether TFIIH is involved in this process. To accomplish this, we needed an assay capable of distinguishing the requirement for TFIIH in synthesis of the first phosphodiester bond of promoter-specific transcripts from its potential roles in subsequent transcriptional stages. We therefore took advantage of the artificial AdML promoter derivative Ad(−9/−1), which contains a premelted transcriptional start site (Fig. 1) and does not require TFIIH for initiation (10, 21–23).
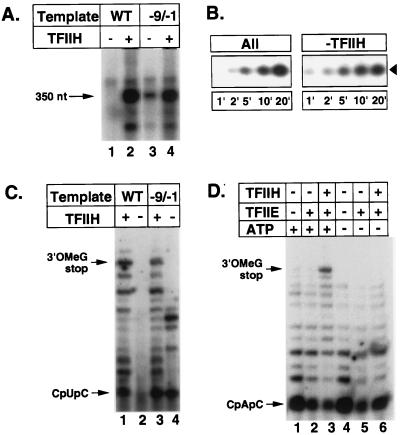
In the experiment of Fig. 2A, transcription was reconstituted, in the presence or absence of TFIIH, with recombinant TBP, TFIIB, TFIIE, TFIIF, purified RNA polymerase II, TFIIH from rat liver, and DNA fragments containing either the wild-type AdML promoter or the Ad(−9/−1) promoter. Consistent with previous results (1), synthesis of full-length run-off transcripts by RNA polymerase II from the wild-type AdML promoter was dependent on TFIIH, whereas synthesis of run-off transcripts from the Ad(−9/−1) promoter was observed in the absence of TFIIH. Notably, synthesis of full-length run-off transcripts by RNA polymerase II from the Ad(−9/−1) promoter was significantly reduced in the absence of TFIIH. This reduction in synthesis of full-length run-off transcripts from the Ad(−9/−1) promoter was not due to an effect of TFIIH on initiation, since, as measured by the dinucleotide-primed abortive initiation assay (4, 24–26), similar levels of initiation from the Ad(−9/−1) promoter were observed in the presence or absence of TFIIH (Fig. 2B).
Figure 2.
ATP-dependent suppression of promoter-proximal arrest requires TFIIH and TFIIE. (A) Run-off transcription from the AdML and Ad(−9/−1) promoters in the presence or absence of TFIIH. Transcription reactions were carried out. Transcription was initiated with 100 μM ATP, 100 μM GTP, 100 μM UTP, and 10 μM [α-32P]CTP and was carried out for 30 min at 28°C. Approximately 350-nt run-off transcripts were analyzed by electrophoresis through a 6% acrylamide/0.8% N,N′-methylenebisacrylamide/7.0 M urea gel. WT, wild type; −9/−1, Ad(−9/−1). (B) Kinetics of transcription initiation at the Ad(−9/−1) promoter. Dinucleotide-primed abortive initiation assays were performed as described (11) for the times indicated with 200 μM CpU and 0.5 μM [α-32P]CTP. Trinucleotide transcripts were analyzed as described (11) by electrophoresis through a 25% acrylamide/3% N,N′-methylenebisacrylamide/7.0 M urea gel. Lanes labeled All contained the complete set of general initiation factors. Lanes labeled −TFIIH did not contain TFIIH. (C) TFIIH dependence of transcription of the wild-type AdML and Ad(−9/−1) promoters. Transcription of the wild-type AdML and Ad(−9/−1) promoters were performed in the presence or absence of TFIIH as indicated. Transcripts were analyzed as described (12) by electrophoresis through a 25% acrylamide/3% N,N′-methylenebisacrylamide/7.0 M urea gel. WT, wild type; −9/−1, Ad(−9/−1); 3′OMeG stop, 3′-O-MeG-terminated transcripts. (D) ATP dependence of transcription of the Ad(−9/−1) promoter. Transcription reactions were performed in the presence or absence of either TFIIH or TFIIE and TFIIH. Transcription was initiated by addition of 200 μM CpA, 5 μM UTP, 0.5 μM [α-32P]CTP, and 100 μM 3′-O-MeGTP, with or without 5 μM ATP. Transcripts were analyzed as described (12) by electrophoresis through a 25% acrylamide/3% N,N′-methylenebisacrylamide/7.0 M urea gel. 3′OMeG stop, 3′-O-MeG-terminated transcripts.
Further analysis revealed that reduction in synthesis of full-length run-off transcripts from the Ad(−9/−1) promoter was due in large part to premature arrest by very early RNA polymerase II elongation complexes. In these experiments, preinitiation complexes were assembled at the wild-type AdML promoter or the Ad(−9/−1) promoter in the presence or absence of TFIIH, and transcription was carried out with 200 μM of the initiating dinucleotide CpU, 5 μM ATP, 5 μM UTP, 0.5 μM [α-32P]CTP, and 100 μM 3′-O-methylguanosine 5′-triphosphate (3′-O-MeGTP), which prevents most transcription beyond the first guanosine at position +18 in the AdML transcript (Fig. 1). As shown in Fig. 2C, when transcription was carried out in the presence of TFIIH, a substantial fraction of RNA polymerase II elongation complexes at both the wild-type AdML and the Ad(−9/−1) promoters successfully synthesized 18-nt 3′-O-MeG-terminated transcripts. As expected, when transcription was carried out in the absence of TFIIH, only the Ad(−9/−1) promoter supported detectable transcription by RNA polymerase II. The vast majority of RNA polymerase II elongation complexes at the Ad(−9/−1) promoter, however, suffered premature arrest at promoter-proximal sites within 14 bp of the transcriptional start site. To determine whether ATP is required for TFIIH-dependent suppression of arrest, it was necessary to eliminate the requirement for ATP as a substrate in synthesis of 3′-O-MeG-terminated transcripts. To accomplish this, we substituted CpA for CpU as the priming dinucleotide, thereby eliminating the requirement for ATP as a substrate for RNA chain elongation. As shown in Fig. 2D, results of this experiment revealed that TFIIH-dependent relief of arrest by very early RNA polymerase II elongation complexes at the Ad(−9/−1) promoter depends on ATP (Fig. 2D). We note that the reduction in synthesis of 18-nt 3′-O-MeG-terminated transcripts from the Ad(−9/−1) promoter in the absence of TFIIH (Fig. 2C) is greater than the reduction in synthesis of full-length run-off transcripts from the Ad(−9/−1) promoter in the absence of TFIIH (Fig. 2A). This is due at least in part to a difference in the ribonucleoside triphosphate concentrations used in the two experiments, since we consistently observe that increasing the ribonucleoside triphosphate concentrations decreases the fraction of early RNA polymerase II elongation complexes that suffer arrest at promoter-proximal sites. Thus, these results indicate that, just as a fraction of very early RNA polymerase II elongation complexes deprived of ATP suffer premature arrest at promoter-proximal sites 10–14 bp downstream of the wild-type AdML promoter (12), RNA polymerase II elongation complexes suffer premature arrest at promoter-proximal sites a similar distance downstream of the Ad(−9/−1) promoter in the absence of either ATP or TFIIH.
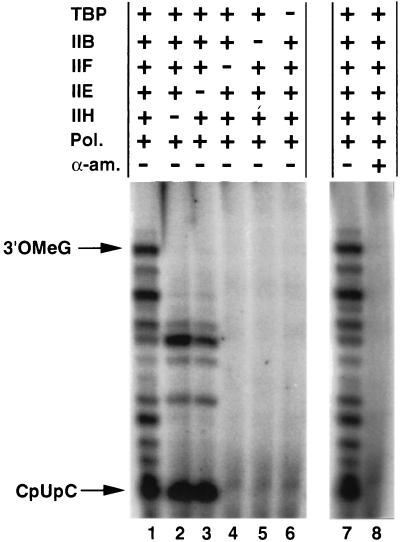
TFIIH- and ATP-dependent activation of the preinitiation complex depends strongly on TFIIE (7, 10, 11, 22). To determine whether TFIIE is also required for TFIIH- and ATP-dependent suppression of arrest by early RNA polymerase II elongation complexes, transcription from the Ad(−9/−1) promoter was carried out in the presence of RNA polymerase II and various combinations of initiation factors. In agreement with previous studies (10, 23), transcription initiation by RNA polymerase II at the Ad(−9/−1) promoter was dependent only on TBP, TFIIB, and TFIIF and was inhibited by α-amanitin at 1 μg/ml (Fig. 3). RNA polymerase II elongation complexes suffered arrest at promoter-proximal sites when transcription was carried out in the absence of either TFIIE or TFIIH. Thus, like TFIIH- and ATP-dependent activation of the preinitiation complex, TFIIH- and ATP-dependent suppression of arrest by very early RNA polymerase II elongation complexes is strongly dependent on TFIIE.
Figure 3.
Transcription from the Ad(−9/−1) promoter depends on general initiation factors and is sensitive to inhibition by α-amanitin. Transcription was performed as described in Fig. 2C in the presence of the indicated transcription factors, with or without α-amanitin at 1 μg/ml. Transcripts were analyzed as described (12) by electrophoresis through a 25% acrylamide/3% N,N′-methylenebisacrylamide/7.0 M urea gel. IIB, TFIIB; IIE, TFIIE; IIH, TFIIH; Pol., RNA polymerase II; α-am., α-amanitin; 3′OMeG; 3′-O-MeG-terminated transcripts.
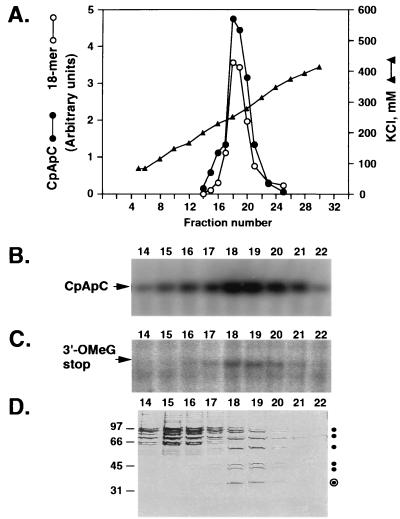
Further investigation of the mechanism of TFIIH-dependent suppression of arrest revealed that TFIIH can prevent arrest even when added to transcription reactions after synthesis of the first four to six phosphodiester bonds of nascent transcripts. In these experiments, RNA polymerase II elongation complexes containing 5- to 7-nt transcripts were synthesized from the Ad(−9/−1) promoter in the presence of TBP, TFIIB, TFIIE, and TFIIF and were purified by gel filtration. Transcripts associated with purified elongation complexes were then chased by addition of 100 μM UTP, 200 μM CTP, and 100 μM 3′-O-MeGTP into longer products in the presence or absence of various concentrations of TFIIH and either 100 μM ATP or 100 μM adenosine 5′-[γ-thio]triphosphate (ATP[γS]). As shown in Fig. 4, in the presence of ATP, addition of TFIIH to very early elongation complexes significantly increased the fraction of 5- to 7-nt transcripts that could be chased into 18-nt 3′-O-MeG-terminated transcripts. Furthermore, consistent with our previous results (12) indicating that ATP[γS] inhibits ATP-dependent suppression of arrest at promoter-proximal sites, we observe that ATP[γS] is a potent inhibitor of TFIIH-dependent suppression of promoter-proximal arrest. To address the possibility that an activity other than TFIIH in our TFIIH preparations might be responsible for suppressing arrest by RNA polymerase II, we tested chromatographic fractions from the final step in purification of the TFIIH used in this study for their abilities to suppress promoter-proximal arrest. As shown in Fig. 5, “arrest-suppressing” activity copurified closely with both TFIIH transcriptional activity and TFIIH subunits during high-resolution TSK SP-5-PW HPLC, strongly supporting the proposal that TFIIH is indeed capable of controlling the activity of early RNA polymerase II elongation complexes.
Figure 5.
Cochromatography of TFIIH and ATP-dependent arrest suppressing activity. (A) Elution profile of TFIIH initiation activity and arrest suppressing activity during ion-exchange chromatography of purified TFIIH on TSK SP-5-PW. TSK SP-5-PW HPLC purification of TFIIH was performed as described (15). The amounts of dinucleotide-primed trinucleotide CpApC and 18-nt 3′-O-MeG-terminated transcripts synthesized are expressed in arbitrary units determined by PhosphorImager analysis of polyacrylamide gels. (B) TFIIH initiation activity measured using the abortive initiation assay performed as described in Fig. 2B. Trinucleotide transcripts were analyzed as described (11) by electrophoresis through a 25% acrylamide/3% N,N′-methylenebisacrylamide/7.0 M urea gel. (C) Pulse–chase assay for arrest suppressing activity. Transcription reactions were performed. Preinitiation complexes were assembled at the Ad(−9/−1) promoter in the absence of TFIIH. Transcription was initiated with 200 μM CpU, 5 μM ATP, 0.01 μM UTP, and 0.5 μM [α-32P]CTP. After a 15-min incubation at 28°C, short transcripts were chased into longer transcripts in the presence of the indicated TSK SP-5-PW fractions and 100 μM ATP, 100 μM UTP, 200 μM CTP, and 100 μM 3′-O-MeGTP. Transcripts were analyzed as described (12) by electrophoresis through a 25% acrylamide/3% N,N′-methylenebisacrylamide/7.0 M urea gel. (D) Polypeptide composition of TSK SP-5-PW fractions. TSK SP-5-PW fractions were analyzed by electrophoresis through an SDS/8% polyacrylamide gel and silver staining. •, TFIIH subunits; ⊙, two TFIIH subunits that were not well resolved in this gel (data not shown).
DISCUSSION
TFIIH was initially identified as an essential RNA polymerase II general transcription factor (3, 27–29). Further characterization revealed that TFIIH is a multifunctional protein possessing closely associated DNA-dependent ATPase, DNA helicase, and CTD kinase activities (1). Biochemical studies have shown that promoter-specific transcription initiation by RNA polymerase II requires a hydrolyzable ATP cofactor in addition to TFIIH and remaining general transcription factors TFIIB, TFIID, TFIIE, and TFIIF. Substantial evidence argues that TFIIH plays a critical role in initiation by entering the preinitiation complex and catalyzing ATP-dependent formation of the open complex prior to synthesis of the first phosphodiester bond of nascent transcripts.
In this report, to our knowledge, we provide the first direct evidence that TFIIH is also capable of controlling the activity of RNA polymerase II at a postinitiation stage of transcription and, moreover, that TFIIH need not assemble into the preinitiation complex to accomplish this. In a previous study, we found that RNA polymerase II elongation complexes at the AdML promoter are highly susceptible to arrest at promoter-proximal sites 10–14 bp downstream of the transcriptional start site when elongation is carried out in the absence of a hydrolyzable ATP cofactor (12). By exploiting the premelted AdML promoter derivative Ad(−9/−1), we discovered that early RNA polymerase II elongation complexes formed at the Ad(−9/−1) promoter in the absence of TFIIH, TFIIE, or both are also prone to arrest at promoter-proximal sites a similar distance downstream of the transcriptional start site. Furthermore, we showed that arrest by RNA polymerase II can be suppressed even when TFIIH is added to transcription reactions after synthesis of the first few phosphodiester bonds of nascent transcripts as long as ATP is also present. Thus, TFIIH and TFIIE play dual roles in transcription: they are essential for ATP-dependent open complex formation prior to first bond synthesis, and they are critical for ATP-dependent suppression of premature arrest by very early RNA polymerase II elongation complexes prior to their transition to stably elongating complexes.
Exactly how TFIIH and TFIIE function together to prevent premature arrest by RNA polymerase II is presently unknown. A reasonable hypothesis is that the TFIIH DNA helicase is involved in this process, in light of evidence that ATP-dependent open complex formation and initiation, as well as ATP-dependent suppression of arrest, are blocked by ATP[γS], an inhibitor of the TFIIH DNA helicase but not of the TFIIH CTD kinase (refs. 5 and 10–12 and data not shown). In addition, on the basis of evidence that TFIIE is capable of stabilizing the association of TFIIH with both polymerase (30, 31) and the preinitiation complex in vitro (3, 32), it is possible that TFIIE prevents premature arrest by RNA polymerase II at least in part by stablilizing the interaction of TFIIH with early elongation complexes.
Finally, what role TFIIH-mediated suppression of arrest by early RNA polymerase II elongation complexes might play in mRNA synthesis in eukaryotic cells is not known. Based on results of experiments carried out in vivo (33–35), it has been suggested that TFIIH may be involved in regulation of elongation by a variety of DNA binding transactivators. From our biochemical experiments, it is clear TFIIH and ATP can substantially suppress promoter-proximal arrest and, therefore, have the capacity to control a potentially rate limiting step in transcription. We note that, although TFIIH and ATP are essential for promoter-specific transcription initiation under most conditions, they do not appear to be essential for promoter escape, since a fraction of polymerases are capable of exiting the promoter in the absence of TFIIH and ATP. Nevertheless, under conditions where promoter escape is physically blocked by, for example, a promoter-proximal nucleosome or DNA-bound protein, the efficiency of ATP-dependent promoter escape might become critical for transcription.
Acknowledgments
We are grateful to Siyuan Tan for assistance in preparation of the premelted Ad(−9/−1) DNA template, to Jean-Marc Egly for helpful discussions, to Kenneth Jackson of the Molecular Biology Resource Center at the Oklahoma Center for Molecular Medicine for oligonucleotide synthesis, and to Dewan Haque for expert technical assistance. This work was supported by National Institutes of Health Grant GM-41628 (R.C.C. and J.W.C.), by Oklahoma Center for the Advancement of Science and Technology Grant HN5-043 (A.D.), and by funds provided to the Oklahoma Medical Research Foundation by the H. A. and Mary K. Chapman Charitable Trust.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: CTD, carboxyl-terminal domain; AdML, adenovirus 2 major late; 3′-O-MeGTP, 3′-O-methylguanosine 5′-triphosphate; ATP[γS], adenosine 5′-[γ-thio]triphosphate; TBP, TATA box-binding protein.
References
- 1.Svejstrup J Q, Vichi P, Egly J M. Trends Biochem Sci. 1996;21:346–350. [PubMed] [Google Scholar]
- 2.Conaway R C, Conaway J W. J Biol Chem. 1990;265:7559–7563. [PubMed] [Google Scholar]
- 3.Flores O, Lu H, Reinberg D. J Biol Chem. 1992;267:2786–2793. [PubMed] [Google Scholar]
- 4.Luse D S, Jacob G A. J Biol Chem. 1987;262:14990–14997. [PubMed] [Google Scholar]
- 5.Conaway R C, Conaway J W. J Biol Chem. 1988;263:2962–2968. [PubMed] [Google Scholar]
- 6.Wang W, Carey M, Gralla J D. Science. 1992;255:450–453. doi: 10.1126/science.1310361. [DOI] [PubMed] [Google Scholar]
- 7.Parvin J D, Sharp P A. Cell. 1993;73:533–540. doi: 10.1016/0092-8674(93)90140-l. [DOI] [PubMed] [Google Scholar]
- 8.Parvin J D, Shykind B M, Meyers R E, Kim J, Sharp P A. J Biol Chem. 1994;269:18414–18421. [PubMed] [Google Scholar]
- 9.Timmers H, Th M. EMBO J. 1994;13:391–399. doi: 10.1002/j.1460-2075.1994.tb06273.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holstege F C P, van der Vliet P C, Timmers H, Th M. EMBO J. 1996;15:1666–1677. [PMC free article] [PubMed] [Google Scholar]
- 11.Dvir A, Garrett K P, Chalut C, Egly J M, Conaway J W, Conaway R C. J Biol Chem. 1996;271:7245–7248. doi: 10.1074/jbc.271.13.7245. [DOI] [PubMed] [Google Scholar]
- 12.Dvir A, Conaway R C, Conaway J W. J Biol Chem. 1996;271:23352–23356. doi: 10.1074/jbc.271.38.23352. [DOI] [PubMed] [Google Scholar]
- 13.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 14.Serizawa H, Conaway R C, Conaway J W. Proc Natl Acad Sci USA. 1992;89:7476–7480. doi: 10.1073/pnas.89.16.7476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Conaway R C, Reines D, Garrett K P, Powell W, Conaway J W. Methods Enzymol. 1996;273:194–207. doi: 10.1016/s0076-6879(96)73020-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schmidt M C, Kao C C, Pei R, Berk A J. Proc Natl Acad Sci USA. 1989;86:7785–7789. doi: 10.1073/pnas.86.20.7785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Conaway J W, Hanley J P, Garrett K P, Conaway R C. J Biol Chem. 1991;266:7804–7811. [PubMed] [Google Scholar]
- 18.Tsuboi A, Conger K, Garrett K P, Conaway R C, Conaway J W, Arai N. Nucleic Acids Res. 1992;20:3250. doi: 10.1093/nar/20.12.3250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peterson M G, Inostroza J, Maxon M E, Flores O, Admon A, Reinberg D, Tjian R. Nature (London) 1991;354:369–373. doi: 10.1038/354369a0. [DOI] [PubMed] [Google Scholar]
- 20.Tan S, Conaway R C, Conaway J W. BioTechniques. 1994;16:824–828. [PubMed] [Google Scholar]
- 21.Tantin D, Carey M. J Biol Chem. 1994;269:17397–17400. [PubMed] [Google Scholar]
- 22.Holstege F, Tantin D, Carey M, van der Vliet P C, Timmers H, Th M. EMBO J. 1995;14:810–819. doi: 10.1002/j.1460-2075.1995.tb07059.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pan G, Greenblatt J. J Biol Chem. 1994;269:30101–30104. [PubMed] [Google Scholar]
- 24.McClure W R, Cech C L, Johnston D E. J Biol Chem. 1978;253:8941–8948. [PubMed] [Google Scholar]
- 25.Carpousis A J, Gralla J D. Biochemistry. 1980;19:3245–3253. doi: 10.1021/bi00555a023. [DOI] [PubMed] [Google Scholar]
- 26.Jiang Y, Yan M, Gralla J D. J Biol Chem. 1995;270:27332–27338. doi: 10.1074/jbc.270.45.27332. [DOI] [PubMed] [Google Scholar]
- 27.Conaway R C, Conaway J W. Proc Natl Acad Sci USA. 1989;86:7356–7360. doi: 10.1073/pnas.86.19.7356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feaver W J, Gileadi O, Kornberg R D. J Biol Chem. 1991;266:19000–19005. [PubMed] [Google Scholar]
- 29.Gerard M, Fischer L, Moncollin V, Chipoulet J M, Chambon P, Egly J M. J Biol Chem. 1991;266:20940–20945. [PubMed] [Google Scholar]
- 30.Serizawa H, Conaway J W, Conaway R C. In: Transcription: Mechanisms and Regulation. Conaway R C, Conaway J W, editors. New York: Raven; 1994. pp. 27–43. [Google Scholar]
- 31.Maxon M E, Goodrich J A, Tjian R. Genes Dev. 1994;8:515–524. doi: 10.1101/gad.8.5.515. [DOI] [PubMed] [Google Scholar]
- 32.Conaway J W, Bradsher J N, Conaway R C. J Biol Chem. 1992;267:10142–10148. [PubMed] [Google Scholar]
- 33.Akhtar A, Faye G, Bentley D L. EMBO J. 1996;15:4654–4664. [PMC free article] [PubMed] [Google Scholar]
- 34.Blau J, Xiao H, McCracken S, O’Hare P, Greenblatt J, Bentley D. Mol Cell Biol. 1996;15:2044–2055. doi: 10.1128/mcb.16.5.2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yankulov K Y, Pandes M, McCracken S, Bouchard D, Bentley D L. Mol Cell Biol. 1996;16:3291–3299. doi: 10.1128/mcb.16.7.3291. [DOI] [PMC free article] [PubMed] [Google Scholar]