Abstract
Complete resolution of the amide resonances in a three-dimensional solid-state NMR correlation spectrum of a uniformly 15N-labeled membrane protein in oriented phospholipid bilayers is demonstrated. The three orientationally dependent frequencies, 1H chemical shift, 1H–15N dipolar coupling, and 15N chemical shift, associated with each amide resonance are responsible for resolution among resonances and provide sufficient angular restrictions for protein structure determination. Because the protein is completely immobilized by the phospholipids on the relevant NMR time scales (10 kHz), the linewidths will not degrade in the spectra of larger proteins. Therefore, these results demonstrate that solid-state NMR experiments can overcome the correlation time problem and extend the range of proteins that can have their structures determined by NMR spectroscopy to include uniformly 15N-labeled membrane proteins in phospholipid bilayers.
More than 30% of all protein sequences encoded in the genomes of organisms, ranging in complexity from the simplest bacteria to humans, appear to correspond to hydrophobic membrane proteins (1). Understanding the functions of this important class of proteins requires the same kind of high-resolution structural analysis that is now routinely applied to globular proteins. However, determining the three-dimensional structures of membrane proteins is proving to be a difficult problem because the lipids associated with these proteins impede crystallization for x-ray diffraction as well as rapid overall reorientation for solution NMR spectroscopy. As a result, there are only a few selected examples of membrane proteins with their structures determined at atomic resolution by x-ray diffraction (2). And, although in favorable cases multidimensional solution NMR methods can be successfully applied to membrane proteins in detergent micelles (3), it is highly desirable to determine their structures in phospholipid bilayers where solution NMR methods fail completely.
Solid-state NMR spectroscopy is well suited for peptides and proteins immobilized in phospholipid bilayers, and several complementary approaches are under development (4–8). However, up until now experimental studies have required samples labeled with stable isotopes in one or a few selected sites. This not only makes the determination of complete structures laborious, but limits applications to peptides prepared by solid-phase synthesis, or small proteins with favorable distributions of amino acids, or selected regions of large proteins. Uniform 15N labeling of proteins was first implemented for solid-state NMR (9) and subsequently applied to solution NMR (10) studies where structure determination of expressed proteins has been facilitated by the resolution available in three-dimensional spectra (11, 12). The use of uniformly 15N-labeled samples shifts the burden from sample preparation to spectroscopy, where complete spectral resolution is the essential starting point for structure determination.
The solid-state NMR method for determining the structures of proteins that we are developing takes advantage of the favorable spectroscopic properties exhibited by uniaxially oriented samples (4). When the direction of molecular orientation is parallel to that of the applied magnetic field, the resulting spectra are characterized by single line resonances in all frequency dimensions (13). Both mechanically (14) and magnetically (15–17) oriented samples of membrane proteins in phospholipid bilayers are ideal for this approach. Because the observed resonance frequencies depend on the orientations of the molecular sites relative to the axis of orientation, they provide the basis for a method of structure determination. This method is independent of multidimensional solution NMR spectroscopy that relies instead on properties of the isotropic resonances observed from rapidly reorienting molecules (18). The spin interactions present at 15N-labeled backbone sites in proteins include the 1H chemical shift of the amide hydrogen, the 15N chemical shift of the amide nitrogen, and the 1H–15N heteronuclear dipolar coupling of these two directly bonded nuclei. Generally, the strong 1H–1H dipolar couplings are decoupled and the weak 15N–15N dipolar couplings ignored in these experiments, following the principles of dilute spin NMR spectroscopy of uniformly 15N-labeled proteins (9, 19). In separate experiments these homonuclear couplings can provide distance measurements and an approach to making sequential resonance assignments (20–22).
We have recently developed a family of two-, three-, and four-dimensional solid-state NMR experiments (21–24) for high resolution spectroscopy and structure determination of proteins (25, 26). Here we demonstrate that a fully resolved three-dimensional solid-state NMR correlation spectrum can be obtained from a uniformly 15N-labeled membrane protein in phospholipid bilayers. In these spectra each resonance associated with an amide site is characterized by three frequencies, the 1H chemical shift, 15N chemical shift, and 1H–15N dipolar coupling, that are responsible for resolution among resonances and provide sufficient angular restrictions for the determination of the orientation of each peptide plane. Complete three-dimensional structures are then assembled from the orientations of contiguous peptide planes (4, 27, 28).
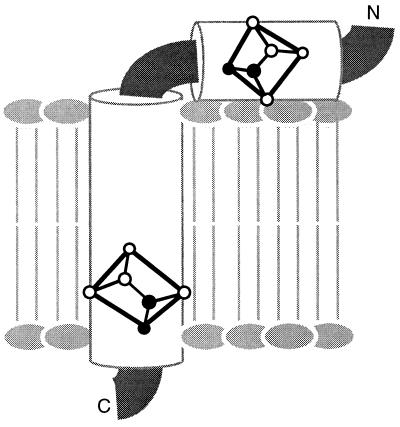
The major coat protein of fd bacteriophage has the primary sequence N-AEGDD PAKAA FDSLQ ASATE YIGYA WAMVV VIVGA TIGIK LFKKF TSKAS-C. Newly synthesized copies of the protein are stored in the membrane of infected Escherichia coli bacteria prior to incorporation into virus particles as they are extruded through the membrane. The structure of the membrane bound form of this protein has been determined in micelles by solution NMR spectroscopy (29–32). It has an amphipathic helix that lies in the plane of the membrane (residues 7–16), and a longer hydrophobic membrane spanning helix (residues 27–44). A loop, showing some evidence of mobility, connects the two helices, and residues near the N and C termini are mobile.
MATERIALS AND METHODS
Samples of fd Coat Protein in Bilayers.
15N-labeled fd coat protein was prepared as described (29), starting with the isolation of bacteriophage particles by polyethylene glycol precipitation from the supernatant, following the growth of fd-infected E. coli (K3300F+) in a medium containing either 15N ammonium sulfate for uniform labeling, or 15N Leu (Cambridge Isotope Laboratories, Cambridge, MA) for selective labeling. The coat protein was purified by size exclusion chromatography. Fifty milligrams of fd bacteriophage was vortex mixed and bath sonicated for 1 hr in 5 ml of buffer (50 mM phosphate, pH 7.0) containing 100 mM SDS and 200 μl of chloroform. The resulting solution was applied to a 2.6 × 100 cm Sephacryl S-200 (Pharmacia) column and eluted with the same buffer containing 5 mM SDS. The fractions containing coat protein were pooled and concentrated by ultrafiltration in an Amicon cell using a YM10 membrane. SDS was exchanged for cholate by passing the protein solution through a Sephacryl S-200 column equilibrated in buffer (10 mM Tris⋅Cl, pH 7.0/0.1 mM EDTA/200 mM KCl) containing 12 mM cholate instead of SDS. The cholate-solubilized protein was concentrated to 2.5 mg/ml by ultrafiltration.
Protein reconstitution in phospholipid vesicles was accomplished using the freeze-thaw method of Bayer and Feigenson (33). The samples were prepared at a protein/lipid molar ratio of 1.2:100 and contained 16 mg of coat protein plus 170 mg of combined palmitoyl-oleoyl-phosphatidylcholine/palmitoyl-oleoyl-phosphatidylglycerol (POPC/POPG) lipids in a molar ratio of 80:20. The lipids were mixed in chloroform, and the solvent was evaporated under a stream of nitrogen followed by high vacuum for 1 hr. The dry lipid mixture was then suspended in 9 ml of buffer and sonicated to transparency using a Branson sonifier equipped with a micro tip. To facilitate subsequent steps the vesicle suspension was divided into two 85 mg aliquots, and 8 mg of 2.5 mg/ml coat protein in cholate buffer was added to each. After vortex mixing, the lipid protein mixtures were each diluted by the addition of 9 ml of buffer, rapidly frozen in liquid nitrogen, allowed to thaw at room temperature, and bath sonicated for 30 sec. Each preparation was transferred to a 10,000 molecular weight cutoff dialysis bag (Spectrum Scientific, Dallas) and dialyzed against six 12-hr changes of 4 liters of buffer, followed by four 5-hr changes of 4 liters of water. The two reconstituted vesicle preparations were combined and concentrated by ultrafiltration.
For the unoriented sample the protein containing vesicles were simply transferred to a 7 mm × 2 cm cylindrical glass tube (Wilmad, Buena, NY). For the oriented samples the vesicle suspension was evenly distributed over the surface of forty glass cover slides (Marienfeld Glassware, Bad Margentheim, Germany) with dimensions 11 × 20 × 0.07 mm. After allowing the bulk water to evaporate, the slides were stacked and placed in a sealed chamber together with a saturated ammonium phosphate solution which provided a 93% relative humidity atmosphere. Oriented bilayers formed after equilibrating the sample in this chamber at 42°C for 12 hr. Before insertion into the coil of the NMR probe, the stack of glass plates was wrapped in a thin layer of parafilm and then placed in thin polyethylene tubing, which was heat sealed at both ends, to maintain sample hydration during the NMR experiment. The orientation of the lipids was confirmed with 31P NMR spectra, whereas the narrow single line resonances of the one-dimensional 15N NMR spectra verified the protein orientation. The spectra presented in Figs. 1, 2, 3, 4 were obtained from a single oriented sample containing 16 mg of uniformly 15N-labeled fd coat protein in phospholipid bilayers.
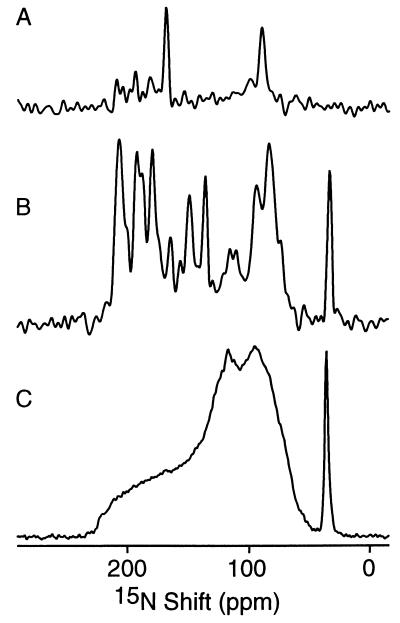
Figure 1.
One-dimensional 15N chemical shift NMR spectra of 15N-labeled fd coat protein in phospholipid bilayers. (A) Selectively 15N Leu-labeled protein in oriented bilayers. (B) Uniformly 15N-labeled protein in oriented bilayers. (C) Uniformly 15N-labeled protein in unoriented bilayer vesicles. Two hundred and fifty-six data points were acquired using CPMOIST cross polarization with a mix time of 1 ms and a recycle delay of 3 sec.
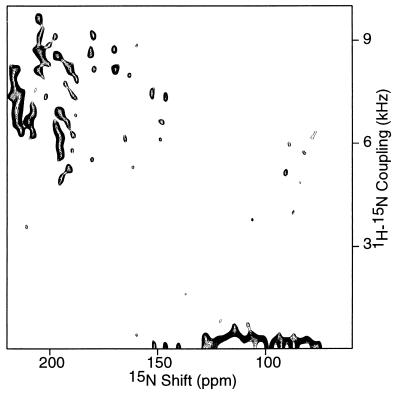
Figure 2.
Two-dimensional 1H–15N dipolar coupling/15N chemical shift PISEMA spectrum of an oriented sample of uniformly 15N-labeled fd coat protein in phospholipid bilayers. Two hundred and fifty-six transients were acquired for each of 64 t1 values incremented by 40.8 μsec. The cross-polarization mix time was 1 msec and the recycle delay was 3 sec.
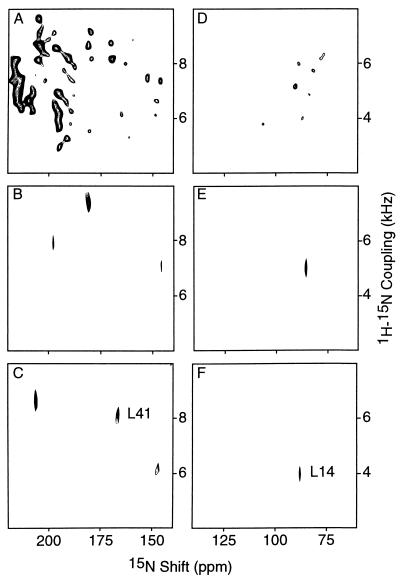
Figure 3.
Two-dimensional 1H–15N heteronuclear dipolar coupling/15N chemical shift spectral planes from spectra of an oriented sample of uniformly 15N-labeled fd coat protein in phospholipid bilayers. (A and D) Expansions of the two-dimensional PISEMA spectrum shown in Fig. 2; A is the trans-membrane helix region and D is the in-plane helix region. (B) Plane extracted from the three-dimensional correlation spectrum at 1H chemical shift 7.4 ppm. (C) Plane extracted from a three-dimensional correlation spectrum at 1H chemical shift 11.0 ppm; the resonance assigned to Leu-41 in the hydrophobic trans-membrane helix is marked. (E) Plane extracted from a three-dimensional correlation spectrum at 1H chemical shift 12.5 ppm. (F) Plane extracted from a three-dimensional correlation spectrum at 1H chemical shift 11.6 ppm; the resonance assigned to Leu-14 in the amphipathic in-plane helix is marked. The three-dimensional correlation spectrum was obtained with 16 t1 and 16 t2 experiments with respective dwell times of 32.7 and 40.8 μsec. The two leucine resonances in the three-dimensional spectrum were assigned by comparison with the two-dimensional PISEMA spectrum of an oriented sample of selectively 15N Leu-labeled fd coat protein in phospholipid bilayers.
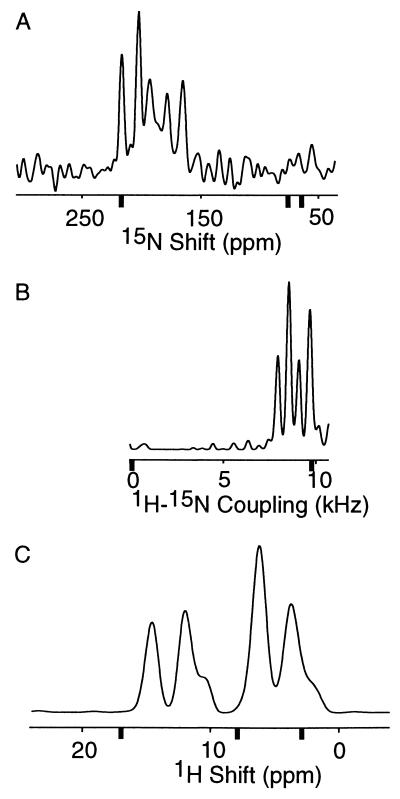
Figure 4.
One-dimensional spectral slices along the three frequency axes taken from two-dimensional spectra. The magnitudes of the principal elements of the relevant spin-interactions are shown with bold tick marks. (A) 15N chemical shift slice taken from the PISEMA spectrum in Fig. 2 at the 1H–15N dipolar coupling frequency of 8.0 kHz. The 15N amide chemical shift tensor elements, referenced to liquid ammonia at 0 ppm, are 64, 77, and 217 ppm for σ11, σ22, and σ33, respectively (40). (B) 1H–15N dipolar coupling slice taken from the PISEMA spectrum in Fig. 2 at a 15N chemical shift value of 205.0 ppm. The maximum 1H−15N dipolar coupling frequency at ν∥ is 9.7 kHz. (C) 1H chemical shift slice taken from a two-dimensional heteronuclear correlation spectrum at a 15N shift value of 205.0 ppm. The 1H amide chemical shift tensor elements, referenced to tetramethylsilane at 0 ppm, are 3, 8, and 17 ppm for σ11, σ22, and σ33, respectively (40).
Solid-State NMR Spectroscopy.
The NMR experiments were performed on a Chemagnetics–Otsuka Electonincs (Ft. Collins, CO) CMX 400 spectrometer with a wide-bore Oxford Magnet Technology (Oxford, U.K.) 400/89 magnet. The home-built single coil probes were double-tuned to the resonance frequencies of 1H at 400.5 MHz and 15N at 40.6 MHz. The stack of glass plates was placed in a rectangular, five-turn coil with inner dimensions of 20 × 11 × 5 mm. This high power “square” coil version of the original flat coil probe (34) was shown previously to have adequate rf homogeneity for the homonuclear line-narrowing procedures used here (25). The unoriented sample was placed in a 7-mm solenoidal coil. The temperature was monitored with a thermocouple located near the sample and maintained at 22°C by flowing large volumes of cooled nitrogen gas over the sample.
The one-dimensional 15N chemical shift spectra were obtained with single-contact CPMOIST (cross-polarization with mismatch-optimized IS transfer) cross-polarization (19, 35) to generate 15N magnetization that was acquired in the presence of continuous 1H irradiation to decouple the heteronuclear dipolar interactions. The two-dimensional PISEMA (polarization inversion with spin exchange at the magic angle) (23) and three-dimensional solid-state NMR correlation (24) experiments utilize flip-flop, phase- and frequency-switched, Lee–Goldburg homonuclear decoupling (36–38) to provide line-narrowing in the 1H chemical shift and 1H–15N dipolar coupling dimensions. A two-dimensional 1H chemical shift/15N chemical shift heteronuclear correlation spectrum was obtained by fixing the SEMA (spin exchange at the magic angle) period, which is incremented as t2 in the pulse sequence that generates a three-dimensional correlation spectrum (24). The sensitivity enhancement available from cross-polarization (19) is so great that multidimensional solid-state and solution NMR spectra can be obtained on similar amounts of uniformly 15N-labeled protein in comparable time periods. Typically, high quality two-dimensional spectra are obtained in overnight runs, whereas the three-dimensional data shown in Fig. 3 were obtained in 3 days, the same length of time as the corresponding solution nuclear Overhauser effect spectroscopy–heteronuclear multiple quantum correlation (NOESY-HMQC) spectrum from a sample in micelles (32).
The 1H rf field strength of 1.2 mT (150 W) dictates 1H frequency jumps of 35.4 kHz to fulfill the Lee–Goldburg off-resonance condition for magic angle rf irradiation (36). During the SEMA period the 15N rf field strength was increased to 1.4 mT (1.45 kW) to match the effective 1H rf field strength. In all experiments the 1H rf field strength was increased to 1.7 mT (350 W) to provide adequate decoupling during data acquisition. The 1H and 15N chemical shifts were referenced to 0 ppm for external liquid samples of tetramethylsilane and ammonia, respectively. The NMR data were processed using the program felix (Biosym Technology, San Diego) on Silicon Graphics computer workstations.
RESULTS AND DISCUSSION
All of the spectral features in the one-dimensional spectra in Fig. 1 result from the 15N chemical shift interaction. The spectrum in Fig. 1C was obtained from an unoriented sample of uniformly 15N-labeled fd coat protein in phospholipid bilayers. The narrow peak near 30 ppm results from the amino groups of the lysine sidechains and the N terminus and is not of concern here. Solid-state NMR spectra are strongly affected by molecular motion. Most of the backbone sites are structured and immobile on the time scale of the 15N chemical shift interaction (10 kHz) and contribute to the characteristic amide powder pattern between about 220 and 60 ppm. Several backbone sites of the membrane bound form of fd coat protein including those near the N and C termini and some in the loop region between the two helices are mobile and unstructured, resulting in the narrower resonance band centered at the isotropic resonance frequency near 120 ppm. Additional details about the frequencies and amplitudes of intramolecular motions can be described through lineshape analysis and relaxation measurements. No resolution among the resonances is feasible in the spectrum of an unoriented sample with multiple labeled sites without additional sample manipulations. Magic angle sample spinning narrows the resonances significantly and, in favorable cases, it is possible to resolve individual resonances in proteins, albeit at the expense of the orientational information. In contrast, sample orientation gives well-resolved spectra that retain the orientational information used for structure determination. In both magic angle sample spinning and oriented sample approaches it is possible to utilize dipole–dipole couplings to make distance measurements.
The orientational dependence of the 15N chemical shift interaction serves as a means for obtaining spectral resolution and as a source of structural information. The solid-state NMR spectrum of an oriented bilayer sample is strikingly different from that of an unoriented sample. The spectrum in Fig. 1B, obtained from an oriented sample of uniformly 15N-labeled fd coat protein, displays significant resolution with identifiable peaks at frequencies throughout the range of the 15N amide chemical shift anisotropy powder pattern. There are two resonances in the spectrum of selectively 15N Leu-labeled fd coat protein in oriented phospholipid bilayers (Fig. 1A). We have observed 15N resonance linewidths narrower than 3 ppm from these samples, consistent with mosaic spreads of sample orientation of less than about 2°. The large 80 ppm frequency difference between the resonances from Leu-41, near the downfield end of the spectrum, and Leu-14, near the upfield end, demonstrates the effect of protein structure on these spectra. Leu-41, in the trans-membrane helix, has its N—H bond approximately parallel to the field and to σ33 of the chemical shift tensor, while Leu-14, in the amphipathic in-plane helix, has its N—H bond perpendicular (29).
The two-dimensional PISEMA spectrum of uniformly 15N-labeled fd coat protein in Fig. 2 has many resolved resonances, each of which is characterized by a 1H–15N dipolar coupling frequency and a 15N chemical shift frequency. There are some resonances with zero dipolar coupling frequencies. Those backbone sites that are mobile on the time scales of the 1H–15N dipolar and 15N chemical shift interactions have resonances with zero dipolar and isotropic chemical shift frequencies. Some orientations of amide groups in helices perpendicular to the field have very small or zero dipolar coupling frequencies associated with chemical shift frequencies near σ11 and σ22 (25, 39, 40). Also, any magnetization transferred from the 1H spin reservoir to the 15N spin reservoir through cross-polarization before polarization inversion, which does not participate in spin exchange at the magic angle, appears at zero dipolar frequency.
The three-dimensional correlation spectrum provides both greatly increased resolution and the simultaneous measurement of three different resonance frequencies for each amide site of the protein. Fig. 3 displays the trans-membrane and in-plane helical regions of 1H–15N dipolar coupling/15N chemical shift planes. The spectra in Fig. 3 A and D are expansions of the two-dimensional PISEMA shown in Fig. 2. The two-dimensional planes in Figs. 3 B, C, E, and F were extracted from the three-dimensional 1H chemical shift/1H–15N dipolar coupling/15N chemical shift correlation spectrum of uniformly 15N-labeled fd coat protein in phospholipid bilayers. These planes contain only those resonances with the selected 1H chemical shift frequencies. In particular, the planes in Fig. 3 C and G were selected at 1H chemical shifts of 11.0 ppm and 11.6 ppm so that they include the resonances of Leu-14 and Leu-41, respectively, as marked. Examination of two-dimensional planes throughout the 1H chemical shift range, demonstrates that all of the resonances of this uniformly 15N-labeled protein are fully resolved. This enables the measurement of the 1H chemical shift, 1H–15N dipolar coupling, and 15N chemical shift frequencies for all amide sites in the protein.
The spectral resolution available with this approach is illustrated in Fig. 4. The linewidths observed in the one-dimensional spectral slices taken from two-dimensional spectra are 1.2 ppm for the 1H chemical shift, 300 Hz for the 1H–15N dipolar coupling, and 3 ppm for the 15N chemical shift dimensions. These compare favorably with the values (0.8 ppm, 180 Hz, and 2–8 ppm) observed in spectra of single crystal samples of 15N-labeled dipeptides obtained in solenoidal coil probes. With further spectroscopic development, and especially the use of high-field spectrometers, even better resolution will be observed in the spectra of proteins. Nonetheless, the current level of resolution is impressive because the resonances are spread across the full frequency spans of three different spin interactions. The bold ticks mark the frequencies of the principal elements of the spin-interaction tensors in the spectral slices shown in Fig. 4. A useful index of spectral resolution is the ratio of the total spectral range available to the resonance linewidth. The observed ratios are about 10, 30, and 50 for the 1H chemical shift, 1H–15N dipolar coupling, and 15N chemical shift dimensions, respectively. These ratios are comparable to those observed along the 1H and 15N frequency axes of multidimensional solution NMR spectra of the same protein in detergent micelles (32); even though the linewidths are considerably narrower in solution NMR spectra, the range of isotropic chemical shift frequencies are correspondingly smaller as well. Note that three independent parameters contribute to the resolution in three-dimensional solid-state NMR correlation spectra compared with only two in typical three-dimensional solution NMR spectra, for example NOESY-HMQC spectra (11, 12).
The data in the three-dimensional correlation spectrum of an oriented sample of a uniformly 15N-labeled protein provide sufficient information for complete structure determination (4). The three frequencies measured for each resonance depend on the magnitudes and orientations of the principal elements of the spin interaction tensors in the molecular frame and on the orientation of the molecular site with respect to the direction of the applied magnetic field. The peptide plane orientations derived from the data in Table 1 for the amide sites of Leu-14 and Leu-41 are shown in Fig. 5. By placing these planes in the context of trans-membrane and in-plane helices, the angular ambiguities are resolved. In general, this is part of the process of assembling the structure of a protein from the orientations of contiguous planes (27).
Table 1.
Values of the spectral parameters for Leu-14 and Leu-41 measured from the spectra in Fig. 3 F and C, respectively
| Residue | 1H shift, ppm | 1H–15N coupling frequency, kHz | 15N shift, ppm |
|---|---|---|---|
| Leu-14 | 11.6 | 4.5 | 90.0 |
| Leu-41 | 11.0 | 8.1 | 170.0 |
Figure 5.
The orientations of the peptide planes for Leu-14 and Leu-41 of fd coat protein in phospholipid bilayers, as determined from the data in Fig. 3 F and C. The planes are placed in the context of the structure determined by solution NMR spectroscopy of this membrane protein in micelles, which has a hydrophobic trans-membrane helix and an amphipathic in-plane helix.
Once the amide resonances are resolved and their frequencies measured, the final necessary step is to assign them to individual residues in the protein. The nuclear Overhauser effect is a valuable tool in solution NMR spectroscopy where it is used to measure internuclear distances as well as for resonance assignment (18). Likewise, homonuclear spin exchange experiments using either 1H or 15N nuclei provide a general assignment strategy for solid-state NMR spectra of uniformly 15N-labeled proteins. Abundant spin exchange occurring among nearby 1H nuclei in model peptides (22), and dilute spin exchange among 15N sites in both model peptides and proteins (20, 21) have been demonstrated. Alternative assignment strategies that utilize uniformly 13C and 15N-labeled proteins (41), analogous to those used in solution NMR spectroscopy (42), are also under development.
CONCLUSIONS
The seminal developments of multiple-pulse methods (43) for narrowing the resonances of abundant (1H) spins, and double-resonance methods (19) for sensitivity enhancement and high-resolution spectroscopy of dilute (15N) spins, provide the fundamental basis for high resolution NMR spectroscopy of immobile molecules, including those as complex as proteins (44). The data in Fig. 3 show that these methods can be combined with sample orientation to yield completely resolved spectra of a uniformly 15N-labeled protein.
The analysis of orientationally dependent spectral frequencies in terms of structure (4, 27) is particularly straightforward for the majority of amide sites of fd coat protein in phospholipid bilayers because they are completely ordered, as shown by each having a single orientation and a maximal order parameter of 1. No structural information is available on these time scales for sites with isotropic resonances, for example the N- and C-terminal residues, because of the effects of complete motional averaging. However, it is possible to obtain crucial details about structure and dynamics of sites with limited motional averaging, whether due to local segmental or residue motions or because the sample itself undergoes some motions as seen in magnetically oriented bicelles (15–17).
Although this approach is still at an early stage in its development, the resolution and sensitivity observed in bilayer samples are comparable to those in solution NMR spectra for the same uniformly 15N-labeled protein in detergent micelles. The 1H chemical shift resolution, in particular, can be improved by performing the experiments in higher field spectrometers, with the added bonus of greater sensitivity. Even with current instrumentation these methods can be applied to polypeptides with substantially more than 50 residues, because the data in Fig. 3 indicate that there is room for many additional resolved resonances in each of the two-dimensional planes at selected 1H chemical shift frequencies. More importantly, even though the fd coat protein is relatively small, it is completely immobilized in the phospholipid bilayers (witness the full breadth of the 15N chemical shift powder pattern in Fig. 1C); thus the linewidths will not degrade in spectra of larger proteins or complexes. This is an important difference between the effects of size of the polypeptides on these solid state NMR experiments and on multidimensional solution NMR experiments. As the proteins get larger there will be additional resonances in both types of spectra. However, in solid-state NMR spectra increased size is not accompanied by changes in relaxation parameters or linewidths, so that sensitivity will only be diminished because of the smaller number of molecules in a fixed volume of material. This is in contrast to solution NMR where not only there will be more resonances, but also the shorter relaxation times and broader lines render it difficult to make correlations and to transfer magnetization. Thus, the ability to obtain completely resolved solid-state NMR spectra of immobile proteins extends the range of proteins that can have their structures determined by NMR spectroscopy to include membrane proteins in phospholipid bilayers.
Acknowledgments
We thank Chia-Shang Jason Liu for his assistance in the preparation of the samples used in these experiments. This research was supported by Grants R37 GM24266, RO1 AI20770, and RO1 GM29754 from the National Institutes of Health, and utilized the Resource for Solid-State NMR of Proteins at the University of Pennsylvania supported by Grant P41 RR09793 from the Biomedical Resource Technology Program, Division of Research Resources, National Institutes of Health. F.M.M. was supported by postdoctoral fellowship 9304FEN-1004-43344 from the Medical Research Council of Canada.
ABBREVIATIONS
- PISEMA
polarization inversion with spin exchange at the magic angle
- SEMA
spin exchange at the magic angle
- NOESY-HMQC
nuclear Overhauser effect spectroscopy–heteronuclear multiple quantum correlation
References
- 1.Fraser C M, Gocayne J D, White O, Adams M D, Clayton R A, et al. Science. 1995;270:397–403. doi: 10.1126/science.270.5235.397. [DOI] [PubMed] [Google Scholar]
- 2.Garavito R M, Picot D, Loll P J. J Bioenerg Biomembr. 1996;28:13–27. [PubMed] [Google Scholar]
- 3.McDonnell P A, Opella S J. J Magn Reson Ser B. 1993;102:120–125. [Google Scholar]
- 4.Opella S J, Stewart P L, Valentine K G. Q Rev Biophys. 1987;19:7–49. doi: 10.1017/s0033583500004017. [DOI] [PubMed] [Google Scholar]
- 5.Gullion T, Schaefer J. J Magn Reson. 1989;81:196–200. [Google Scholar]
- 6.Levitt M H, Raleigh D P, Creuzet F, Griffin R G. J Chem Phys. 1990;92:6347–6364. [Google Scholar]
- 7.Cross T A, Opella S J. Curr Opin Struct Biol. 1994;4:574–581. [Google Scholar]
- 8.Smith S O. Curr Opin Sruct Biol. 1993;3:755–759. [Google Scholar]
- 9.Cross T A, Di Verdi J A, Opella S J. J Am Chem Soc. 1982;104:1759–1761. [Google Scholar]
- 10.Bogusky M J, Tsang P, Opella S J. Biochem Biophys Res Commun. 1985;127:540–545. doi: 10.1016/s0006-291x(85)80193-5. [DOI] [PubMed] [Google Scholar]
- 11.Fesik S W, Zeiderweg E R P. J Magn Reson. 1988;78:588–593. [Google Scholar]
- 12.Marion D, Driscoll P C, Kay L E, Wingfield P E, Bax A, Gronenborn A M, Clore G M. Biochemistry. 1989;29:6150–6156. doi: 10.1021/bi00441a004. [DOI] [PubMed] [Google Scholar]
- 13.Opella S J, Waugh J S. J Chem Phys. 1977;66:4919–4924. [Google Scholar]
- 14.Bechinger B, Kim Y, Chirlian L E, Gesell J, Neumann J-M, Montal M, Tomich J, Zasloff M, Opella S J. J Biomol NMR. 1991;1:167–173. doi: 10.1007/BF01877228. [DOI] [PubMed] [Google Scholar]
- 15.Howard K P, Opella S J. J Magn Reson Ser B. 1996;112:91–94. doi: 10.1006/jmrb.1996.0116. [DOI] [PubMed] [Google Scholar]
- 16.Sanders C R, Hare B, Howard K P, Prestegard J H. Prog NMR Spectrosc. 1994;26:421–444. [Google Scholar]
- 17.Prosser R S, Hunt S A, DiNatale J A, Vold R R. J Am Chem Soc. 1996;118:269–270. [Google Scholar]
- 18.Wuthrich K. NMR of Proteins and Nucleic Acids. New York: Wiley; 1986. [Google Scholar]
- 19.Pines A, Gibby M G, Waugh J S. J Chem Phys. 1973;59:569–590. [Google Scholar]
- 20.Cross T A, Frey M H, Opella S J. J Am Chem Soc. 1983;105:7471–7473. [Google Scholar]
- 21.Ramamoorthy A, Gierasch L M, Opella S J. J Magn Reson Ser B. 1995;109:112–116. doi: 10.1006/jmrb.1995.1157. [DOI] [PubMed] [Google Scholar]
- 22.Ramamoorthy A, Gierasch L M, Opella S J. J Magn Reson Ser B. 1996;111:81–84. doi: 10.1006/jmrb.1996.0063. [DOI] [PubMed] [Google Scholar]
- 23.Wu C H, Ramamoorthy A, Opella S J. J Magn Reson Ser A. 1994;109:270–272. [Google Scholar]
- 24.Ramamoorthy A, Wu C H, Opella S J. J Magn Reson Ser B. 1995;107:88–90. doi: 10.1006/jmrb.1995.1063. [DOI] [PubMed] [Google Scholar]
- 25.Ramamoorthy A, Marassi F M, Zasloff M, Opella S J. J Biomol NMR. 1995;6:329–334. doi: 10.1007/BF00197814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jelinek R, Ramamoorthy A, Opella S J. J Am Chem Soc. 1995;117:12348–12349. [Google Scholar]
- 27.Opella S J, Stewart P L. Methods Enzymol. 1989;176:242–275. doi: 10.1016/0076-6879(89)76015-8. [DOI] [PubMed] [Google Scholar]
- 28.Ketchem R R, Hu W, Cross T A. Science. 1993;261:1457–1460. doi: 10.1126/science.7690158. [DOI] [PubMed] [Google Scholar]
- 29.McDonnell P A, Shon K, Kim Y, Opella S J. J Mol Biol. 1993;233:447–463. doi: 10.1006/jmbi.1993.1523. [DOI] [PubMed] [Google Scholar]
- 30.Henry G D, Sykes B D. Biochemistry. 1992;31:5284–5297. doi: 10.1021/bi00138a007. [DOI] [PubMed] [Google Scholar]
- 31.van de Ven F J M, Os J W M, Aelen J M A, Wymenga S S, Remerowski M L, Konings R N H, Hilbers C W. Biochemistry. 1993;32:8322–8328. doi: 10.1021/bi00083a036. [DOI] [PubMed] [Google Scholar]
- 32.Almeida F, Opella S J. J Mol Biol. 1997;270:481–495. doi: 10.1006/jmbi.1997.1114. [DOI] [PubMed] [Google Scholar]
- 33.Bayer R, Feigenson G W. Biochim Biophys Acta. 1985;815:369–379. doi: 10.1016/0005-2736(85)90363-3. [DOI] [PubMed] [Google Scholar]
- 34.Bechinger B, Opella S J. J Magn Reson. 1991;95:585–588. [Google Scholar]
- 35.Levitt M H. J Chem Phys. 1986;94:30–38. [Google Scholar]
- 36.Lee M, Goldburg W I. Phys Rev A. 1965;140:1261–1271. [Google Scholar]
- 37.Mehring M, Waugh J S. Phys Rev B. 1972;5:3459–3471. [Google Scholar]
- 38.Bieleki A, De Groot H J M, Griffin R G, Levitt M H. Adv Magn Reson. 1990;14:111–124. [Google Scholar]
- 39.Bechinger B, Zasloff M, Opella S J. Protein Sci. 1993;2:2007–2084. doi: 10.1002/pro.5560021208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu C H, Ramamoorthy A, Gierasch L M, Opella S J. J Am Chem Soc. 1995;117:6148–6149. [Google Scholar]
- 41.Schneider D M, Tycko R, Opella S J. J Magn Reson. 1987;73:568–573. [Google Scholar]
- 42.Kay L E, Ikura M, Tschudin R, Bax A. J Magn Reson. 1990;89:496–514. doi: 10.1016/j.jmr.2011.09.004. [DOI] [PubMed] [Google Scholar]
- 43.Waugh J S, Huber L M, Haeberlen U. Phys Rev Lett. 1968;20:180–182. [Google Scholar]
- 44.Ramamoorthy A, Marassi F M, Opella S J. In: Dynamics and the Problem of Recognition in Biological Macromolecules. Jardetzky O, Lefevre J, editors. New York: Plenum; 1996. pp. 237–255. [Google Scholar]