Abstract
SMAD2 is a member of the transforming growth factor β and activin-signaling pathway. To examine the role of Smad2 in postgastrulation development, we independently generated mice with a null mutation in this gene. Smad2-deficient embryos die around day 7.5 of gestation because of failure of gastrulation and failure to establish an anterior–posterior (A-P) axis. Expression of the homeobox gene Hex (the earliest known marker of the A-P polarity and the prospective head organizer) was found to be missing in Smad2-deficient embryos. Homozygous mutant embryos and embryonic stem cells formed mesoderm derivatives revealing that mesoderm induction is SMAD2 independent. In the presence of wild-type extraembryonic tissues, Smad2-deficient embryos developed beyond 7.5 and up to 10.5 days postcoitum, demonstrating a requirement for SMAD2 in extraembryonic tissues for the generation of an A-P axis and gastrulation. The rescued postgastrulation embryos showed malformation of head structures, abnormal embryo turning, and cyclopia. Our results show that Smad2 expression is required at several stages during embryogenesis.
The transforming growth factor β (TGF-β) superfamily is a group of signaling molecules that play essential roles in development and tissue homeostasis in vertebrates and invertebrates (1–3). Members of this family include the TGF-βs, activins, and bone morphogenetic proteins.
A family of proteins known as SMADs has been shown to function downstream of TGF family type I receptors (reviewed in ref. 4). The TGF-β and activin type I receptors interact with SMAD2 and the closely related SMAD3 (5–9). After being phosphorylated by type I receptors, pathway-restricted SMADs associate with SMAD4 (10), translocate to the nucleus, and activate, in conjunction with several specific transcription factors, their respective target genes (4, 11).
The SMAD2 protein consists of three domains (4), the Mad homology domain 1 (MH1) and the MH2 domain that harbors the DNA-binding site (12). The third linker region domain connects the MH1 and MH2 domains, is phosphorylated, and may serve as a molecular hinge. The MH2 domain interacts with the type I receptors and is phosphorylated on ligand activation. The SMAD2-type I receptor interaction is also regulated by a newly discovered protein named SARA (13). The MH2 domain also exhibits transactivation properties.
The human SMAD2 gene is located on chromosome 18q21, in close proximity to the tumor suppressors DCC and SMAD4/DPC4 (14–16). Mutations that disrupt the function of SMAD2 have been identified in colorectal cancers (16, 17) and lung cancer (18), suggesting its role as a tumor suppressor.
Smad2 is expressed almost ubiquitously and has an important role in early development (19–22). Targeted inactivation of Smad2 resulted in early lethality before gastrulation. Though all these studies showed that Smad2-deficient embryos die because of gastrulation failure, two studies did not describe mesoderm formation (21, 22), whereas the other reported transient mesoderm induction but failure to form the anterior–posterior (A-P) axis (20).
We also generated mice with a targeted mutation in the Smad2 gene. Smad2dex2 homozygous embryos also die because of failure to establish an A-P axis. To assess the postgastrulation functions of SMAD2, we rescued the early lethality by using the tetraploid embryo complementation assay. Homozygous Smad2−/− embryos derived by injection of mutant embryonic stem cells (ES cells) into tetraploid blastocysts overcame the early embryonic lethality and developed beyond days postcoitum (dpc)7.5 and up to dpc10.5. These experiments revealed a requirement for SMAD2 in the visceral endoderm development for proper gastrulation and establishment of an A-P axis. Our results also show that SMAD2 signaling is important for embryo turning, heart tube looping, and anterior development.
Materials and Methods
Isolation of the Murine Smad2 Gene.
The murine Smad2 gene was cloned by using the human IMAGE Consortium Clone ID 148042 (23) as a probe to screen a mouse-brain cDNA library (Stratagene). The Smad2 cDNA was used to screen a mouse genomic 129/Ola λ phage library (24).
The targeting vector pSmad2 was prepared by using a 10-kbp genomic BglII/EcoRI fragment, in which a 2.5-kbp BglII/PstI fragment containing exon 2 was replaced by a 1.8-kbp phosphoglycerate kinase-neomycin expression cassette.
Generation of Smad2 Mutant ES Cells and Mice.
The target vector pSmad2 (40 μg in each of the two experiments) was linearized at the unique NotI site and electroporated into 2 × 107 WW6 ES cells (25). Stable transfected ES cells were selected as described (24, 26) and screened for homologous recombination events PCR assay. Primers used for detection of the dex2 allele were J52 5′-GCCAAGCTGAGTATGCAACA-3′ and J55 5′-TGCCCAGTCATAGCCGAATA-3′. Three positive clones that yielded the expected 2.6-kbp PCR product were obtained (644, 6D3, and 3A1). Correct targeting events were verified by Southern blot hybridization.
Chimeric mice were generated by injecting C57/BL6 blastocysts with ES cells from clones 644, 6D3, and 3A1. Chimeras from all three cell lines transmitted the mutated allele through the germline and were used to establish the mouse lines. ES cells of all genotypes were established from F1 heterozygous intercrosses as described (27). All animals and ES cells analyzed were on a C57/BL6 × 129V mixed genetic background.
Genotyping Mice and Embryos.
Tail biopsies from F1 animals were analyzed by PCR and the results confirmed by Southern blot analysis. F2 animals were mostly analyzed by PCR. The presence of the mutant allele was detected with primers J68 5′-TACTCTGTGCAGATGAAGTGC-3′ and J76 5′-TGTCCATCTGCACGAGACTA-3′, resulting in a 293-bp PCR product. The wild-type allele was detected by using primers J71 5′-TGAATGGCAAGATGGACGAC-3′ and J73 5′-TAAAGACAGCATCGTCATCAG-3′, resulting in a 225-bp product.
Western Blot Analysis.
SMAD2 protein expression in ES cells was detected by immunoprecipitation followed by immunoblot analysis (28). Polyclonal goat anti-SMAD2 antibodies raised against a peptide sequence at the N-terminal end of human SMAD2 (S20, Santa Cruz Biotechnology) and the rabbit pan-SMAD antibody 367 (kindly provided by Robert Lechleider, National Cancer Institute, Bethesda, MD) were used for detection of SMAD2 protein. A second anti-Smad2 antibody was also used (Transduction Laboratories, Lexington, KY, no. S66220, epitope aa142–263). To probe for the C terminus of SMAD2, an affinity-purified rabbit polyclonal antibody that detects the phosphorylated serine residues in the C terminus was used (kindly provided by Peter ten Dijke, Ludwig Institute, Uppsala, Sweden).
Formation of Teratomas.
Wild-type (1.0 × 106) and Smad2 −/− ES cells were injected subcutaneously into nude mice. Tumors formed were recovered, fixed in neutral formalin, and paraffin embedded. Sections were stained with hematoxlin/eosin for histological analysis.
Differentiation of ES Cells into Embryoid Bodies (EBs).
EBs were obtained as described (27). The expression patterns of several differentiation markers in wild-type and mutant EBs were analyzed by using a semiquantitative reverse transcription—PCR approach. One microgram of total RNA was reverse transcribed by using the 1st Strand cDNA Synthesis Kit (Boehringer Mannheim). Between 1 and 3 μl of the cDNA reaction was used for amplification. Samples of PCR products were taken in the linear phase (between 25 and 30 cycles), separated in 1.75% agarose gels, and Southern blot analysis performed by use of 32P end-labeled internal probes.
Production of ES Cell-Derived Embryos by Tetraploid or Diploid Blastocyst Injection and Whole-Mount in Situ Hybridization.
Tetraploid blastocysts from ROSA26 (30), B6C3H, or CAF1 females were derived by using a described protocol (29). Standard blastocyst injection of mutated ES cells was performed, and embryos were recovered from equivalent 7–13.5 gestation days. β-Galactosidase staining was performed as described (31).
Dissected embryos were fixed and processed for in situ hybridization as described elsewhere (31). Antisense RNA probes for Brachyury and Shh were utilized (32, 33). Hex sense and antisense probes were synthesized by using a 533-bp PCR fragment corresponding to nucleotides 100–652 of the sequence deposited in GenBank.
Results
Generation of Mice with a Targeted Smad2 Allele.
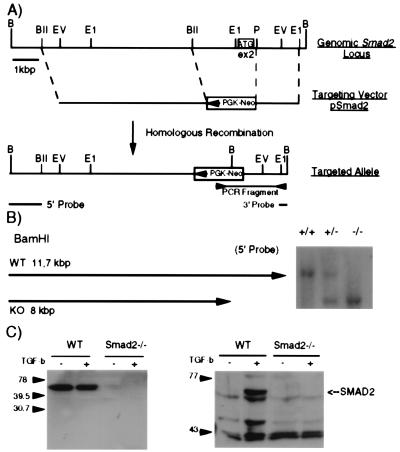
WW6 ES cells were transfected with the linearized target vector pSmad2. pSmad2 contains a 6-kb BglII fragment 5′ of exon 2, a phosphoglycerate kinase-neomycin expression cassette in opposite transcriptional orientation replacing exon 2, and a 1.5-kb PstI/EcoRI fragment 3′ of exon 2 (Fig. 1A). Correct integration of the targeting vector into the Smad2 locus results in the deletion of a 2.5-kb fragment, including exon 2, depriving the Smad2 gene of its original ATG codon (34). Three clones were identified to have the correct targeting event (Fig. 1B), were injected into C57/BL6 blastocysts, and all three clones gave rise to chimeras, which transmitted the modified allele through the germline. We named the mutated Smad2 allele Smad2dex2. We also established ES cell lines of all three genotypes from blastocysts obtained from F1 heterozygous intercrosses (Fig. 1B).
Figure 1.
Generation of Smad2-deficient mice. (A) Gene-targeting strategy. A part of the genomic Smad2 locus containing exon 2, which harbors the ATG codon (white box with ATG), is shown. The targeting vector pSmad2 is shown in the corresponding location below the genomic map. Homologous recombination yields the dex2 allele, where exon 2 is deleted and replaced by a phosphoglycerate kinase-neomycin cassette in opposite transcriptional orientation. Primers and probes used for PCR and Southern analysis, respectively, are depicted below the dex2 allele. (B) Southern blot analysis of ES cells derived from Smad2 heterozygous intercrosses. Probing the BamHI-digested DNA with the 5′ probe yields an 11.7 wild-type band and 8-kb band for the dex2 allele (+/+ wild type, +/− heterozygous, and −/− homozygous). (C) Western blot analysis of protein extracts from Smad2dex2 mutant and wild-type ES cells. Antibodies against the linker domain of SMAD2 (Left) and against the C-terminal domain were used (Right).
To determine whether the Smad2dex2 mutation results in a null allele, we performed Western blot analysis of protein extracts from Smad2dex2 homozygous and wild-type ES cells (Fig. 1C). In +/+ ES, cells the SMAD2 protein was detected, whereas in −/− ES cells no SMAD2 protein, or a truncated form, was detectable. These results were obtained with three different antibodies, one directed against MH1 domain epitope (Santa Cruz Biotechnology), one against the linker domain epitope (Transduction Laboratories), and one against the phosphorylated C-terminal MH2 domain. We conclude that the Smad2dex2 allele is a null allele.
The Smad2dex2 Mutation Is Homozygous Embryonic Lethal.
To understand the biological consequences of the Smad2dex2 mutation, we generated F2 offspring from F1 heterozygous intercrosses. We analyzed 281 offspring and detected 102 wild-type, 179 heterozygous, and no homozygous mutant offspring. These results show that the Smad2dex2 mutation in the homozygous state also leads to embryonic lethality, as seen in the other Smad2 mutants (20–22). Heterozygous were phenotypically indistinguishable from wild-type littermates. The Smad2dex2 mutation strongly affected the development of homozygous embryos. Analysis of Smad2dex2 −/− concepti at day 8.5 revealed absence of embryonic ectoderm derivatives but apparently normal yolk-sac membrane (Fig. 2A) and tissues of mesoderm origin such as blood islands (not shown). This observation suggested that a transient induction of mesoderm occurred, which led to normal contribution of mesoderm derivatives to extraembryonic tissues. Supporting the latter idea was the expression of the early mesoderm marker Brachyury in day 7.0 mutant embryos. In these, a weak signal distributed within the embryonic ectoderm region was observed, in contrast with the strong and polarized expression seen in wild-type or heterozygous embryos (Fig. 2B). Moreover, a well-organized columnar embryonic ectoderm epithelium was undistinguishable in these embryos (Fig. 2B). We conclude that homozygous Smad2dex2 embryos die around day 7.5 of gestation because of abnormal embryonic ectoderm development, although transient induction of mesoderm occurs. This phenotype was accompanied by the loss of brachyury expression in a polarized fashion. The phenotype produced by our mutation is identical to that obtained with the robm1 mutant allele of Smad2 (20).
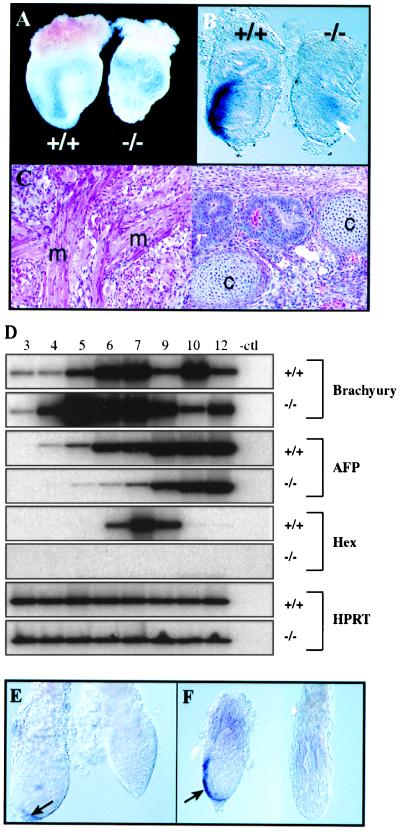
Figure 2.
Smad2−/− phenotype and differentiation of Smad2−/− ES cells into mesoderm derivatives. (A) Concepti recovered at day 8.5 of gestation. A wild-type U-shaped embryo (+/+) can be clearly seen through the yolk-sac membrane, whereas the Smad2−/− conceptus consists of an empty yolk sac with no embryo visible inside. (B) Brachyury in situ hybridization in day 7.0 embryos. Although in the wild-type embryo the expression of this mesoderm marker is confined to the posterior region in the primitive streak area, in the mutant embryo a weak signal (arrow) can be observed in the interior of a not well-organized embryonic ectoderm. (C) Teratomas produced with Smad2dex2−/− ES cells. Areas showing differentiation of muscle (m) and cartilage (c) are indicated. (D) Expression analysis of markers during EBs differentiation. Semiquantitative reverse transcription–PCR analysis of the indicated markers is shown at time points (in days) as indicated above the lanes. The loading control is the Hprt gene (Bottom). (E) Hex in situ hybridization in day 5.5 embryos. Although Hex expression was observed in the distal tip of wild-type embryos (Left, arrow), no staining was observed in mutant embryos. (F) Hex in situ hybridization in day 7.0 embryos. Hex expression in the anterior visceral endoderm of wild-type or heterozygous embryos (Right, arrow) was absent in mutant embryos (Left).
Hex Expression Is Affected in Smad2dex2 Mutant Embryos.
Visceral endoderm has been implicated in the patterning of the early embryo (35). In particular, it has been suggested that Smad2 deficiency in the extraembryonic tissues disrupts the process of A-P specification. (20). To further investigate the molecular defect(s) in this tissue because of Smad2 deficiency, we studied the expression of several visceral endoderm markers during EBs differentiation. The homeobox gene Hex is the earliest known visceral endoderm marker showing an A-P asymmetric expression in pregastrulation embryos (35). Gata-4 and α-fetoprotein are markers of early and later differentiation of visceral endoderm, respectively. While Gata-4 showed no dramatic differences in wild-type vs. mutant EBs (not shown), the expression of α-fetoprotein was delayed and/or reduced in comparison to the wild-type control during the first few days of culture (Fig. 2D).
Hex expression peaked around day 7 in cultures of wild-type EBs, whereas in Smad2-deficient EBs, Hex expression was completely absent (Fig. 2D). To analyze the relevance of this observation in vivo, we performed in situ hybridization of day 5.5–7.5 embryos from heterozygous intercrosses by using Hex as a probe. In wild-type or heterozygous embryos, Hex expression was first seen at the distal tip of visceral endoderm and in older embryos extended asymmetrically along the anterior visceral endoderm (Fig. 2 E–F). However, Hex expression in Smad2-deficient embryos was absent (Fig. 2 E–F). Taken together, these results show that Smad2, directly or indirectly, regulates the expression of several visceral endoderm genes, including the homeobox gene Hex. Despite the normal appearance of the extraembryonic structures, important functions of the visceral endoderm may be impaired in Smad2-deficient embryos.
Potentiality of Smad2dex2−/− ES Cells to Form Mesoderm Derivatives.
To test the potential of Smad2dex2 ES cells to differentiate into mesoderm, we generated teratomas by using Smad2−/− ES cells. Analysis of the tumors showed that these cells were able to differentiate into several mesoderm tissues, including muscle and cartilage (Fig. 2C). Smad2+/+ and −/− ES cells were also differentiated in vitro into EBs. After 10 days in culture, about 50% of the Smad2−/− and +/+ EBs formed cystic structures, many with contracting cardiac muscle. Histologic analysis of wild-type and mutant EBs showed that both were formed by visceral endoderm, mesenchyme, blood islands, and epithelia (not shown). Moreover, expression of the mesoderm marker Brachyury, and the mesoderm inducers BMP-4 and nodal were very similar in wild-type and mutant EBs at all time points analyzed (Fig. 2D and data not shown). These results suggest that mesoderm differentiation is independent of Smad2 function.
Wild-Type Extraembryonic Tissue Rescues the Smad2−/− Gastrulation Phenotype.
The abnormal expression of several developmental markers in the visceral endoderm, as well as the inability of wild-type ES cells to develop an embryo in the presence of Smad2-deficient extraembryonic tissues (this work and ref. 20), suggest that the embryo lethality is related to defective extraembryonic tissue development. To test this hypothesis, we produced chimeric embryos by injection of homozygous Smad2−/− ES cells into tetraploid wild-type blastocysts. In this kind of assay, tetraploid cells contribute to the trophoblast and extraembryonic endoderm (making little contribution to the proper embryo) providing normal extraembryonic signaling (29, 37). Homozygous Smad2dex2 ES cells were injected into B6C3H or ROSA26 tetraploid blastocysts. The contribution of tetraploid wild-type cells was monitored by glucose 6-phosphate isomerase assay (not shown) or 5-bromo-4-chloro-3-indolyl β-d-galactoside (X-gal) staining. Embryos with recognizable anterior and posterior structures were recovered from tetraploid ⇔ Smad2−/− chimeras at day 7.5–10.5 (Table 1 and Fig. 3). X-gal staining confirmed that the embryos were formed almost exclusively by tissues derived from Smad2−/− ES cells (Fig. 3 C and D). This result clearly demonstrates that Smad2 is required in the extraembryonic tissues for proper gastrulation and establishment of an A-P axis. Another relevant observation derived from these experiments was that Smad2dex2−/− ES cells were able to differentiate in vivo into mesoderm derivatives, as indicated by the expression of brachyury in the region of the primitive streak (Fig. 3A) and the expression of sonic hedgehog (shh) in the axial mesoderm (Fig. 3B). This observation strongly supports that mesoderm induction and differentiation is independent of Smad2 function.
Table 1.
Phenotypes in rescued Smad2−/− embryos
| Phenotypes | E7.5 | E8.5 | E9.5 | E10.5 | E13.5 |
|---|---|---|---|---|---|
| Tetrapoloid ⇔ Smad2−/− chimeras | (n = 22) | (n = 24) | (n = 17) | (n = 10) | ND |
| Normal appearance | 27 | 37 | 12 | 10 | ND |
| Defects in embryo turning | — | — | 59 | 60 | ND |
| Anterior truncation | — | 12* | 24† | 70‡ | ND |
| Abnormal yolk-sac/egg cylinder | 73§ | 50 | 52 | 40 | ND |
| Diploid ⇔ Smad2−/− chimeras | (n = 9) | (n = 6) | |||
| Normal | 55 | 84 | |||
| Defects in embryo turning | 44 | — | |||
| Anterior truncation | 11 | 16¶ |
Numbers in the table represent percent of embryos seen with the particular phenotype. ND, not determined; En, embryonic day.
*†One embryo with holoprosencephaly.
‡One cyclops.
§Three showed multiple egg cylinders per decidua.
¶Cyclops.
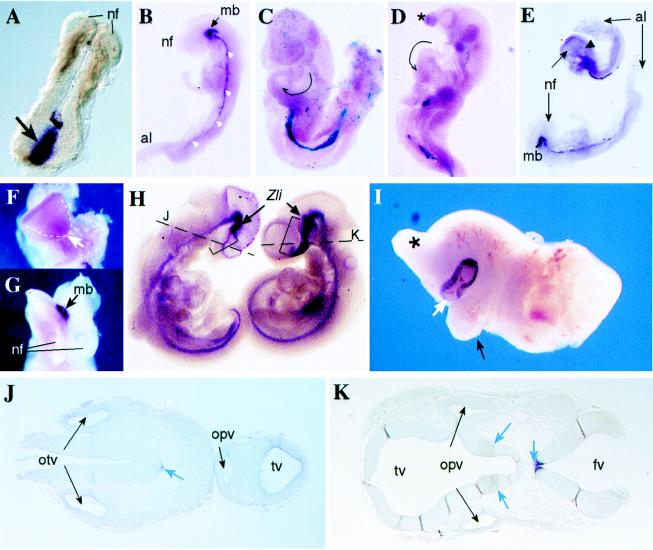
Figure 3.
Rescue of Smad2−/− phenotype and chimeric embryo phenotypes. (A) Brachyury in situ hybridization in a day 7.5 Smad2−/− embryo recovered from a tetraploid ⇔ Smad2−/− chimera. A strong staining of this mesoderm marker is observed in the primitive streak area (arrow). The formation of the neural folds (nf) in the anterior part of the embryo is also evident. (B) Shh in situ hybridization in a day 8.5 Smad2−/− rescued embryo. Staining for this marker is observed in the axial mesoderm (white arrowheads), as well as in the ventral midline of the midbrain (mb). The neural folds (nf, anterior) and the allantois (al, posterior) are indicated. (C) X-gal staining in a day 9.5 tetraploid ROSA26 ⇔ Smad2−/− chimera (viewed from its left side) showing that the embryo is formed almost exclusively by Smad2−/− cells. This particular embryo showed the highest contribution of wild-type tetraploid cells to the embryo (blue cells in the primitive endoderm, the allantois region, and head mesenchyme) and yet the embryo showed turning in clockwise direction (tailbud to the left) and bulbus cordis of the heart looping to the left of the midline (arrow). In normal embryos, the position of the bulbus cordis is at the right of the midline. (D) X-gal staining in a day 10.5 tetraploid ROSA26 ⇔ Smad2−/− chimera viewed from the left showing absence of turning (posterior part U-shaped) and abnormal head formation (∗), but normal turning of the cardiac tube (arrow). Some blue wild-type cells are observed only in the region of the primitive endoderm. (E) Shh in situ hybridization of two rescued embryos at day 8.5. Although the embryo in the bottom shows normal shh expression along the midline and midbrain (mb), in the upper embryo the expression of shh extends just to the hindbrain region (arrowhead). nf, neural folds; al, allantois. (F) Frontal view of the upper embryo shown in E, lacking normal midline separation at the anterior neural folds (arrow). (G) Frontal view of the bottom embryo shown in E exhibiting normal separation of the neural folds (nf). The expression of shh in the ventral midbrain (mb) is indicated. (H) Shh in situ hybridization in a tetraploid ⇔ Smad2−/− chimera (Left) and a wild-type (Right) embryo. The staining in the region of diencephalon and telencephalon (brackets) anterior to the zona limitans (Zli) is absent in the mutant cyclops embryo (see below). Moreover, in the Smad2-deficient embryo, the tip of the tail is positioned to the left, in contrast with the rightwards direction shown by the wild-type embryo. (I) Anterior fragment of a diploid ⇔ Smad2−/− chimera recovered at day 13.5 showing cyclopia. A proboscis (∗) and a protruding remnant of the right branchial arch (black arrow) are observed. The large contribution of mutant ES cells to this chimera is revealed by the strong pigmentation of the single eye (white arrow). (J) Transverse section of the mutant embryo shown in D (at the level of the broken line) shows a single telencephalic vesicle (tv) and a single optic vesicle (opv). A weak Shh staining in the floor plate of the neural tube and notochord (white arrow) and both otic vesicles (otv) are indicated. (K) Transverse section of the wild-type embryo shown in D (at the level of the broken line) shows normal bilobar telencephalic vesicles (tv) and two optic vesicles (opv). Normal Shh expression in the floor plate and ventral diencephalon is shown (blue arrows). The fourth ventricle (fv) is indicated.
Role of SMAD2 Signaling in Anterior Morphogenesis and the Establishment of Left–Right (L-R) Asymmetry.
Embryos rescued from tetraploid ⇔ Smad2−/− chimeras that survived beyond day 7.5 and up to day 10.5 exhibited a spectrum of abnormalities (Table 1). Among the rescued embryos at day 9.5 and day 10.5, 60% showed defects associated with L-R asymmetry. These defects included failure or delay in the sequence of turning and embryo rotation in clockwise direction (instead of anticlockwise) with positioning of the tailbud to the left of the embryo. Leftward (instead of rightward) or ambiguous looping of the cardiac tube was also observed (Table 1; Fig. 3 C and D). Among the embryos with anterior malformations, an evident phenotype was holoprosencephaly (Fig. 3 D, F, and H), an anterior midline defect characterized by facial and central nervous system defects caused by incomplete morphogenesis of the forebrain. To assess the nature of this anterior midline defect, the expression of the midline mesoderm and neuraxis marker shh (33) was examined. Analysis of shh expression in rescued embryos at day 8.5 revealed that in those with undivided neural fold, the most anterior midline mesoderm of the head process was absent (Fig. 3 E and F), in contrast with those with normal anterior development (Fig. 3 E and G). Closer analysis of affected day 10.5 embryos revealed abnormal morphogenesis of the prosencephalon and first branchial arches (Fig. 3 D and H). In the normal forebrain, shh expression in the ventral rostral diencephalon is found in two lateral stripes that merge in the floor of the telencephalon (33). Interestingly, Shh expression was absent in these structures in day 10.5 mutant embryos with severe holopresencephaly (Fig. 3 H–J). Moreover, staining of this marker in the floor plate was weak in some areas along the neuraxis (Fig. 3J). These results suggest that Smad2 is involved in the induction of the forebrain and that mutation of this gene causes the development of holoprosencephaly.
Similar phenotypes were observed in chimeras produced with diploid blastocysts, cells of which can contribute to both extraembryonic and embryonic tissues. In this case, the severity of the phenotypes observed depended on the proportion of wild-type and mutant cells in each embryo. Among embryos recovered at day 9.5 and day 13.5, as many as 85% appeared normal, but defects in embryo turning and in anterior-head morphogenesis were recurrent phenotypes in strong Smad2−/− chimeras (Table 1). The most striking examples were embryos showing several craniofacial abnormalities including cyclopia (Fig. 3I). Taken together, these results show that expression of Smad2 is required after gastrulation for the correct morphogenesis of the forebrain and for the proper embryo turning and looping of the cardiac tube.
Discussion
We have generated, by gene targeting, the Smad2dex2 allele, which in homozygosity results in early embryonic lethality. Smad2dex2 homozygous embryos do not express the earliest marker of A-P polarity Hex and show transient symmetrical expression of the mesoderm posterior marker Brachyury. These observations lead us to conclude that mutant Smad2 embryos lack appropriate A-P polarity and show transient induction of mesoderm. The fact that mesoderm derivatives were produced in vivo (rescued and unrescued embryos) as well as during the in vitro differentiation of Smad2−/− cells and in the formation of teratomas strongly suggests that Smad2 is not required for mesoderm induction in mouse. Similar conclusions were reached by analyzing the Smad2Robm1 mutant allele in which, too, the exon containing the original ATG was deleted (20). Interestingly, homozygous mutants for other Smad2 mutant alleles [mh1- and mh2-lacZ (21); ΔC (22)] fail to show any mesoderm induction. This phenotype, in combination with the fact that 20% of the heterozygous embryos with some of these mutations also displayed an abnormal phenotype (mh1- and mh2-lacZ alleles), something not seen in other Smad2 alleles (dex2, Robm1, ΔC), suggests allelic heterogeneity. One possibility is that the Smad2dex2 allele produces a truncated protein with some biological activity. This possibility is unlikely because we failed to detect the expected 22.6-kDa truncated protein with a C terminus-specific antibody. Different genetic backgrounds may also account for the phenotypic differences.
The use of Smad2dex2−/− ES cells in the formation of chimeras by using tetraploid blastocysts allowed us to show conclusively that Smad2 is required in the extraembryonic tissues for proper gastrulation and A-P axis formation. In the presence of wild-type extraembryonic tissues, mutant embryos are able to overcome the gastrulation defect and give rise to embryos with a clear A-P axis.
Mice with a targeted mutation in the activin receptor IB (ActRIB) die very early during development because of defects in embryonic ectoderm development (38) and Act RIB is required for proper visceral endoderm development. When wild-type ES cells were injected into ActRIB−/− blastocysts, a phenotype similar to that seen in our Smad2−/− embryos was observed (empty yolk sacs). These results are consistent with the role of ActRIB acting upstream of Smad2 to regulate the expression, in the visceral endoderm, of the genes required for the A-P axis specification.
We also showed that Smad2 expression is required at multiple stages during development. Tetraploid ⇔ diploid Smad2−/− chimeras yielded embryos that survived up to day 10.5, showing consistent abnormalities in L-R and anterior patterning. Two early events mark the establishment of L-R asymmetry in the mouse embryo: the consistent looping of the cardiac tube to the right and the turning of the embryo in anticlockwise direction to adopt a fetal position. Around 60% of the rescued Smad2 mutant embryos showed delay or absence of the sequence of turning, clockwise turning, and leftward or ambiguous looping of the cardiac tube. Within the TGF-β super-family, lefty and nodal are known to play important roles in the determination and/or maintenance of the L-R asymmetry (39–41). Together with our results, these observations suggest that lefty and/or nodal might signal through SMAD2, and that the defects in lateral patterning observed in Smad2-deficient embryos may result from the disruption of these pathways.
Defects in anterior patterning were the other set of abnormalities observed in rescued Smad2−/− embryos. The embryos showed holoprosencephaly, which is an anterior midline defect characterized by facial and central nervous system defects caused by incomplete morphogenesis of the forebrain. One of the most severe phenotypes originated by this defect is cyclopia. We observed this particular phenotype in several Smad2−/− embryos. This finding and several other observations suggest that SMAD2 is an essential factor for the establishment of anterior structures. In humans, many chromosomal rearrangements, including those of 18q21, a region where SMAD2 maps, are associated with cyclopia (42, 43). Mutations in Shh also result in holoprosencephaly (44). Here, we show that Smad2 mutant embryos with holoprosencephaly lacked Shh expression in the region anterior to the mesencephalon, indicating abnormal morphogenesis of forebrain. The linkage between Smad2 function and Shh expression remains to be elucidated. However, it is interesting to note that Shh is regulated by members of the forkhead family of transcription factors (45), and that members of this family have been found to interact with Smad2 for the transcriptional activation of genes in response to TGF-β and activins (46).
It has been shown that expression of nodal in the primitive endoderm is needed for the correct anterior patterning (47). About 30% of nodal and Smad2 double heterozygous mice display cyclopia (21). These observations might suggest that the anterior abnormalities we observed could be related to the disruption of nodal signaling. However, the fact that chimeras comprised of nodal−/− ES cells and wild-type blastocysts do not show anterior malformations (47) suggests that Smad2, and not nodal, is the mediator in signaling pathways, disturbance of which leads to cyclopia.
In summary, we have shown that Smad2-deficient embryos fail to gastrulate properly and lack A-P axis. We propose that an important factor contributing to the failure in establishing A-P polarity is the absence of Hex expression in the anterior visceral endoderm. Using a tetraploid embryo complementation assay, we have also shown that the aforementioned defects can be overcome by supplying normal Smad2 signaling in the visceral endoderm. Moreover, this technical approach led us to reveal the postgastrulation participation of Smad2 in several developmental processes, such as anterior head patterning, embryo turning, and cardiac tube looping.
Acknowledgments
The authors thank Teresa Shatzer for her technical assistance, Harry Hou for blastocyst injection, Robert Lechleider for the pan-SMAD antibody 367, and Todd Evans and Matthieu Gerard for the helpful discussion of the manuscript. This research was supported in part by the National Cancer Institute, Department of Health and Human Services, under contract with Advanced BioScience Laboratories, by grant CA67944 to R.K., and by a cancer center grant (CA13330) to the Albert Einstein College of Medicine.
Abbreviations
- A-P
anterior–posterior
- TGF-β
transforming growth factor β
- MH1
Mad homology domain 1
- ES cell
embryonic stem cell
- EB
embryoid bodies
- X-gal
5-bromo-4-chloro-3-indolyl β-d-galactoside
- L-R
left—right
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Hogan B L. Curr Opin Genet Dev. 1996;6:432–438. doi: 10.1016/s0959-437x(96)80064-5. [DOI] [PubMed] [Google Scholar]
- 2.Kingsley D M. Genes Dev. 1994;8:133–146. doi: 10.1101/gad.8.2.133. [DOI] [PubMed] [Google Scholar]
- 3.Roberts A B, Sporn M B. In: Handbook of Experimental Pharmacology. Sporn M B, Roberts A B, editors. Berlin: Springer; 1990. pp. 419–472. [Google Scholar]
- 4.Heldin C-H, Miyazono K, ten Dijke P. Nature (London) 1997;390:465–471. doi: 10.1038/37284. [DOI] [PubMed] [Google Scholar]
- 5.Graff J M, Bansal A, Melton D A. Cell. 1996;85:479–487. doi: 10.1016/s0092-8674(00)81249-0. [DOI] [PubMed] [Google Scholar]
- 6.Baker J C, Harland R M. Genes Dev. 1996;10:1880–1889. doi: 10.1101/gad.10.15.1880. [DOI] [PubMed] [Google Scholar]
- 7.Macias-Silva M, Abdollah S, Hoodless P A, Pirrone R, Attisano L, Wrana J L. Cell. 1996;87:1215–1224. doi: 10.1016/s0092-8674(00)81817-6. [DOI] [PubMed] [Google Scholar]
- 8.Zhang Y, Feng X H, Wu R Y, Derynck R. Nature (London) 1996;383:168–172. doi: 10.1038/383168a0. [DOI] [PubMed] [Google Scholar]
- 9.Nakao A, Roijer E, Imamura T, Souchelnytskyi S, Stenman G, Heldin C H, ten Dijke P. J Biol Chem. 1997;272:2896–2900. doi: 10.1074/jbc.272.5.2896. [DOI] [PubMed] [Google Scholar]
- 10.Lagna G, Hata A, Hemmati-Betarivanlou A, Massague J. Nature (London) 1996;383:832–836. doi: 10.1038/383832a0. [DOI] [PubMed] [Google Scholar]
- 11.Chen X, Rubock M J, Whitman M. Nature (London) 1996;383:691–696. doi: 10.1038/383691a0. [DOI] [PubMed] [Google Scholar]
- 12.Shi Y, Wang Y-F, Jayaraman L, Yang H, Massague J, Pavlevitch N P. Cell. 1998;94:585–594. doi: 10.1016/s0092-8674(00)81600-1. [DOI] [PubMed] [Google Scholar]
- 13.Tsukazaki T, Chiang T A, Davison A F, Attisano L, Wrana J L. Cell. 1998;95:779–791. doi: 10.1016/s0092-8674(00)81701-8. [DOI] [PubMed] [Google Scholar]
- 14.Hahn S A, Schutte M, Shamsul Hoque A T M, Moskaluk C A, da Costa L T, Rozenblum E, Weinstein C L, Fischer A, Yeo C J, Hruban R H, et al. Science. 1996;271:350–353. doi: 10.1126/science.271.5247.350. [DOI] [PubMed] [Google Scholar]
- 15.Thiagalingam S, Lengauer C, Leach F S, Schutte M, Hahn S A, Overhauser J, Willson J K, Markowitz S, Hamilton S R, Kern S E, et al. Nat Genet. 1996;13:343–346. doi: 10.1038/ng0796-343. [DOI] [PubMed] [Google Scholar]
- 16.Eppert K, Scherer S W, Ozcelik H, Pirone R, Hoodless P, Kim H, Tsui L C, Bapat B, Gallinger S, Andrulis I L, et al. Cell. 1996;86:543–552. doi: 10.1016/s0092-8674(00)80128-2. [DOI] [PubMed] [Google Scholar]
- 17.Riggins G J, Thiagalingam S, Rozenblum E, Weinstein C L, Kern S E, Hamilton S R, Willson J K V, Markowitz S D, Kinzler K W, Vogelstein B. Nat Genet. 1996;13:347–349. doi: 10.1038/ng0796-347. [DOI] [PubMed] [Google Scholar]
- 18.Uchida K, Nagatake M, Osada H, Yatabe Y, Kondo M, Mitsudomi T, Masuda A, Takahashi T. Cancer Res. 1996;56:5583–5585. [PubMed] [Google Scholar]
- 19.Dick A, Risau W, Drexler H. Dev Dyn. 1998;211:293–305. doi: 10.1002/(SICI)1097-0177(199804)211:4<293::AID-AJA1>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 20.Waldrip W R, Bikoff E K, Hoodless P A, Wrana J L, Robertson E J. Cell. 1998;92:797–808. doi: 10.1016/s0092-8674(00)81407-5. [DOI] [PubMed] [Google Scholar]
- 21.Nomura M, Li E. Nature (London) 1998;393:786–790. doi: 10.1038/31693. [DOI] [PubMed] [Google Scholar]
- 22.Weinstein M, Yang X, Li C, Xu X, Gotay J, Deng C-X. Proc Natl Acad Sci USA. 1998;95:9378–9383. doi: 10.1073/pnas.95.16.9378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lennon G, Auffray C, Polymeropoulos M, Soares B M. Genomics. 1995;16:151–152. doi: 10.1006/geno.1996.0177. [DOI] [PubMed] [Google Scholar]
- 24.Edelmann W, Yang K, Umar A, Heyer J, Lau K, Fan K, Liedtke W, Cohen P E, Kane M E, Lipford J R, et al. Cell. 1997;91:467–477. doi: 10.1016/s0092-8674(00)80433-x. [DOI] [PubMed] [Google Scholar]
- 25.Ioffe E, Liu Y, Bhaumik M, Poirier F, Factor S M, Stanley P. Proc Natl Acad Sci USA. 1995;92:7357–7361. doi: 10.1073/pnas.92.16.7357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heyer J, Kneitz B, Schuh K, Jankevics E, Siebelt F, Schimpl A, Serfling E. Immunobiology. 1997;198:162–169. doi: 10.1016/S0171-2985(97)80037-X. [DOI] [PubMed] [Google Scholar]
- 27.Robertson E J. In: Manipulating the Mouse Embryo. Robertson E J, editor. Oxford: IRL; 1987. pp. 71–112. [Google Scholar]
- 28.Boettinger E P, Jakubczak J L, Roberts I S D, Mumy M, Hemmati P, Bagnall K, Merlino G, Wakefield L M. EMBO J. 1997;16:2621–2633. doi: 10.1093/emboj/16.10.2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Z Q, Kiefer F, Urbánek P, Wagner E F. Mech Dev. 1997;62:137–145. doi: 10.1016/s0925-4773(97)00655-2. [DOI] [PubMed] [Google Scholar]
- 30.Zambrowicz B P, Imamoto A, Fiering S, Herzenberg L A, Kerr W G, Soriano P. Proc Natl Acad Sci USA. 1997;94:3789–3794. doi: 10.1073/pnas.94.8.3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hogan B, Beddington R, Costantini F, Lacy E. Teratocarcinomas and Embryonic Stem Cells. Plainview, NY: Cold Spring Harbor Lab. Press; 1994. p. 497. [Google Scholar]
- 32.Herrmann B G. Development (Cambridge, UK) 1991;113:913–917. doi: 10.1242/dev.113.3.913. [DOI] [PubMed] [Google Scholar]
- 33.Echelard Y, Epstein D J, St-Jacques B, Shen L, Mohler J, McMahon J A, McMahon A P. Cell. 1993;75:1417–1430. doi: 10.1016/0092-8674(93)90627-3. [DOI] [PubMed] [Google Scholar]
- 34.Takenoshita S, Mogix A, Nagashima M, Yang K, Yagi K, Hanyu A, Nagamachi Y, Miyazono K, Hagiwara K. Genomics. 1998;48:1–11. doi: 10.1006/geno.1997.5149. [DOI] [PubMed] [Google Scholar]
- 35.Thomas P Q, Brown A, Beddington R S P. Development (Cambridge, UK) 1998;125:85–94. doi: 10.1242/dev.125.1.85. [DOI] [PubMed] [Google Scholar]
- 36.Thomas P, Beddington R. Curr Biol. 1996;6:1487–1496. doi: 10.1016/s0960-9822(96)00753-1. [DOI] [PubMed] [Google Scholar]
- 37.Nagy A, Gocza E, Diaz E M, Prideax V R, Ivanyi E, Markkula M, Rossant J. Development (Cambridge, UK) 1990;110:815–821. doi: 10.1242/dev.110.3.815. [DOI] [PubMed] [Google Scholar]
- 38.Gu Z, Nomura M, Simpson B B, Lei H, Feijen A, Eijden-van Raaij J, Donahue P, Li E. Genes Dev. 1998;12:844–857. doi: 10.1101/gad.12.6.844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Collignon J, Varlet I, Robertson E J. Nature (London) 1996;381:155–158. doi: 10.1038/381155a0. [DOI] [PubMed] [Google Scholar]
- 40.Meno C, Saijoh Y, Fujii H, Ikeda M, Yokoyama T, Yokoyama M, Toyoda Y, Hamada H. Nature (London) 1996;381:151–155. doi: 10.1038/381151a0. [DOI] [PubMed] [Google Scholar]
- 41.Meno C, Shimono A, Saijoh Y, Yashiro K, Mochida K, Ohishi S, Noji S, Kondoh H, Hamada H. Cell. 1998;94:287–297. doi: 10.1016/s0092-8674(00)81472-5. [DOI] [PubMed] [Google Scholar]
- 42.Smart R D, Ross J, Amann G, Nelson M M. Am J Med Genet. 1986;24:269–272. doi: 10.1002/ajmg.1320240207. [DOI] [PubMed] [Google Scholar]
- 43.Levy-Mozziconacci A, Piquet C, Scheiner C, Adrai J, Potier A, Pelissier M C, Philip N. Prenatal Diagn. 1996;16:1156–1159. doi: 10.1002/(SICI)1097-0223(199612)16:12<1156::AID-PD13>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 44.Chiang C, Litingtung Y, Lee E, Young K E, Cordon J L, Westphal H, Beachy P A. Nature (London) 1996;383:407–413. doi: 10.1038/383407a0. [DOI] [PubMed] [Google Scholar]
- 45.Epstein D J, McMahon A P, Joyner A. Development (Cambridge, UK) 1999;126:281–292. doi: 10.1242/dev.126.2.281. [DOI] [PubMed] [Google Scholar]
- 46.Labbe E, Silvestri C, Hoodless P A, Wrana J L, Attisano L. Mol Cell. 1998;2:109–120. doi: 10.1016/s1097-2765(00)80119-7. [DOI] [PubMed] [Google Scholar]
- 47.Varlet I, Collignon J, Robertson E J. Development (Cambridge, UK) 1997;124:1033–1044. doi: 10.1242/dev.124.5.1033. [DOI] [PubMed] [Google Scholar]