Abstract
We have studied the kinetics of transcriptional initiation and activation at the malT and malTp1 promoters of Escherichia coli using UV laser footprinting. Contrary to previous studies and because of the very rapid signal acquisition by this technique, we can obtain structural information about true reaction intermediates of transcription initiation. The consequences of adding a transcriptional activator, the cAMP receptor protein/cAMP complex (CRP), are monitored in real time, permitting us to assign specific interactions to the activation of discrete steps in transcription initiation. Direct protein–protein contacts between CRP and the RNA polymerase appeared very rapidly, followed by DNA melting around the −10 hexamer. CRP slightly increased the rate of this isomerization reaction but, more importantly, favored the establishment of additional contacts between the DNA upstream of the CRP binding site and RNA polymerase subsequent to open complex formation. These contacts make a major contribution to transcriptional activation by stabilizing open forms of the promoter complex, thereby indirectly accelerating promoter escape. The ensemble of the kinetic, structural signals demonstrated directly that CRP exerts most of its activating effects on the late stages of transcriptional initiation at the malT promoter.
Initiation of transcription by RNA polymerase can be divided into three successive steps: (i) closed complex formation, (ii) isomerization to an open complex, whereby the region between −10 and the start site of transcription is melted, and (iii) promoter escape (1). In the absence of nucleotides (NTPs), the reaction progresses until the formation of the open complex, which is generally the most stable form of the promoter complexes at 37°C. We call “promoter escape” the steps that lead from the open complex to the elongation complex, leaving the promoter free for the binding of another molecule of RNA polymerase.
Activators or repressors do not, in general, change this reaction pathway. They simply increase or decrease one or several of the rate or equilibrium constants involved in this process. The net effect of a regulator depends on the kinetic parameters characterizing a particular promoter. For example, no change in the steady-state rate of transcription initiation will be observed if the activator affects a step that is not rate-limiting for the overall reaction. An interesting example is provided by the cAMP receptor protein/cAMP complex (CRP), which influences differently the mechanism of transcriptional initiation at different promoters. CRP can favor any of the reaction steps described above: increasing RNA polymerase binding at the lac promoter (2), accelerating isomerization at the gal promoter (3, 4), and improving promoter escape at the malT promoter (5).
Although the effects of activators on the kinetic parameters of transcription initiation have been studied extensively in functional assays (6), the structural underpinnings remain more elusive. Here we present data that measure directly the physical interactions within the transcriptional activation complex during the time course of transcription initiation. We thus determined at which step of the initiation pathway a particular interaction exerts its activating effect. UV laser footprinting (7, 8) is an ideal tool for such a study because the signal is acquired within microseconds, i.e., much faster than typical protein transconformation reactions, which typically proceed on a millisecond time scale (9).
In the experiments presented here, we validated the use of UV laser footprinting as a tool for studying transcription complexes in a kinetic fashion, and we propose a new mechanism of transcription activation affecting the late stages of transcription initiation. We compare the rates of formation of different promoter complexes at the malT promoter in the presence or absence of CRP. The functional aspects of this promoter have been studied extensively (5, 10). However, because classical parameters for binding and isomerization are not modified by CRP (5), the mechanism of transcription activation at this promoter is expected to be quite novel.
MATERIALS AND METHODS
General Methods and Reagents.
Standard methods of molecular biology were used unless otherwise specified (11). Escherichia coli RNA polymerase holoenzyme was purchased from Sigma. The ratio of sigma factor to core enzyme was ≈1:1 (as judged from a Coomassie-stained protein gel). Titration of promoter fragments with RNA polymerase holoenzyme showed that the polymerase preparation was ≈70% active. CRP was purified according to a standard protocol (12).
UV Laser Footprinting.
Linearized SK+malTp1 plasmid (10) at a concentration of 5 nM was incubated for 10 minutes at 30°C with or without 200 μM cAMP/75 nM CRP in a total volume of 420 μl. Two aliquots of 36 μl were removed and UV irradiated with a single pulse of 266-nm laser light of 5-ns duration and an energy equal to or exceeding 30 mJ. These samples, irradiated before the addition of the RNA polymerase, portrayed the conformation of the nucleoprotein complexes at time 0 of the reaction. An aliquot of 324 μl was mixed with 36 μl of 0.9 μM RNA polymerase and further incubated at 30°C. Samples of 40 μl were taken from this reaction mixture at defined time intervals, UV irradiated, and put on ice. All samples were then precipitated with 120 μl of ethanol 96%. The pellets were washed with 70% ethanol and resuspended in 20 μl of H2O. To compare the kinetic signals arising in the upstream and downstream regions of the promoter, the samples were divided in two parts of 10 μl each, and two different primers were used for the extension with T7 DNA polymerase (Pharmacia): an upstream primer hybridizing near the CRP site and a downstream primer hybridizing near the start site of transcription (13). The same procedure was used for the malT promoter [derived from the linearized KS+malT plasmid (10)], but the experiments were carried out at 37°C.
The gels were autoradiographed and scanned with a Molecular Imager (Bio-Rad) for quantification. The lane profiles were quantified by measuring the intensity of a specific peak (deduced from the peak height) for each of the scans corresponding to different reaction times. The data were plotted as a function of time and fitted to an exponential function. The data were normalized: A constant was subtracted from all data to set the smallest value equal to 0, and the data were divided by the amplitude obtained from the fitting procedure. Each series of experiments was repeated three times, and each gave very reproducible results.
The same procedure was used for determining the kinetics of formation of initiating complexes, except that ATP, UTP, and CTP, each at a final concentration of 100 μM, were added to the initial solution. An oligonucleotide with the sequence 5′-GATTAGTTTTGACGGAATCAG-3′, hybridizing downstream of the early transcribed region, was used for the primer extension.
KMnO4 Footprinting.
We used the standard procedure as described (13). NTPs were present at a concentration of 100 μM each. The samples were incubated for 15 min at 37°C before addition of KMnO4. A Δ1malT plasmid (10) at a concentration of 0.5 nM and linearized at the EcoRI site was used as a standard. The standard yielded a primer extension product terminating at position −24 of the promoter.
RESULTS
Kinetics of Open Complex Formation at the malTp1 Promoter: Binding of RNA Polymerase and Promoter Melting.
The principle of UV laser footprinting has been described elsewhere (7, 8, 13). In brief, nucleoprotein complexes are irradiated with a pulse of UV laser light. A series of photoreactions (e.g., thymine dimer formation, protein–DNA crosslinks) takes place as a result of this excitation. The photo-modified bases on the DNA are identified by primer extension using T7 DNA polymerase, an enzyme that ceases polymerization when it encounters a modified base.
We used the malTp1 promoter as a standard to establish the UV laser footprinting patterns that characterize individual reaction intermediates. This promoter-up mutant (a single base pair change at position −12) possesses exactly the same UV footprint of the open complex as the wild-type malT promoter (13), but it reaches almost full activity even in the absence of CRP (14). We initiated the kinetic UV laser footprinting reactions by adding RNA polymerase to the promoter or to the promoter-activator complex. At defined time intervals (between 30 sec and 20 min), aliquots were irradiated. At 37°C, open complex formation was complete after 30 sec at the malTp1 promoter (data not shown). To slow the reaction, we performed the malTp1 experiments at 30°C. At this lower temperature, the transition from the closed to the open complex could be monitored on an optimal time scale for analysis (Fig. 1a).
Figure 1.
Kinetics of open complex formation at the malTp1 promoter in the presence of CRP. (a) UV laser footprints of the core promoter region. The samples at t = 0 min have been irradiated before addition of RNA polymerase. For the other samples, the time shown above the lanes indicates the incubation time before UV irradiation. The intensity of the band at −9 correlates with DNA melting (open complex formation). (b) UV laser footprints of the upstream promoter region. The band at −94 characterizes the interaction of upstream DNA with RNA polymerase; the band at −60 decreases in intensity upon interaction between CRP and RNA polymerase. (c) Time course of open complex formation at the malTp1 promoter. The normalized band intensity is plotted as a function of time. Filled circles, band at −9, +CRP; open circles, band at −9, no CRP; squares, band at −94, +CRP.
Interactions within the ternary complex were established sequentially. The increase of the signal at −32, which characterizes the binding of RNA polymerase to the −35 region (13), was observed at our first time point (30 sec) and remained constant during the further course of the reaction (data not shown). In contrast, the signals at −9 and −4, whose intensities correlated with the extent of open complex formation (13), appeared only gradually. An additional signal located at −19, i.e., within the spacer region between the two recognition hexamers, decreased gradually during the reaction and paralleled the opening of the DNA at −9. We had interpreted this signal to indicate untwisting of the spacer DNA between the −10 and −35 hexamers in the open complex (13).
The disparity in the reaction rates of the different signals showed that RNA polymerase binds very rapidly to the promoter and then slowly isomerizes to the open complex. To deduce the precise rate of open complex formation and to determine whether CRP had an effect on this parameter, we analyzed quantitatively the intensity of the band at −9. Fig. 1c shows a comparison of the time course of open complex formation at the malTp1 promoter in the presence or absence of CRP. The fitted curves are identical, demonstrating that CRP neither affects the rate of open complex formation at malTp1 (Table 1) nor changes detectably the pathway of the reaction.
Table 1.
t1/2 values (in min) of transcription initiation at the malT and malTp1 promoters
| Promoter | CRP | NTPs | Band | t1/2, min |
|---|---|---|---|---|
| malTp1 (30°C) | + | − | −60 | <0.5 |
| − | − | −9 | 1.3 ± 0.2 | |
| + | − | −9 | 1.3 ± 0.2 | |
| + | − | −94 | 4.8 ± 0.2 | |
| malT (37°C) | + | − | −60 | <0.5 |
| + | − | −9 | 2.3 ± 0.2 | |
| + | − | −94 | 3.2 ± 0.6 | |
| + | + | −60 | <0.5 | |
| + | + | +12 | 1.5 ± 0.2 | |
| − | + | +12 | 4.4 ± 0.4 |
Kinetics of CRP-RNA Polymerase Interactions.
A second primer was used to precisely analyze the signals arising upstream of the core promoter (Fig. 1b). Three signals, at −60, −73, and −94, characterized the ternary complex (13). These signals yield information specifically about the interactions (direct or indirect) between CRP and RNA polymerase because none of them is present in any of the binary complexes. Even though the method is not specifically designed to reveal the physical nature of these interactions, these signals probably represent two different kinds of contacts. The most straightforward physical interpretation of the −60 signal attributes it to a contact between the α-subunit of the RNA polymerase and CRP (13). The signals at −73 and −94 may be due to contacts between the upstream DNA and the “back” of RNA polymerase (10, 13).
The two sets of signals displayed very different kinetics. The −60 band disappeared very rapidly, as was observed for the −32 band. This interaction was clearly formed before the isomerization to the open complex. On the contrary, the far upstream signals (at −73 and −94) appeared with identical, very slow rates. The quantification of the band at −94 (Fig. 1c and Table 1) yielded a t1/2 value of 4.8 ± 0.2 min, much greater than the t1/2 value for open complex formation.
Kinetics of Open Complex Formation at the malT Promoter.
The malT promoter is much weaker than the malTp1 mutant, but it is activated to a much greater extent by CRP. In the absence of CRP, very little open complex was formed, and the modifications of the photoreactivity around −10 were too weak to determine the rate of open complex formation. In the presence of CRP, this promoter behaves identically to malTp1. The signals characteristic of RNA polymerase binding (within the −35 hexamer) and of the CRP–RNA polymerase contact (band at −60) were fully present at our first time point and persisted during the entire process (data not shown). We observed open complex formation with a t1/2 value of 2.3 ± 0.2 min whereas the far upstream signals appeared only afterwards (Fig. 2), with a t1/2 value of 3.2 ± 0.6 min (Table 1). Whereas the −60 interaction could affect early steps in transcription initiation, the far upstream interactions, being formed only after promoter melting, could only influence open complex stability or promoter escape, but none of the early steps of initiation. To provide evidence for such a mode of action, we investigated the formation of the transcription complex in the presence of NTPs.
Figure 2.
Time course of open complex formation at the malT promoter in the presence of CRP. Circles, band at −9; Squares, band at −94.
Kinetics of Formation of the Initiating Complex at the malT Promoter: Structure of the Initiating Complexes.
Using KMnO4 footprinting, we examined the transcriptional complexes formed in the presence of limited sets of NTPs. The transcript of the malT promoter starts with the sequence 5′-AUUAAUUACG-3′. Transcription could be arrested at positions +1, +8, and +9 by omitting the appropriate nucleotides from the reaction. We call the complexes formed under these conditions “initiating complexes” (RPinit).
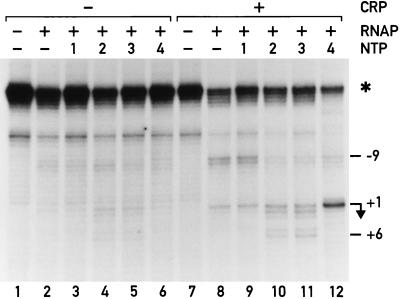
The progression of the transcription bubble could clearly be observed, both in the absence and presence of CRP (Fig. 3). The RNA polymerase could not leave the promoter when only ATP was provided (Fig. 3, lanes 3 and 9), and the footprint was similar to the one obtained in the absence of NTPs (Fig. 3, lanes 2 and 8). When ATP and UTP were added (Fig. 3, lanes 4 and 10), the KMnO4 reactivity downstream of +1 (at positions +2/+3) increases, and a doublet of bands appeared at positions +5/+6. Signals characteristic of the open complex, however, persisted, suggesting that, during abortive initiation, a mixture of complexes were steadily converted into one another. Inclusion of the third nucleotide, CTP, (Fig. 3, lanes 5 and 11) had no further dramatic effect on the reactivity, consistent with the expected movement of the transcription bubble by only 1 nt. CRP influenced these footprinting patterns only quantitatively (compare the left half of Fig. 3 to the right half); all signals were stronger in the presence of CRP. Quantification of the lanes (normalizing to an independent standard, marked with an asterisk) shows that the difference in intensity was even greater than apparent from simple visual inspection of the gel in Fig. 3. CRP clearly stabilized complexes that contain an open region of DNA.
Figure 3.
KMnO4 footprints of transcription complexes at the malT promoter in the presence of limited sets of NTPs. ATP is present in lanes 3 and 9, ATP + UTP are present in lanes 4 and 10, ATP + UTP + CTP are present in lanes 5 and 11, and all four NTPs are present in lanes 6 and 12. CRP is present in lanes 7–12. A linearized plasmid is used for normalization, and the corresponding band is indicated with an asterisk.
When we added all four NTPs, thereby removing the escape limitation, we expected to observe a mixture of transcription complexes yielding a footprint that was dominated by the most stable one. A footprint obtained under these conditions in the absence of CRP looked qualitatively similar to the open complex (compare lane 6 to lane 2). In the presence of CRP, this pattern was modified (compare lanes 8 and 12); CRP provoked a very strong reactivity at position +1 and a ladder of weak bands within the early transcribed region (clearly visible on a radiometric scan of the gel). The complex stabilized by CRP under conditions of steady state transcription thus possessed an open region centered around +1, i.e., further downstream than in the open complex. Rather than relieving an escape limitation (5), CRP appeared to stabilize open forms of promoter complexes.
Kinetics of Formation of the Initiating Complex.
Contrary to the open complex at the malT promoter, the initiating complex formed with two or three NTPs was stable and yielded quantifiable UV footprinting signals, even in the absence of CRP. By adding three NTPs to the reaction, we can therefore measure the influence of CRP on the rate of appearance of the initiating complex. The photoreactivity pattern of the initiating complex was characterized by the increased intensity of bands in the early transcribed region (positions +1, +5, +6, +7, and +12) (Fig. 4a).
Figure 4.
Kinetics of transcription initiation at the malT promoter in the presence of three NTPs. (a) UV laser footprints in the presence of CRP. The bands at +1 and +12 are characteristic of the complex halted at +9 by deprivation of GTP. (b) Time course of formation of the transcription initiation complex. The normalized intensity of the band at +12 is plotted as a function of the time after addition of RNA polymerase. The absolute band intensities of the samples lacking CRP are three times smaller than the corresponding intensities in the presence of CRP. Filled triangles, + CRP; open triangles, no CRP.
In Fig. 4b we compare the kinetics of formation of this RPinit, deduced from the +12 signal, in the presence or absence of CRP. About three times as much initiating complex was formed in the presence of CRP than in its absence. In addition, CRP increased the apparent first order rate constant of formation of the initiating complex by a factor of 3 (t1/2 = 1.5 ± 0.2 min in the presence of CRP vs. t1/2 = 4.4 ± 0.4 min in its absence). CRP may thus accelerate initiation of transcription by increasing both the amount and the rate of formation of the initiating complex.
DISCUSSION
The results presented here have two major implications. First, they introduce the use of UV laser footprinting as a very accurate and generally applicable technique for the kinetic study of protein–DNA interactions. Second, they provide structural evidence for a mechanism of transcriptional activation affecting the last steps of the initiation process.
UV Laser Footprinting.
In contrast to classical footprinting methods, the UV laser technique provides a new way of investigating the role of nucleoprotein intermediates during the uninterrupted course of a reaction. Classical techniques require artificially trapped intermediates to allow extended incubation time of the sample with the footprinting reagent. On the contrary, the photoreactions elicited by the UV laser pulse are completed on a microsecond time scale, and thus the rate of signal acquisition is not limiting for obtaining structural data. In addition, the method is exquisitely sensitive to small variations of the immediate environment of the DNA and can be applied in vivo without modification (ref. 8 and S.D., P.E., and J. G., unpublished work).
Structural Transitions During Open Complex Formation.
To validate the technique, we have used as a model system the formation of an open complex at the activator-independent malTp1 promoter. UV laser footprinting has enabled us to monitor a complete sequence of events and to define at least three successive steps in the course of open complex formation, as revealed by the distinct rates of establishment of the associated interactions. This sequence of events is identical at the related malT and malTp1 promoters and remains unaltered by CRP, confirming the often implicit assumption that an activator merely changes reaction rates without modifying the reaction pathway. The first event, represented by a signal at −32, is extremely rapid and reflects the binding of the RNA polymerase to the promoter to form the closed complex. The second event is characterized by a profound modification of photoreactivity around the −10 hexamer of the promoter and corresponds to the gradual melting of the DNA in this region. The third and last event is observed only in the presence of CRP and consists of the establishment of interactions with the DNA upstream of the CRP binding site, detected by signals at positions −73 and −94. Another CRP-dependent modification of photoreactivity is detected at position −60. This signal corresponds to a direct protein–protein contact between CRP and RNA polymerase that is formed immediately upon promoter binding (see below).
Contrary to inferences based on previous, indirect studies, we can now directly correlate the functional data determined by abortive initiation experiments with the underlying structural rearrangements. The kinetic parameters obtained for the malT promoter are KB ≈ 2 × 107 M−1and k2 ≈ 0.2 min−1 (5). The expected half-time of open complex formation at saturating concentrations of RNA polymerase, 3.5 min, agrees well with the value measured by UV laser footprinting, 2.3 min. The remaining, small difference could be a consequence of the buffer conditions used (glutamate vs. chloride buffers). The close agreement of the functional and structural experiments validates the UV footprinting technique and corroborates our interpretation of the −9 signal as characterizing the open complex (13).
The Mechanism of Transcription Activation.
The footprinting data do more than confirm the functional data; they elucidate the structural basis of transcription activation at the malT promoter. The KMnO4 footprints measure the extent of DNA opening and show that CRP stabilizes the open complex (no NTPs) or the initiating complex (in the presence of the NTPs). The strong hyperreactivity at the +1 position in the presence of all four NTPs proves that the promoter is largely occupied during steady-state transcription in a form that contains an open region of DNA. If the formation of the open complex was rate-limiting, no such open complex form should be observed under steady-state conditions (15) (Fig. 5b). We conclude that the malT promoter is limited at the escape step, even in the presence of CRP.
Figure 5.
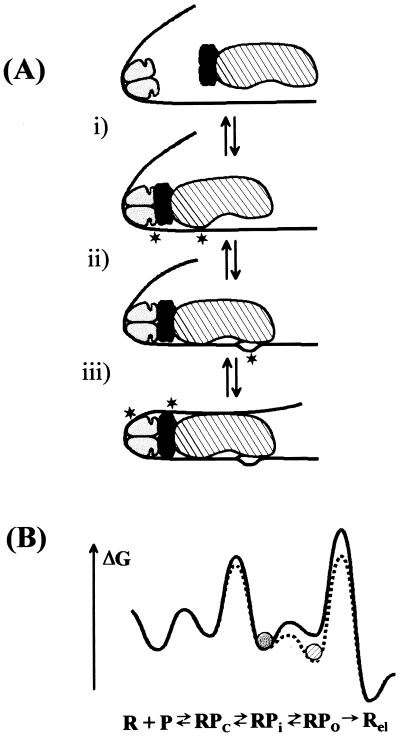
(a) Model of transcriptional activation by CRP at the malT promoter. The CRP dimer is symbolized as a two-domain ellipsoid. The DNA (solid line) is bent around the prebound CRP. Newly appearing signals visible in UV laser footprinting experiments are indicated with an asterisk. The reaction is divided into three steps. (i) RNA polymerase (large ellipsoid) makes a direct contact with the prebound CRP via the carboxyl-terminal domain of the α-subunits (represented by the darker ellipsoids). Signals appear at −32 and at −60. (ii) RNA polymerase opens the DNA between the −10 hexamer and the start site of transcription. (iii) Contacts between the upstream DNA and the back of RNA polymerase stabilize the open region of DNA. The symmetry properties of the α-subunits are arbitrary; the interaction of both α-subunits with the DNA may be restricted to the stretch just upstream of the promoter. (b) Change in free energy during transcription initiation and activation. The solid line represents the malT promoter and the stippled line represents the same promoter in the presence of CRP. The diagram illustrates the redistribution of the most populated state (represented by the circles) in the presence of CRP. The rate-limiting step (highest peak) remains promoter escape under both conditions, and open complex formation is slightly accelerated in the presence of CRP. The diagram is schematic to illustrate how CRP indirectly accelerates promoter escape, and it is not meant to accurately reflect absolute free energies.
The intensity of both the KMnO4 as well as the UV laser footprints of the initiating complexes increased in the presence of CRP, i.e., CRP stabilized promoter complexes that contained an open region of DNA (open and initiating complexes). The stabilization of the open complexes could be due to an increased rate of formation or to a decreased rate of dissociation. Both mechanisms operated at the malT promoter. It has been shown that CRP decreases the rate of dissociation of the open complex (5), and kinetic UV laser footprinting showed directly that CRP accelerated the formation of the initiating complex (Table 1). CRP thus helped to populate open forms of the promoter but did not directly improve the rate-limiting escape step (Fig. 5b). In vitro run-off transcription experiments (data not shown) confirmed that the steady-state rate of transcription was about two times slower than the rate of open complex formation measured by UV footprinting.
The Structural Basis of Transcription Activation.
What physical interactions lead to transcriptional activation by CRP? UV footprinting detected two different kinds of interactions between CRP and RNA polymerase represented by the signal at −60 and the far upstream signals at −94 and −73. Given the promoter geometry and a wealth of genetic and biochemical data (16–20), the −60 signal most likely arises from a direct contact between the downstream subunit of CRP and RNA polymerase (21), more specifically between activating region 1 of CRP and the carboxyl-terminal domain of the α-subunit of RNA polymerase (22–25). Although this interaction is already fully established in the closed promoter complex, it does not alter the affinity (KB) of polymerase for the promoter (5). This interaction is in striking contrast with previous observations at related CRP-dependent promoters, where the direct contact between activating region 1 and the carboxyl-terminal domain has been shown to specifically increase KB (4). Because CRP clearly interacted with RNA polymerase at the malT promoter (UV laser signal at −60), we have to conclude that the favorable contributions of this interaction to promoter binding are completely balanced by the energy needed for their formation, probably because of deformation of the intervening DNA. The direct contact between CRP and RNA polymerase therefore does not appear to contribute to transcriptional activation at the malT promoter.
The mechanism of transcription activation appears to rely entirely on contacts between the DNA upstream of the CRP binding site and the back of RNA polymerase. The functional importance, at the malT promoter, of this far upstream DNA has been shown recently. Removal of this DNA almost completely abolishes transcriptional activation by CRP in vitro (10). The UV laser signal in this region (−94) appears only after open complex formation and is most easily explained by contacts between the upstream DNA and the back of RNA polymerase (13). The signal is present on linear DNA fragments of different lengths and upstream sequence, as well as on circular DNA (ref. 13 and data not shown) and therefore is not an artifact of a second molecule of RNA polymerase binding specifically or nonspecifically to the upstream DNA or to DNA ends. Because these upstream contacts form late during transcription initiation, they can only affect the open or initiating complexes. Our data suggest that these interactions are responsible for stabilizing open forms of the promoter complex.
A Model of Transcription Activation at the malT Promoter.
All functional and structural data concur to show that the malT promoter is limited at promoter escape. The experimental results are reconciled by the following model (Fig. 5a) of transcriptional activation by CRP at the malT promoter: (i) Binding of RNA polymerase and interaction with CRP (most likely via activating region 1 and the carboxyl-terminal domain of α) is very rapid and probably in equilibrium with free RNA polymerase. (ii) The subsequent isomerization to the open complex possesses an intrinsic rate on the order of a minute, and this step is slightly accelerated by CRP. Because the only detectable interactions between CRP and RNA polymerase at this stage of the reaction are direct contacts, we propose that these contacts improve the rate of isomerization. (iii) The resulting open complex is relatively unstable, and synthesis of short RNA transcripts as well as contacts between the DNA upstream of the CRP binding site and RNA polymerase stabilize the open form of the promoter complex. The rate-limiting step (the step with the highest molar activation free energy) remains promoter escape. CRP mainly helps populating the state just before the rate-limiting step. The initiating complex that constitutes the substrate for promoter escape may be in rapid equilibrium with a closed form of the complex, possibly a complex akin to the RPi complex identified at several promoters (26, 27). CRP would shift this equilibrium toward complexes that contain a melted region of DNA, thus constituting a substrate for promoter escape (Fig. 5b).
Because UV laser footprinting can be applied to in vivo samples without modification, we are in the process of validating the proposed mechanism under the most physiologically relevant conditions, i.e., within the living cell. Preliminary experiments only confirm the model but also point to complexities introduced by additional cellular proteins that compete for binding to the promoter.
Acknowledgments
We thank especially Dany Rifat and Pascal Damay for invaluable help with laboratory procedures. We gratefully acknowledge Manuel Engelhorn and Axel Zotter for helpful suggestions concerning the footprinting procedures. We thank Nicolas Roggli for his expert photographic work. This work was supported by grants 31-32582.91 and 31-42501.94 from the Swiss National Fund.
ABBREVIATIONS
- CRP
cAMP/cAMP receptor protein complex
- NTPs
ribonucleoside triphosphates
References
- 1.McClure W R. Annu Rev Biochem. 1985;54:171–204. doi: 10.1146/annurev.bi.54.070185.001131. [DOI] [PubMed] [Google Scholar]
- 2.Malan T P, Kolb A, Buc H, McClure W R. J Mol Biol. 1984;180:881–909. doi: 10.1016/0022-2836(84)90262-6. [DOI] [PubMed] [Google Scholar]
- 3.Herbert M, Kolb A, Buc H. Proc Natl Acad Sci USA. 1986;83:2807–2811. doi: 10.1073/pnas.83.9.2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Niu W, Kim Y, Tau G, Heyduk T, Ebright R E. Cell. 1996;87:1123–1134. doi: 10.1016/s0092-8674(00)81806-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Menendez M, Kolb A, Buc H. EMBO J. 1987;6:4227–4234. doi: 10.1002/j.1460-2075.1987.tb02771.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Record M T, Reznikoff W S, Craig M L, McQuade K L, Schlax P J. In: Escherichia coli and Salmonella. 2nd ed. Neidhardt F C, editor. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 792–820. [Google Scholar]
- 7.Buckle M, Geiselmann J, Kolb A, Buc H. Nucleic Acids Res. 1991;19:833–840. doi: 10.1093/nar/19.4.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Engelhorn M, Boccard F, Murtin C, Prentki P, Geiselmann J. Nucleic Acids Res. 1995;23:2959–2965. [PubMed] [Google Scholar]
- 9.Hockensmith J W, Kubasek W L, Vorachek W R, Evertsz E M, von Hippel P H. Methods Enzymol. 1991;208:211–236. doi: 10.1016/0076-6879(91)08015-a. [DOI] [PubMed] [Google Scholar]
- 10.Déthiollaz S, Eichenberger P, Geiselmann J. EMBO J. 1996;15:5449–5458. [PMC free article] [PubMed] [Google Scholar]
- 11.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 12.Ghosaini L R, Brown A M, Sturtevant J M. Biochemistry. 1988;27:5257–5261. doi: 10.1021/bi00414a046. [DOI] [PubMed] [Google Scholar]
- 13.Eichenberger P, Déthiollaz S, Fujita N, Ishihama A, Geiselmann J. Biochemistry. 1996;35:15302–15312. doi: 10.1021/bi961377d. [DOI] [PubMed] [Google Scholar]
- 14.Chapon C, Kolb A. J Bacteriol. 1983;156:1135–1143. doi: 10.1128/jb.156.3.1135-1143.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ellinger T, Behnke D, Bujard H, Gralla J D. J Mol Biol. 1994;239:455–465. doi: 10.1006/jmbi.1994.1388. [DOI] [PubMed] [Google Scholar]
- 16.Busby S, Ebright R H. Cell. 1994;79:743–746. doi: 10.1016/0092-8674(94)90063-9. [DOI] [PubMed] [Google Scholar]
- 17.Zhou Y, Zhang X, Ebright R H. Proc Natl Acad Sci USA. 1993;90:6081–6085. doi: 10.1073/pnas.90.13.6081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou Y, Busby S, Ebright R H. Cell. 1993;73:375–379. doi: 10.1016/0092-8674(93)90236-j. [DOI] [PubMed] [Google Scholar]
- 19.Williams R, Bell A, Sims G, Busby S. Nucleic Acids Res. 1991;19:6705–6712. doi: 10.1093/nar/19.24.6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bell A, Gaston K, Williams R, Chapman K, Kolb A, Buc H, Minchin S, Williams J, Busby S. Nucleic Acids Res. 1990;18:7243–7250. doi: 10.1093/nar/18.24.7243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou Y, Pendergrast P S, Bell A, Williams R, Busby S, Ebright R H. EMBO J. 1994;13:4549–4557. doi: 10.1002/j.1460-2075.1994.tb06776.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tang H, Severinov K, Goldfarb A, Fenyo D, Chait B, Ebright R H. Genes Dev. 1994;8:3058–3067. doi: 10.1101/gad.8.24.3058. [DOI] [PubMed] [Google Scholar]
- 23.Chen Y, Ebright Y W, Ebright R H. Science. 1994;265:90–92. doi: 10.1126/science.8016656. [DOI] [PubMed] [Google Scholar]
- 24.Ishihama A. J Bacteriol. 1993;175:2483–2489. doi: 10.1128/jb.175.9.2483-2489.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zou C, Fujita N, Igarashi K, Ishihama A. Mol Microbiol. 1992;6:2599–2605. doi: 10.1111/j.1365-2958.1992.tb01437.x. [DOI] [PubMed] [Google Scholar]
- 26.Buc H, McClure W R. Biochemistry. 1985;24:2712–2723. doi: 10.1021/bi00332a018. [DOI] [PubMed] [Google Scholar]
- 27.Roe J-H, Burgess R R, Record M T., Jr J Mol Biol. 1985;184:441–453. doi: 10.1016/0022-2836(85)90293-1. [DOI] [PubMed] [Google Scholar]