Abstract
Rhodopsin is a prototypical G protein-coupled receptor that is activated by photoisomerization of its 11-cis-retinal chromophore. Mutant forms of rhodopsin were prepared in which the carboxylic acid counterion was moved relative to the positively charged chromophore Schiff base. Nanosecond time-resolved laser photolysis measurements of wild-type recombinant rhodopsin and two mutant pigments then were used to determine reaction schemes and spectra of their early photolysis intermediates. These results, together with linear dichroism data, yielded detailed structural information concerning chromophore movements during the first microsecond after photolysis. These chromophore structural changes provide a basis for understanding the relative movement of rhodopsin’s transmembrane helices 3 and 6 required for activation of rhodopsin. Thus, early structural changes following isomerization of retinal are linked to the activation of this G protein-coupled receptor. Such rapid structural changes lie at the heart of the pharmacologically important signal transduction mechanisms in a large variety of receptors, which use extrinsic activators, but are impossible to study in receptors using diffusible agonist ligands.
Keywords: retinal, rhodopsin mutants, linear dichroism, photointermediates, visual transduction
Rhodopsin is the light-triggered membrane receptor in rod outer segments responsible for dim-light vision. Absorption of light transforms rhodopsin (rho) into an activated state, which interacts with a heterotrimeric G protein to initiate an enzyme cascade leading to visual transduction. To reach this active state, called metarhodopsin (meta) II (1, 2), rho passes through a number of intermediates that have been characterized by UV-visible, Fourier-transform infrared (FTIR), Raman, and NMR spectroscopies, in addition to spin-labeling and biochemical studies (3–10). One way to characterize the intermediates is to trap them at low temperatures. This approach led to the classical “bleaching” scheme (λmax values in nanometers):
rho (498) → bathorhodopsin (batho) (542) → lumirhodopsin (lumi) (497) → meta I (480) → meta II (380)
Scheme I
However, at more physiologically relevant temperatures, time-resolved measurements up to 1 msec after photolysis reveal a new intermediate and a different kinetic scheme (3, 11). The following reaction scheme was proposed to occur during the nanosecond-to-microsecond time regime (3):
rho (498) → batho (529) ⇋ bsi (477) → lumi (492) → …
Scheme II
The new intermediate, blue-shifted intermediate of rho (bsi), is entropically favored and thus not trapped at low temperatures (11).
The role of various structural features of the retinal chromophore in early rho photointermediates has been studied through time-resolved spectral measurements of artificial rho, in which the native chromophore is replaced by synthetically modified retinals (12). In studies of bacteriorhodopsin, time-resolved absorption spectroscopy has been shown to be particularly valuable when combined with site-directed mutagenesis to probe the roles of specific protein residues in different intermediates (13). Until now, this has been impossible for studies of visual pigments, because of the difficulty of expressing sufficient quantities of protein for laser photolysis studies. The recent development of a microscale apparatus (14) makes it possible to measure sets of UV-visible difference spectra on samples as small as 300 μg from a few nanoseconds to microseconds following pigment photolysis. This opens the possibility of conducting kinetic measurements on site-directed rho mutants to probe the role of the chromophore and its interactions with protein residues in terms of trigger mechanisms leading to the active state. To date, spin-label studies have provided the most detailed time-resolved information about protein motion following pigment photolysis (8, 9), though only with millisecond time resolution. The combination of site-directed mutagenesis with nanosecond spectroscopy used in this study thus provides unique information about early structural changes in proteins in a noncrystalline environment.
In this report, we describe nanosecond photolysis studies of counterion mutants E113D (Glu-113, the carboxylic acid counterion to the protonated Schiff base (PSB) was replaced by the shorter Asp residue) and E113A/A117E (Glu-113 was moved one helix turn toward the center of the membrane-embedded domain, ≈5.4 Å, 400°) and show that this information can be used to elucidate chromophore structural changes in the early batho, bsi, and lumi intermediates.
MATERIALS AND METHODS
Preparation of Rho Mutants.
Site-directed mutagenesis of bovine opsin was carried out as previously reported (15–17). Opsin genes were expressed in COS-1 cells and reconstituted with 11-cis-retinal. The resulting pigments were purified in dodecylmaltoside buffer solution [10 mM Tris⋅HCl, pH 7.0/30 mM NaCl/60 mM KCl/2 mM MgCl2/0.1% (wt/vol) dodecylmaltoside] and concentrated as described previously (15–18).
Time-Resolved Spectroscopy.
All laser photolysis measurements were performed in a microapparatus described previously (14). Spectral measurements were carried out at 20°C for rho and mutant pigment E113D, and at 10°C for mutant pigment E113A/A117E. The latter mutant was studied at a lower temperature because at 20°C most of the batho intermediate already had decayed at the first time point investigated (20 ns).
Data Analysis.
The data were analyzed using global exponential fitting following singular value decomposition as described elsewhere (11, 19, 20). This procedure fits the entire set of absorbance data at all wavelengths and delay times simultaneously.
Molecular Modeling.
We used an IBM compatible Pentium computer equipped with the program chemsite to simulate the orientations of the chromophore in its different states. Because the position and orientation of polar residues within the chromophore pocket are unknown and likely to change during the progression of the intermediates, we did not calculate the transition dipole moments of the intermediates. To model chromophore orientations we adopted the measured orientation of the main transition dipole moment with respect to the C6—C12 axis of the chromophore from ref. 21. This information, together with the angles of the transition dipole moments of the intermediates with respect to each other (22), and additional information from NMR, FTIR, and mutant work, was used to deduce the absolute direction of chromophore motions proceeding through the early intermediates.
RESULTS AND DISCUSSION
Time-Resolved UV-Visible Spectroscopy of Glu-113 Mutants.
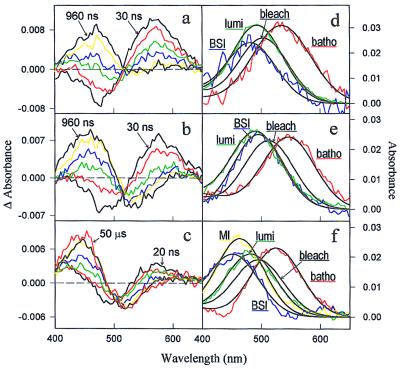
We measured difference spectra from 400 nm to 650 nm in a time range from 20 ns to 50 μs for recombinant rho (COS rho) and both mutants (Fig. 1). The recombinant rho data set (Fig. 1a) could be fit to Scheme II and agreed in λmax values (Fig. 1d, Table 1) as well as in time constants (Table 2) with the intermediates of native rho measured previously under identical conditions (11). The difference spectra of mutant E113D are shown in Fig. 1b. This mutant also generally followed reaction Scheme II, but with λmax values and time constants that differ from recombinant rho (Fig. 1e, Tables 1 and 2). In the case of mutant E113A/A117E, batho decayed into an equilibrium with bsi on a time scale that was too fast to resolve. Therefore, the first spectrum at 20 ns represents a batho/bsi mixture decaying to lumi, which subsequently decays to a meta I-like absorber (Fig. 1c). To fit the E113A/A117E data set, Scheme II had to be modified as follows:
Figure 1.
(a) Difference spectra for COS rho after photolysis with 7-ns full width at half maximum 477-nm laser pulses at time points 30 ns (black), 60 ns (red), 120 ns (green), 240 ns (blue), 480 ns (yellow), and 960 ns (black) (T = 20°C). (b) Difference spectra for E113D; times as in a. (c) Difference spectra for E113A/A117E at time points 20 ns (black), 60 ns (red), 200 ns (green), 800 ns (blue), 3 μs (yellow), 10 μs (black), and 50 μs (red) (T = 10°C). (d–f) COS rho-, E113D-, E113A/A117E-intermediate spectra, respectively. Intermediate spectra require the kinetic scheme to be known, and the bleach (the amount and Gaussian shape of the photolyzed sample determined in a separate experiment, data not shown) must be added back to the difference spectra (23).
Table 1.
Summary of the λmax values and shifts relative to parent pigment spectrum (in parentheses) in nanometers and in cm−1 (bottom lines) of the intermediates of the two counterion mutants, recombinant rhodopsin (COS rho) and rhodopsin
| Species | Rhodopsin* | COS rho† | E113D† | E113A/A117E† |
|---|---|---|---|---|
| Pigment | 498 20,080 | 500 20,000 | 509 19,646 | 490 20,408 |
| Batho | 529 (+31) 18,904 (1,176) | 531 (+31) 18,832 (1,168) | 540 (+31) 18,519 (1,127) | 525 (+35) 19,048 (1,360) |
| BSI | 477 (−21) 20,964 (884) | 479 (−21) 20,877 (877) | 495 (−14) 20,202 (556) | 453 (−37) 22,075 (1,667) |
| Lumi | 492 (−6) 20,325 (245) | 490 (−10) 20,408 (408) | 486 (−23) 20,576 (930) | 482 (−8) 20,747 (339) |
| Meta I | NA | NA | NA | 466 (meta I-like) 21,459 (1,051) |
Table 2.
Summary of the decay times of the intermediates of the two counterion mutants, recombinant rhodopsin (COS rho) and rhodopsin
rho → batho/bsi ⇋ lumi ⇋ meta I-like → … .
Scheme III
The spectra, λmax values, and kinetic time constants of the intermediates are presented in Fig. 1f, Table 1, and Table 2, respectively.
In the dark state of rhodopsin it is largely accepted that increasing the counterion–PSB distance red shifts, while decreasing the distance blue shifts, the absorption maximum (25–30). In batho, the newly introduced twist in the chromophore complicates calculation of the counterion–PSB distance. However, in bsi and lumi the chromophore twists have largely relaxed (31, 32) and we assume that the relative shifts (numbers in parentheses in Table 1) of the λmax values, in proceeding from the dark state to bsi and from bsi to lumi, can be interpreted qualitatively in terms of counterion − PSB distance (24). This hypothesis rests on the assumption that during these transitions the relative orientations of the counterion and other polar residues in the binding site of rho and the mutants do not change dramatically, since it has been shown that the absorption spectra can be modulated up to 20 nm in changing the orientation of the counterion and other polar residues (33). For the mutation E113D this is clearly true, but for the more uncertain displacement of E113A/A117E counterion orientation changes may contribute to the spectral shifts. The net effect would be a quantitative error in the distance from the PSB to Glu-117. This would not be significant for our model, however, since the spectral shifts are used to deduce direction of motion rather than to calculate exact distances.
Using the relative spectral shifts to approximate changes in the PSB–counterion distance constrains linear dichroism data previously obtained (22) such that the direction of chromophore movements progressing through the batho, bsi and lumi intermediates can be deduced. The linear dichroism data from ref. 22 theoretically allows for 16 different pathways of transition dipole orientation changes proceeding from the dark state through batho, bsi, and lumi, if all mirror images represented in figure 4 of ref. 22 (including mixing of open and closed symbols) are included. One main goal of this study was to find out which of these possible pathways the chromophore takes.
The Dark State of Rho.
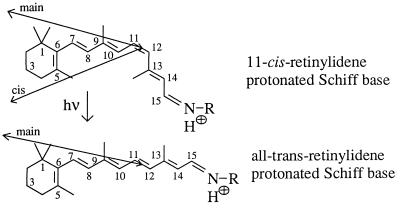
The geometry of 11-cis-retinal in native rho is derived from crystal structure data (34). The basic structure and carbon numbering scheme of the chromophore are shown in Fig. 2. To reduce steric interaction of the 5- and the 13-methyl groups with their adjacent hydrogens (8-H and 10-H, respectively), 11-cis-retinal is twisted ≈30° around both the C6—C7 and C12—C13 single bonds, the latter angle being rotated clockwise when viewed from C12 to C13 (ref. 35 and nomenclature from ref. 36). Although the direction of rotation of the 5-methyl group in the pigment has not yet been measured, we propose that the C6—C7 angle is positive (clockwise viewed from C6 to C7), because after isomerization this orientation “loads” the 5-methyl group of the retinal against 8-H (3, 37).
Figure 2.
Basic structure and carbon numbering of the chromophore before (11-cis-retinal) and after (all-trans-retinal) isomerization. The transition dipole orientations of the main and the cis bands are indicated by arrows.
The orientation of the main (498 nm) transition dipole moment of the chromophore is 16° from the membrane plane (38). It is important to note that the transition dipole of the main band, defined by the in-phase oscillation of all five ethylenic oscillators, C7=C8, C9=C10, C11=C12, C13=C14, and C15=N16, does not point exactly along the retinal axis, defined as the horizontal line from C6 to C12 in Fig. 2, but is inclined ≈16° (21). The cis band transition dipole, defined by the out-of-phase oscillation of the C7=C8, C9=C10, and C11=C12 oscillators with respect to the C13=C14 and C15=N16 oscillators, forms an angle of 38° with the main band transition dipole (22). Fig. 2 shows the orientations of the transition dipole of the main bands (deduced from crystal structure data from ref. 21) and the cis band (deduced from linear dichroism measurements from ref. 22). Bond twists are not shown in Fig. 2. Further information on the position of the chromophore in the protein comes from the fact that the β-ionone ring occupies a position in which a photoactivatable group attached to C3 is able to crosslink to Trp-265 and Leu-266 in helix 6 (39, 40). This information, with the arrangement of the helices from Fig. 4a (see below), provides enough constraints on the 11-cis-retinal chromophore structure and position that it can be modeled into its binding site. We rotated 11-cis-retinal along the retinal axis to a position where the main band, looking from the Schiff base (SB) end to the β-ionone ring, points 16° out of the membrane plane toward the intradiscal side and the cis band points 22° out of the membrane plane in the opposite direction (toward the cytoplasmic side). This results in a rotation angle of ≈40–50° of the retinal plane (defined as C6 to C12) relative to the membrane plane and places Glu-113 below the retinal axis in a position proposed previously (6, 7). This initial position of 11-cis-retinal in the dark state reduces the 16 different possible configurations determined in ref. 22 to four possible configurations. The distance of C12 to O1 of Glu-113 was taken to be ≈3 Å (6, 7, 41).
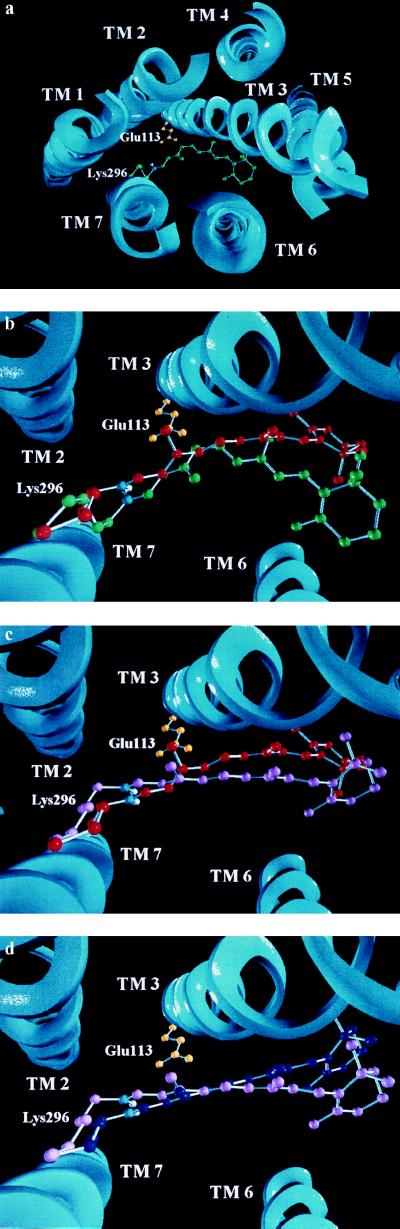
Figure 4.
(a) Graphics model of the hypothetical arrangement of the seven helices in rho. The view is from the cytoplasmic to the intradiscal side. The membrane plane is the paper plane. 11-cis-retinal (green) is attached to Lys-296 (green) via a protonated SB (proton, white; nitrogen, blue). Lys-296 is pointing down to the intradiscal side and Glu-113 (yellow) is pointing up to the cytoplasmic side. (b) Expanded view into the protein. Linear dichroism (22) and λmax values of rho and rho mutants (Table 1) were used to model the chromophore into its binding pocket showing the rho (green) to batho (red) transition. (c) Batho (red) to bsi (violet) transition structural changes deduced as in b. (d) bsi (violet)-to-lumi (blue) transition structural changes deduced as in b and c.
Figs. 3 and 4a show the orientation of 11-cis-retinal in the dark state of rho (green). The view is down from the cytoplasmic to the intradiscal side in Figs. 3a and 4a and from within the plane of the membrane in Fig. 3b. The hypothetical positions of Glu-113 in rho (yellow), Asp-113 in E113D (blue), and Glu-117 in E113A/A117E (green), as deduced qualitatively from the pigment absorption, the blue shifts of the bsi, and lumi intermediates, and from the arrangement of the helices in Fig. 4a (see below) are shown as well. Structural water forming hydrogen bonds from the PSB to O2 of Glu-113 (not shown in Fig. 3) is likely to be present (7, 42).
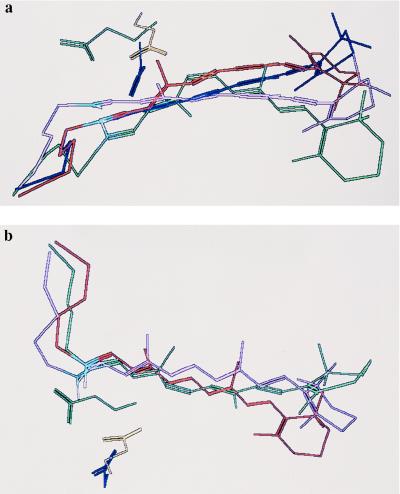
Figure 3.
(a) View of chromophore motions from the cytoplasmic to the intradiscal side. The movement of the chromophore (attached to Lys-296) from rho (green), to batho (red), to bsi (violet), and to lumi (blue), is deduced from linear dichroism data (22) and the relative blue shifts of the bsi and lumi spectra (Table 1). The pathway that fits the spectral data is represented by the open symbols used in figure 4 of ref. 22, if that figure is rotated by 180° along the plane containing the main-band and the cis-band transition dipole orientations. The relative angles of the transition dipole orientation changes given in ref. 22 could be reproduced within the experimental error. The hypothetical positions of the counterions Glu-113 (yellow), Asp-113 (blue), and Glu-117 (green), as deduced qualitatively from the blue shifts of the pigment and the arrangement of the helices in Fig. 4a, are shown as well. The distance of the Glu-113 oxygen closest to C12 (referred to as O1) was kept at 3 Å in rho and in batho (7, 41). It is likely that structural water forms a hydrogen bond network from O2 of Glu-113 to the SB proton (white) (not shown in this figure) (7, 42). In rho, the distances of the SB proton to O2 of Glu-113 in the dark state, batho, bsi, and lumi are 4.6, 4.4, 3.6, and 3.9 Å, respectively. In E113D, the respective SB proton–O2 of Asp-113 distances are 4.9, 4.7, 4.2, and 4.0 Å and in E113A/A117E the respective SB proton–O2 of Glu-117 distances are 3.9, 3.9, 3.0, and 3.3 Å. These numbers were deduced from the orientation of the chromophore guided by the blue shifts of the bsi and lumi intermediates. Thus, they should be taken as a qualitative measure of the relative PSB–counterion distance changes. (b) Same molecular arrangement as in Fig. 3a but without lumi (for clarity of the figure) and viewed from within the plane of the membrane.
Starting from the transition dipole orientation of 11-cis-retinal and the position of Glu-113 proposed in ref. 7, four different configurations of the transition dipoles of batho and lumi are consistent with the linear dichroism results (22). Two configurations are possible with both batho and lumi on the same side of the plane containing the cis and main band transition dipole moments, and two others are possible with batho and lumi on opposite sides of this plane. All four configurations were tested for agreement with the relative movements of the SB end progressing from the dark state to bsi and from bsi to lumi. If configurations were used with batho and lumi transition dipoles on opposite sides of the cis-main band plane (solid and open symbols from figure 4 in ref. 22), the transition dipole motions would be too large to be modeled into the chromophore binding site.
The Batho Intermediate.
In batho, the SB environment remains unchanged (5, 32), but the transition dipole orientation differs from the dark state by 8 ± 2° (22). Taking the steric hindrance between the 13-methyl group and 10-H into account, isomerization must involve a rotation with the 13-methyl group moving away from the adjacent 10-H. To accomplish this, we moved the C12—H and C11—H bonds in opposite directions as previously proposed (7). The displacement of the chromophore was reduced by introducing twists along the single bonds C14—C15 (40°), C10—C11 (20°), and C8—C9 (20°); whereas, the C12—C13 twist was kept constant at 30° and the C6—C7 angle was reduced to 20° to load the 5-methyl group against 8-H (3, 37). These twist assignments agree well with Raman and FTIR data (5, 32). CCC bends of the chromophore in this intermediate (accompanied by CCCC torsions), leading to protein–retinal steric interactions, are thought to represent the main contribution to energy storage in the batho intermediate (7, 43, 44).
From recent UV-visible data attributing the loss of intensity at 275 nm in the batho intermediate to a reduced hyperchromism (45) involving Trp-265 and Tyr-268 (55), it is likely that the β-ionone ring is positioned differently after isomerization. This is also consistent with recent crosslinking studies (39, 40). Fig. 3 a and b shows the different conformations of the retinal in going from the dark state (green) to batho (red) as deduced from the above information and transition dipole orientations (22). The change of position and orientation of the SB proton after isomerization was minimized as required from FTIR and resonance Raman measurements (5, 32). As a result, the largest chromophore movement is represented by the ring portion of the retinal. Only one of the two remaining configurations consistent with the linear dichroism results (open symbols in figure 4 of ref. 22) could account for the relatively large movement of the β-ionone ring parallel to the membrane plane.
The bsi Intermediate.
FTIR results show a change in PSB environment in bsi (31). Taking the relative blue shifts induced by all counterions into account (Table 1), PSB movement must be more toward Glu-117 than toward Glu-113 and Asp-113 in the mutants (24). Inspection of Table 1 reveals a relatively large blue shift for E113A/A117E-bsi (37 nm), which cannot entirely be accounted for by a possible reorientation of the Glu-117 counterion. For the mutant E113D, the SB stretching frequency as well as the “fingerprint” region of the chromophore are very similar to rho (46), so the orientation of Asp-113 in the mutant must be similar to Glu-113 in rho. Thus, we conclude that the major contribution to the blue shifts of the mutant spectra stem from a decreased counterion–PSB distance. Partial positive charges are located on the SB proton and adjacent odd numbered carbons, C15 and C13, as a result of close proximity to the negative counterion. We therefore interpret SB movements as movements of the SB end of the chromophore. In general, a distance change of the positive charges with respect to the counterion can be evoked by rotational and translational movements. Since the chromophore is fixed on both ends by means of a SB linkage and a bulky β-ionone ring, respectively, we assume rotational movements to be dominant. The transition dipole orientation relative to the dark state in bsi is 11 ± 2° (22). The chromophore movement that is consistent with this angle and with the bsi blue shifts (Table 1) is shown in Fig. 3 a and b. Looking from the cytoplasmic to the intradiscal side, the bsi transition dipole has moved to a lateral position close to that in the original 11-cis conformation (22) (Fig. 3a), though oriented more into the membrane plane compared with rho and batho (22) as shown in Fig. 3b.
In bsi, the retinal also assumes a more relaxed conformation, as indicated by reduced FTIR hydrogen out-of-plane modes (31). We thus propose relaxations of the C14—C15 and C12—C13 twists and reductions of the C10—C11 and C8—C9 twist angles. The 5-methyl group, however, passes 8-H and forms an ≈60° angle on the opposite side of the retinal plane. This relatively large angle places the β-ionone ring into a position where the ring orientations in bsi and the dark state are similar, as required from time-resolved spectral measurements in the UV (55).
The Lumi Intermediate.
In lumi, the spectral blue shifts (Table 1) show that the SB moves away from Glu-117 and toward Asp-113 (24), as indicated in Fig. 3a. This movement agrees with the transition dipole orientation in lumi (12 ± 2° with respect to the dark state) (22), where the chromophore has to move into a side-to-side position similar to that in batho (Fig. 3a), though it is less inclined relative to the membrane plane. All twists in lumi are relaxed except for the ≈45° C6—C7 twist, in agreement with the largely reduced hydrogen out-of-plane modes in this intermediate (32).
Modeling of the Chromophore into Its Binding Site.
The chromophore structures of Fig. 3 were used to model the seven-helix receptor structure showing the rho → batho → bsi → lumi transitions. A model for the seven-helix structure was designed based on a projection structure (47), with assignments of the helices as reported in ref. 48. Fig. 4a shows the arrangement of the seven helices with 11-cis-retinal attached to Lys-296 in an orientation deduced from Fig. 3. The SB nitrogen is blue, its proton white, and the counterion Glu-113 yellow. Fig. 4 b–d shows expanded views into the receptor showing the rho → batho, batho → bsi, and bsi → lumi transitions, respectively.
Relative movements of transmembrane helices 3 and 6 have recently been identified as requirements for rho activation (8, 9, 49). Since the chromophore resides between transmembrane helices 3 and 6 and moves quite substantially during the conversions from rho → batho → bsi → lumi (Fig. 4), it is most likely that these early triggers are the precursors of previously observed motions of those helices (8, 9, 49). This agrees with FTIR spectra, which show that the 9-methyl group, 13-methyl group, and β-ionone ring affect the early intermediates (32, 50, 51). In addition, for retinal binding to the opsin apoprotein, the positioning of the β-ionone ring and the 9-methyl group have been shown to be crucial (52, 53).
Kinetics of the Rho → Batho → bsi → Lumi Transitions.
As expected, repositioning of the counterion had an effect not only on the λmax values of the intermediates but on the reaction kinetics as well. In mutant E113D, the ≈1 Å increase in PSB − counterion distance resulted in a forward shifted batho ⇋ bsi equilibrium and in a slowing of the bsi-to-lumi transition. It seems that bsi is favored in this mutant relative to wild type, possibly because the shorter side chain gives more freedom to the chromophore, increasing the entropy. In mutant E113A/A117E there exist back reactions for all intermediates. Obviously, the energy barriers for the back reactions have decreased. This agrees with previous studies that find that specific structural features of rho exist in their respective active (“on”) or inactive (“off”) states, respectively (54).
In conclusion, time-resolved spectral measurements of the early photointermediates of rho mutants, together with time-resolved linear dichroism studies, have enabled us to determine chromophore structural changes in the first microsecond of visual transduction and to link them to proposed trigger mechanisms. Even though a high-resolution structure of the dark state of rho is not available, these time-resolved spectral measurements of rho and rho mutants allow us to determine relative chromophore structural changes of short-lived intermediates, which lead to the activation of the protein. Future studies will enable us to explore additional structural features of photointermediates and probe protein chromophore interactions of this receptor; most importantly, measured under physiologically relevant conditions. In an extension of the method described here, the use of photoactivatable ligands bound to other G protein-coupled receptors could provide new insights into the early processes of activation of a large class of receptors that would not be amenable to other techniques.
Acknowledgments
This work was supported in part by National Institutes of Health Grant EY00983 to D.S.K. T.P.S is an Associate Investigator the Howard Hughes Medical Institute. T.A.Z. is supported by National Institutes of Health Training Grant EV07138.
ABBREVIATIONS
- FTIR
Fourier-transform infrared
- rho
rhodopsin
- batho
bathorhodopsin
- bsi
blue-shifted intermediate of rho
- lumi
lumirhodopsin
- meta
metarhodopsin
- SB
Schiff base
- PSB
protonated Schiff base
References
- 1.Hofmann K P. Photochem Photobiophys. 1986;13:309–338. [Google Scholar]
- 2.Hofmann K P, Jäger S, Ernst O P. Isr J Chem. 1995;35:339–355. [Google Scholar]
- 3.Kliger D S, Lewis J W. Isr J Chem. 1995;35:289–307. [Google Scholar]
- 4.Siebert F. Isr J Chem. 1995;35:309–323. [Google Scholar]
- 5.Palings I, Pardoen J A, Van den Berg E, Winkel C, Lugtenburg J, Mathies R M. Biochemistry. 1987;26:2544–2556. doi: 10.1021/bi00383a021. [DOI] [PubMed] [Google Scholar]
- 6.Han M, DeDecker B S, Smith S O. Biophys J. 1993;65:899–906. doi: 10.1016/S0006-3495(93)81117-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Han M, Smith S O. Biochemistry. 1995;34:1425–1432. doi: 10.1021/bi00004a037. [DOI] [PubMed] [Google Scholar]
- 8.Farahbakhsh Z T, Hideg K, Hubbell W L. Science. 1993;262:1416–1418. doi: 10.1126/science.8248781. [DOI] [PubMed] [Google Scholar]
- 9.Farrens D L, Altenbach C, Yang K, Hubbell W L, Khorana H G. Science. 1996;274:768–770. doi: 10.1126/science.274.5288.768. [DOI] [PubMed] [Google Scholar]
- 10.Sakmar T P, Fahmy K. Isr J Chem. 1995;35:325–337. [Google Scholar]
- 11.Hug S J, Lewis J W, Einterz C M, Thorgeirsson T E, Kliger D S. Biochemistry. 1990;29:1475–1485. doi: 10.1021/bi00458a019. [DOI] [PubMed] [Google Scholar]
- 12.Randall C E, Lewis J W, Hug S J, Björling S C, Eisner-Shanas I, Friedman N, Ottolenghi M, Sheves M, Kliger D S. J Am Chem Soc. 1991;113:3473–3485. [Google Scholar]
- 13.Lanyi J K, Váró G. Isr J Chem. 1995;35:365–385. [Google Scholar]
- 14.Lewis J W, Kliger D S. Rev Sci Instrum. 1993;64:2828–2833. [Google Scholar]
- 15.Zvyaga T A, Fahmy K, Sakmar T P. Biochemistry. 1994;33:9753–9761. doi: 10.1021/bi00198a046. [DOI] [PubMed] [Google Scholar]
- 16.Sakmar T P, Franke R R, Khorana H G. Proc Natl Acad Sci USA. 1989;86:8309–8313. doi: 10.1073/pnas.86.21.8309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zvyaga T A, Min K C, Beck M, Sakmar T P. J Biol Chem. 1993;268:4661–4667. [PubMed] [Google Scholar]
- 18.Fahmy K, Sakmar T P. Biochemistry. 1993;32:9165–9171. doi: 10.1021/bi00086a023. [DOI] [PubMed] [Google Scholar]
- 19.Thorgeirsson T E, Lewis J W, Wallace-Williams S E, Kliger D S. Biochemistry. 1993;32:13861–13872. doi: 10.1021/bi00213a015. [DOI] [PubMed] [Google Scholar]
- 20.Lewis J W, Liang J, Ebrey T G, Sheves M, Kliger D S. Biochemistry. 1995;34:5817–5823. doi: 10.1021/bi00017a013. [DOI] [PubMed] [Google Scholar]
- 21.Drikos G, Rüppel H. Photochem Photobiol. 1984;40:93–104. [Google Scholar]
- 22.Lewis J W, Einterz C M, Hug S J, Kliger D S. Biophys J. 1989;56:1101–1111. doi: 10.1016/S0006-3495(89)82758-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Albeck A, Friedman N, Ottolenghi M, Sheves M, Einterz C M, Hug S J, Lewis J W, Kliger D S. Biophys J. 1989;55:233–241. doi: 10.1016/S0006-3495(89)82798-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jäger S, Lewis J W, Zvyaga T A, Szundi I, Sakmar T P, Kliger D S. Biochemistry. 1997;36:1999–2009. doi: 10.1021/bi962320u. [DOI] [PubMed] [Google Scholar]
- 25.Blatz P E, Mohler J H, Navangul H V. Biochemistry. 1972;11:848–855. doi: 10.1021/bi00755a026. [DOI] [PubMed] [Google Scholar]
- 26.Honig B, Greenberg A, Dinur U, Ebrey T G. Biochemistry. 1976;15:4593–4599. doi: 10.1021/bi00666a008. [DOI] [PubMed] [Google Scholar]
- 27.Kliger D S, Milder S J, Dratz E A. Photochem Photobiol. 1977;25:277–286. doi: 10.1111/j.1751-1097.1977.tb06911.x. [DOI] [PubMed] [Google Scholar]
- 28.Arnaboldi M, Motto M G, Tsujimoto K, Balogh-Nair V, Nakanishi K. J Am Chem Soc. 1979;101:7082–7084. [Google Scholar]
- 29.Koutalos Y, Ebrey T G, Tsuda M, Odashima K, Lien T, Park M H, Shimizu N, Derguini F, Nakanishi K, Gilson H R, Honig B. Biochemistry. 1989;28:2732–2739. doi: 10.1021/bi00432a055. [DOI] [PubMed] [Google Scholar]
- 30.Birge R R. Biochim Biophys Acta. 1990;1016:293–327. doi: 10.1016/0005-2728(90)90163-x. [DOI] [PubMed] [Google Scholar]
- 31.Ganter U M, Kashima T, Sheves M, Siebert F. J Am Chem Soc. 1991;113:4087–4092. [Google Scholar]
- 32.Ganter U M, Schmid E D, Perez-Sala D, Rando R R, Siebert F. Biochemistry. 1989;28:5954–5962. doi: 10.1021/bi00440a036. [DOI] [PubMed] [Google Scholar]
- 33.Beppu Y, Kakitani T. Photochem Photobiol. 1994;59:660–669. [PubMed] [Google Scholar]
- 34.Gilardi R D, Karle I L, Karle J. Acta Crystallogr B. 1972;28:2605–2612. [Google Scholar]
- 35.Nakanishi K, Borhan B, Karnaukhova E, Lou J, Tan Q, Sakai N, Ren R. Seventh International Conference on Retinal Proteins. Ramat-Gan, Israel: Dan Knassim; 1996. p. 40. [Google Scholar]
- 36.Klyne W, Prelog V. Experientia. 1960;16:521–523. [Google Scholar]
- 37.Mah T L, Lewis J W, Sheves M, Ottolenghi M, Kliger D S. Photochem Photobiol. 1995;62:356–360. doi: 10.1111/j.1751-1097.1995.tb05282.x. [DOI] [PubMed] [Google Scholar]
- 38.Liebman P A. Biophys J. 1962;2:161–178. doi: 10.1016/s0006-3495(62)86847-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang H, Lerro K A, Yamamoto T, Lien T H, Sastry L, Gawinowicz M A, Nakanishi K. J Am Chem Soc. 1994;116:10165–10173. [Google Scholar]
- 40.Nakayama T A, Khorana H G. J Biol Chem. 1990;265:15762–15769. [PubMed] [Google Scholar]
- 41.Honig B, Dinur U, Nakanishi K, Balogh-Nair V, Gawinowicz M A, Arnaboldi M, Motto M G. J Am Chem Soc. 1979;101:7084–7086. [Google Scholar]
- 42.Rafferty C N, Shichi H. Photochem Photobiol. 1981;33:229–234. doi: 10.1111/j.1751-1097.1981.tb05329.x. [DOI] [PubMed] [Google Scholar]
- 43.Birge R R, Einterz C M, Knapp H M, Murray L P. Biophys J. 1988;53:367–385. doi: 10.1016/S0006-3495(88)83114-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smith S O, Courtin J, de Groot H, Gebhard R, Lugtenburg J. Biochemistry. 1991;30:7409–7415. doi: 10.1021/bi00244a007. [DOI] [PubMed] [Google Scholar]
- 45.Ebrey T G, Honig B. Q Rev Biophys. 1975;8:129–184. doi: 10.1017/s0033583500001785. [DOI] [PubMed] [Google Scholar]
- 46.Jäger F, Fahmy K, Sakmar T P, Siebert F. Biochemistry. 1994;33:10878–10882. doi: 10.1021/bi00202a005. [DOI] [PubMed] [Google Scholar]
- 47.Schertler G F X, Hargrave P A. Proc Natl Acad Sci USA. 1995;92:11578–11582. doi: 10.1073/pnas.92.25.11578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Baldwin J M. EMBO J. 1993;12:1693–1703. doi: 10.1002/j.1460-2075.1993.tb05814.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sheikh S P, Zvyaga T A, Lichtarge O, Sakmar T P, Bourne H R. Nature (London) 1996;383:347–349. doi: 10.1038/383347a0. [DOI] [PubMed] [Google Scholar]
- 50.Ganter U M, Gärtner W, Siebert F. Eur Biophys J. 1990;18:295–299. [Google Scholar]
- 51.Jäger F, Jäger S, Kräutle O, Friedman N, Sheves M, Hofmann K P, Siebert F. Biochemistry. 1994;33:7389–7397. doi: 10.1021/bi00189a045. [DOI] [PubMed] [Google Scholar]
- 52.Han M, Lin S W, Smith S O, Sakmar T P. J Biol Chem. 1996;271:32330–32336. doi: 10.1074/jbc.271.50.32330. [DOI] [PubMed] [Google Scholar]
- 53.Han M, Lin S W, Minkova M, Smith S O, Sakmar T P. J Biol Chem. 1996;271:32330–32336. doi: 10.1074/jbc.271.50.32330. [DOI] [PubMed] [Google Scholar]
- 54.Fahmy K, Siebert F, Sakmar T P. Biophys Chem. 1995;56:171–181. doi: 10.1016/0301-4622(95)00030-2. [DOI] [PubMed] [Google Scholar]
- 55.Lewis, J. W., Jäger, S. & Kliger, D. S. (1997) Photochem. Photobiol., in press. [DOI] [PubMed]