Abstract
The signal transduction and activation of RNA (STAR) family of RNA-binding proteins, whose members are evolutionarily conserved from yeast to humans, are important for a number of developmental decisions. For example, in the mouse, quaking proteins (QKI-5, QKI-6, and QKI-7) are essential for embryogenesis and myelination , whereas a closely related protein in Caenorhabditis elegans, germline defective-1 (GLD-1), is necessary for germ-line development. Recently, GLD-1 was found to be a translational repressor that acts through regulatory elements, called TGEs (for tra-2 and GLI elements), present in the 3′ untranslated region of the sex-determining gene tra-2. This gene promotes female development, and repression of tra-2 translation by TGEs is necessary for the male cell fates. The finding that GLD-1 inhibits tra-2 translation raises the possibility that other STAR family members act by a similar mechanism to control gene activity. Here we demonstrate, both in vitro and in vivo, that QKI-6 functions in the same manner as GLD-1 and can specifically bind to TGEs to repress translation of reporter constructs containing TGEs. In addition, expression of QKI-6 in C. elegans wild-type hermaphrodites or in hermaphrodites that are partially masculinized by a loss-of-function mutation in the sex-determining gene tra-3 results in masculinization of somatic tissues, consistent with QKI-6 repressing the activity of tra-2. These results strongly suggest that QKI-6 may control gene activity by operating through TGEs to regulate translation. In addition, our data support the hypothesis that other STAR family members may also be TGE-dependent translational regulators.
Keywords: quaking, myelination, mouse, translation control, tra-2 and GLI elementTGE
The STAR family of RNA-binding proteins is required for a number of important developmental decisions in both vertebrates and invertebrates (1). STAR family members contain a conserved KH domain as well as two conserved domains called QUA1 and QUA2. STAR proteins are necessary for embryogenesis and myelination in the mouse (2, 3), muscle development in Drosophila (4, 5), notochord differentiation in Xenopus (6), and germ-line development in Caenorhabditis elegans (7, 8). Although over 30 STAR family members have been identified, for most, it is not clear how they function to regulate gene activity.
Recently, GLD-1 was shown to inhibit the translation of the C. elegans sex-determining gene tra-2 (9). C. elegans has two sexes: hermaphrodites and males. Hermaphrodites are essentially female animals that make sperm and then oocytes. tra-2 is required for female development and is predicted to encode a large transmembrane protein, called TRA-2A, that is necessary to inhibit downstream male determinants (10–12). GLD-1 is germ line specific and plays multiple roles in hermaphrodite germ cell development. In the hermaphrodite, GLD-1 is essential for oogenesis (7) and is necessary for hermaphrodite spermatogenesis and inhibition of premeiotic proliferation (7).
tra-2 is translationally regulated by two elements, called TGEs (for tra-2 and GLI elements), located in the 3′ untranslated region (3′UTR) (13, 14). Translational repression of tra-2 is required for hermaphrodite spermatogenesis (13, 15). GLD-1 specifically binds the TGEs and represses translation of TGE-containing RNAs both in vivo and in vitro (9). These findings suggest that GLD-1 controls hermaphrodite spermatogenesis by repressing tra-2 translation (9). The TGE control is a conserved process that is present in nematodes and mammals (14), raising the possibility that other STAR family members act through TGE-like elements to control translation. We addressed this idea by asking whether the mouse quaking proteins (QKI) are TGE-dependent translational regulators.
The qk gene produces at least three major transcripts that are 5 (qkI-5), 6 (qkI-6), and 7 (qkI-7) kb long (2). The three transcripts produce very similar proteins, QKI-5, QKI-6, and QKI-7, respectively, that differ only at the carboxyl terminus. All three proteins contain the identical KH RNA-binding domain and QUA1 and QUA2 domains. Northern blot analyses indicate that qkI-5 mRNA is the major form present in early embryos, and that qkI-6 and qkI-7 mRNA are found in late development when myelination begins (16). Immunocytochemistry experiments indicate that QKI proteins are expressed in oligodendrocytes and astrocytes in the central nervous system as well as Schwann cells in the peripheral nervous system. QKI-5 is nuclear, although it shuttles between the nucleus and cytoplasm, whereas QKI-6 and QKI-7 are predominantly cytoplasmic (16, 17).
Recessive viable mutations in quaking (qkv) result in hypomyelination, and as a result the qkv homozygotes develop a rapid tremor by postnatal day 10. Homozygotes also suffer chronic/tonic seizures (18). The myelin deficiency is most pronounced in the brain, less in the spinal cord, and even less in the peripheral nervous system (19). In the qkv allele, QKI-6 and QKI-7 are absent from oligodendrocytes and Schwann cells (16), possibly explaining the tremors in mutant animals. Other qk alleles, embryonic lethal mutations induced by ethylnitrosourea (qkkt4), have been isolated (20–22). The terminal phenotype is one of arrested growth with generalized abnormalities by embryonic day 9, but the precise cause of lethality is not known (3). The mice die more than 10 days before myelination begins, indicating that QKI proteins may affect multiple cellular processes.
In this paper, we find that like GLD-1, the QKI-6 protein can act through TGEs to repress translation. This finding is supported by the observation that QKI-6 specifically binds TGEs, represses translation of reporter RNAs both in vivo and in vitro, and promotes the fate of male cells in C. elegans. Our findings indicate that a number of STAR family proteins may control gene expression by repressing translation through TGEs.
Methods
General Procedures and Strains.
Routine maintenance was as described in ref. 23. All strains were raised at 20°C unless otherwise noted. The following mutant alleles were used in this study: LGII, tra-2(e2020 gf) (15); LGIV, unc-24(e138)fem-3(e1996lf); LGIV, tra-3(e1107)/nT1 (24). The balancer nT1 (previously called DnT1) suppresses recombination over much of chromosome IV (25).
RNA Gel Shift Analysis.
A construct encoding the entire QKI-6 or a mutant QKI-6 that carries the qkkt4 mutation (amino acid number 48 has been changed from glutamic acid to glycine) with an amino-terminal His tag (His-QKI-6 or His-QKkt4), were expressed in bacteria and purified by using the His-Binding Buffer Kit (Novagen) (17). RNA gel shifts were performed as described (13). 32P-labeled and unlabeled tra-2(+) and tra-2(−108) 3′UTR probes were made by standard methods as described (26). Small 32P-labeled RNAs (EBG-9, EBG-11, EJ-19, EJ-32, EJ-35, EJ-38, EMP-9, and EMP-10) were produced by using the method of Milligan and Uhlenbeck (27). See below for sequences.
Transgenic Analysis.
hsp∷QKI-6 (also called pBG102) was made by amplifying the qkI-6 coding region by PCR with primers EMP-7 and EMP-8. The PCR product was cut with SalI and cloned into the same site of pBG518. pBG518 contains the C. elegans-inducible heat shock promoter, hsp16.41, and the C. elegans unc-54 3′UTR (9). The construction of pBG3 [lacZ∷Ce-tra-2(-32)3′UTR] and pBG4 [lacZ∷Ce-tra-2(−60)3′UTR] are described elsewhere (28).
Transgenic C. elegans animals were generated by using standard methods (29): 25 ng/μl of hsp∷QKI-6 with 50 ng/μl of emb-9∷GFP were injected into animals carrying either lacZ∷tra-2(−32)3′UTR or lacZ∷tra-2(−60)3′UTR. emb-9∷GFP contains the coding region of green fluorescent protein (GFP) controlled by the emb-9 promoter. For analysis of the ability of QKI-6 to promote the fate of male cells in the soma, tra-3(e1107)/nt1 hermaphrodites were injected with 100 ng/μl of hsp∷QKI-6 and 100 ng/μl of pRF4 which encodes the collagen rol-6 gene.
In Vitro Yeast Translational Assay.
Yeast lysates were produced as described (30) with modifications (31). The in vitro translation assay was performed as described (30). Capped RNAs containing the luciferase (luc) gene, and different 3′UTRs were produced by a standard in vitro transcription reaction (26). The ratio of cap analog to GTP was 5 to 1. pBG51 and pBG53 are described in ref. 9 and correspond to constructs containing the luc gene and wild-type tra-2(+) 3′UTR or mutant tra-2(−108) 3′UTR in which both TGEs and some flanking sequences were deleted, respectively. The two plasmids contain a poly(A)65 tract downstream of the 3′UTRs followed by an NsiI site. The constructs were linearized by using NsiI and transcribed by using a Promega SP6 Ribomax kit. RNAs were quantitated by spectrophotometry and checked for purity on an agarose gel. RNA was added to the yeast lysates to a final concentration of 6 nM, and His-QKI-6 or His-QKkt4 was added to a final concentration of 60 nM. At specific time points, aliquots were taken. Each aliquot was divided into two; half was frozen in liquid nitrogen and processed for luciferase activity and the other half added to TRIZOL (GIBCO/BRL) for Northern analysis. Luciferase activity was assayed by using a Monolight 2010 Luminometer. RNA levels were quantitated by Northern blot analysis (Storm 860 phosphoimager).
Oligonucleotide Sequences.
EMP-9: 5′-TGCAGCTCCCCCAATTTTTCTGGAAGGATCCCCTTAGGAAATGCGATCTGTGATGGATGAGATTCCCTCGCCCTATAGTGAGTCGTATTA-3′.
EMP-10: 5′-TGCAGCTCCCCCAATTTTTCTGGAAGGATCCCCTTAGGAAATGCGATCTGTGATGGCACAGATTCCCTCGCCCTATAGTGAGTCGTATTA-3′.
EBG-9: 5′-TGGACGATTAGATATGAGATGATAAGAAATTAAATATGAGTAGATATGAGTAGATAAGAAATTAAATAATGAAATGGAAATTGTCGCCCTATAGTGAGTCGTATTA-3′.
EBG-11: 5′-TGGACGATTATGAAATGGAAATTGTACAAATAATAGAAACGAAAATGAGTAAGAAATGAAATTTTGGAACCAAATTCTCGCCCTATAGTGAGTCGTATTA-3′.
EJ-19: 5′-TGTGTTCAGAAAACTAGGCAGGAAAGTAGGAAAAGATCTGTTAATCGCCCTATAGTGAGTCGTATTA-3′.
EJ-24: 5′-GGAAGGATAGAAACCCCTTAGGAAATGCGATCTGTGATGGATGAGATTCCCTCGCCCTATAGTGAGTCGTATTA-3′.
EJ-32: 5′-TACAAGATCTGTGTTCCTAGGCAGGAAAGTAGGAAACACAGATCTGTTCGCCCTATAGTGAGTCGTATTA-3′.
EJ-35: 5′-GAATTCTCGAGTACAAGATCTGTGTTCCTAGGCAGGAAAGTAGGAAAGTGAGATCTGTTCGCCCTATAGTGAGTCGTATTA-3′.
EJ-38: 5′-TGCAGCTCCCCCAATTTTTCTGGAAGGATAGAAACCCCTTAGGAAATGCGATCTGTGATGGATGAGATTCCCTCGCCCTATAGTGAGTCGTATTA-3′.
Results and Discussion
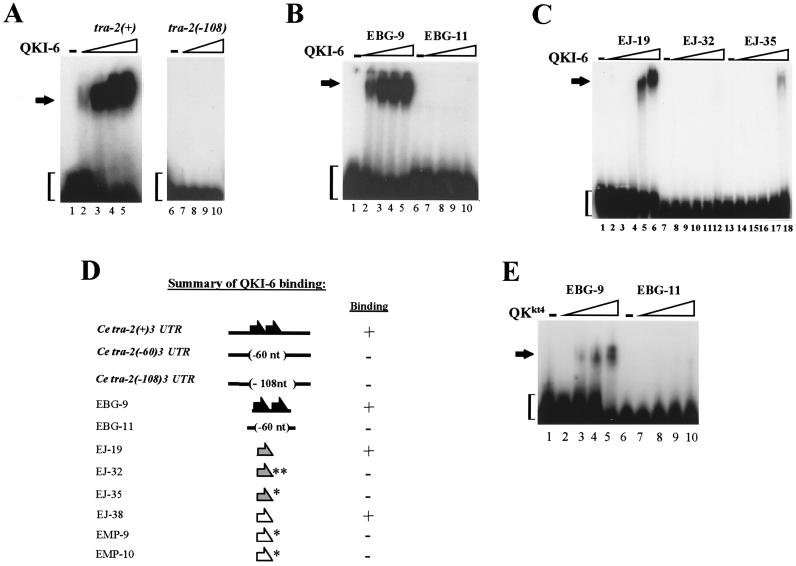
To test whether other STAR proteins, like GLD-1, are translational repressors, we asked whether the murine QKI proteins could control translation in a TGE-dependent manner. By using RNA gel shift analysis, we examined the different QKI proteins (QKI-5, QKI-6, and QKI-7) for specific binding to the C. elegans tra-2 TGEs. Purified bacterial-expressed proteins were incubated with radiolabeled RNAs that contain either the wild-type tra-2 3′UTR [tra-2(+)3′UTR] or a mutant 3′UTR in which both TGEs were deleted [tra-2(−108)3′UTR] and electrophoresed on a nondenaturing polyacrylamide gel. The QKI proteins contained a His tag at their amino terminus and encoded the entire QKI protein. We found that QKI-6 bound to RNAs that carried TGEs but not to ones in which the TGEs were removed (Fig. 1 A and D). The binding of QKI-6 to the TGEs was specific because complex formation was abolished by an excess of unlabeled tra-2(+) 3′UTR RNA but not by an excess of unlabeled tra-2(−108) 3′UTR RNA (data not shown). We also examined the binding of QKI-5 and QKI-7 to the different RNAs in gel shift assays and found little difference in complex formation (data not shown). Because the cytoplasmic localization of QKI-6 and QKI-7 is more consistent with a role in translational control, and there is little sequence difference between QKI-6 and QKI-7 protein, we focused our studies on QKI-6.
Figure 1.
QKI-6 as well as QKkt4 specifically bind the TGEs. The binding of QKI-6 and QKkt4 to the TGEs was tested by using RNA gel shift analysis. (A) One femtomole of 32P-labeled C. elegans tra-2(+) 3′UTR (lanes 1–5) or mutant tra-2(−108) 3′UTR (lanes 6–10) in which the TGEs have been deleted were incubated alone or with increasing amounts of purified bacterially expressed His-QKI-6 protein (0, 0.25, 0.5, 0.75, or 1 μg). Reactions were electrophoresed on a 3.75% nondenaturing polyacrylamide gel. Slower migrating bands represent complex formation (arrow); the faster migrating bands indicate free probe (brackets). (B) One femtomole of 32P-labeled C. elegans tra-2 TGE (EBG-9; lanes 1–5) or mutant tra-2 TGE (EBG-11; lanes 6–10; see Methods for sequences) were incubated alone or with increasing amounts of purified bacterially expressed His-QKI-6 protein (0, 0.25, 0.5, 0.75, or 1 μg). (C) One femtomole of 32P-labeled C. briggsae tra-2 TGE (EJ-19; lanes 1–6) or mutant C. briggsae tra-2 TGEs (EJ-32, lanes 7–12 and EJ-35, lanes 13–18) were incubated with increasing amounts of His-QKI-6 (0, 0.16, 0.32, 0.64, 0.80, or 3.5 μg). (D) Summary of QKI-6 binding. Binding of His-QKI-6 was determined by using RNA gel shifts. (Left) Names of RNAs (for sequences, see Methods). (Middle) Diagrams of RNAs. Black arrows represent C. elegans tra-2 TGEs, stippled arrows represent C. briggsae tra-2 TGEs, and white arrows represent the GLI TGEs. Asterisks indicate mutant C. briggsae tra-2 or GLI TGE sequences. Deletions are indicated in brackets. (Right) RNAs were scored for the ability (plus sign) or inability (minus sign) to bind QKI-6. (E) One femtomole of 32P-labeled C. elegans tra-2 TGEs (EBG-9, lanes 1–5) or mutant tra-2 TGEs (EBG-11, lanes 6–10; see Methods for sequences) were incubated alone or with increasing amounts of purified bacterially expressed His-QKkt4 protein (0, 0.25, 0.5, 0.75, or 1 μg). In every case, QKI-6 and QKkt4 formed complexes with RNAs containing wild-type TGEs but not those with mutant elements.
To further test the binding specificity of QKI-6, we assayed for binding of the protein to a small RNA that contained the C. elegans tra-2 TGEs (EBG-9) or to one in which the TGEs were deleted (EBG-11; see Methods for sequences). We found that similar to GLD-1, QKI-6 bound EBG-9 but not EBG-11 (Fig. 1 B and D). Previously, we identified TGEs in the 3′UTRs of the Caenorhabditis briggsae tra-2 and the human GLI genes, and demonstrated that GLD-1 specifically associates with these elements (9); therefore, we asked whether QKI-6 also bound the wild-type C. briggsae tra-2 (EJ-19) and wild-type GLI (EJ-38) TGEs but not mutant TGEs (for mutant C. briggsae tra-2: EJ-32 and EJ-35 and for mutant GLI: EMP-9 and EMP-10). EJ-35 and EMP-9 contain a 6-nt deletion, and EJ-32 and EMP-10 contain the same 6-nt deletion plus three altered nucleotides. Similar to GLD-1, QKI-6 binds EJ-19 and EJ-38 but not EJ-32, EJ-35, EMP-9, or EMP-10 (Fig. 1 C and D). These results show that similar to GLD-1, QKI-6 specifically associates with wild-type TGEs.
As mentioned above, the qkkt4 mutation is embryonic lethal and likely represents a strong loss-of-function mutation. This mutation changes a glutamic acid at position 48 in the QUA1 domain to glycine (2). QKI-5 protein carrying this mutation binds RNA but does not form a homodimer, raising the possibility that the interaction of the protein with itself or another protein is required for function (32). We tested whether the QKkt4 protein could bind TGEs by assaying the ability of this protein to bind EBG-9 but not EBG-11. Interestingly, we find that QKkt4 bound EBG-9, though less efficiently than wild type, and that QKkt4 did not bind EBG-11 (Fig. 1E). These findings are consistent with earlier findings that indicated this mutation in the QUA1 domain does not abolish RNA binding.
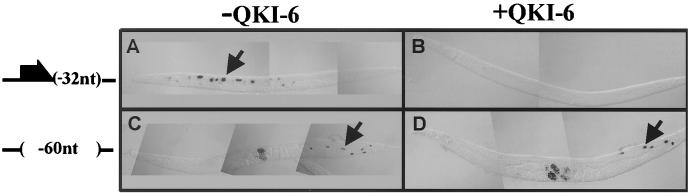
To test whether QKI-6 can act as a TGE translational repressor in vivo, we assayed for its ability to repress the activity of a reporter transgene that contains the TGEs or one in which the TGEs have been deleted. We expressed QKI-6 in C. elegans animals carrying a reporter transgene that contained the tra-2 3′UTR lacking one TGE [lacZ∷tra-2(−32)3′UTR] or a 3′UTR missing both TGEs [lacZ∷tra-2(−108)3′UTR]. The presence of a single TGE within the 3′UTR only partially controls translation and provides a sensitive assay for regulation. Consequently, for analysis of the ability of QKI-6 to regulate translation through the TGEs, we used the lacZ∷tra-2(−32)3′UTR construct (28). Both reporter transgenes encode β-galactosidase (β-gal) and are controlled by the inducible heat-shock promoter (hsp16–41).
The expression of QKI-6 dramatically decreased the number of animals with intestinal β-gal staining. In the absence of QKI-6, 51% of transgenic animals carrying lacZ∷tra-2(−32)3′UTR had β-gal staining in intestinal cells (Fig. 2A, Table 1). In contrast, when QKI-6 was expressed in the soma, 8% of lacZ∷tra-2(−32)3′UTR transgenic animals had intestinal β-gal staining (Fig. 2B, Table 1). Ectopic expression of QKI-6 had little effect on the β-gal expression of lacZ∷tra-2(−108)3′UTR animals (Fig. 2 C and D, Table 1). These results indicate that QKI-6 can repress the activity of the reporter transgene in a TGE-dependent manner.
Figure 2.
QKI-6 represses the activity of reporter transgenes through the TGEs in vivo. Lateral views of C. elegans adult animals with anterior to the left and ventral down. Two reporter transgenes were used. Both encode the lacZ gene and are driven by the C. elegans heat shock promoter (hsp16-41) (37). lacZ∷tra-2(−32)3′UTR carries a tra2 3′UTR lacking one TGE, and lacZ∷tra-2(−60)3′UTR has a tra-2 3′UTR in which both TGEs have been deleted. Somatic expression of QKI-6 (hsp∷QKI-6) was accomplished by expressing the entire QKI-6 cDNA by using the heat shock promoter. For lacZ(−32)3′UTR, (A) in the absence of QKI-6, more animals had intestinal β-gal staining [β-gal activity is detected in 11 intestinal cells (arrow)] than (B) in the presence of QKI-6. For lacZ(−60)3′UTR, (C) the absence or (D) presence of QKI-6 did not affect the number of animals with intestinal β-gal staining [β-gal staining is detected in seven and five intestinal cells, respectively (arrows)]. (Left) Cartoon of the different tra-2 3′UTRs inserted downstream of the lacZ reporter gene.
Table 1.
QKI-6 represses TGE control in vivo
| Reporter transgene* | QKI-6 transgene† | Animals with intestinal β-gal staining, %‡ |
|---|---|---|
| lacZ∷tra-2(−32)3′UTR | None hsp∷QKI-6 | 51 (n = 47) 8 (n = 37) |
| lacZ∷2(−108)3′UTR | None hsp∷QKI-6 | 62 (n = 58) 69 (n = 51) |
Reporter transgene containing the C. elegans heat shock promoter (hsp-16.41) controlling the lacZ gene. Mutant tra-2 3′UTRs were inserted downstream of the lacZ gene. In all experiments, adult transgenic worms were heat-shocked 2 h at 33°C and allowed to recover for an additional 2 h at 20°C before being fixed and stained for β-gal activity.
Transgenic C. elegans animals containing different reporter transgenes were injected with hsp∷QKI-6. hsp∷QKI-6 is controlled by the heat-shock promoter (hsp-16-41) and carries the unc-54 3′UTR and the coding region for QKI-6.
Trangenic animals were scored as positive if blue precipitate was detectable in intestinal cells at ×630 magnification. Intestinal cells were scored genetic evidence indicates the TGE regulation is present in these cells (15). Percentiles represent the values of one typical transgenic line. Other lines gave similar results. n = total number of animals scored from at least four different experiments.
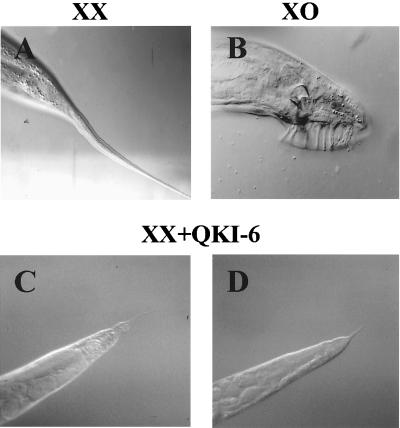
As discussed above, the TGE regulation is necessary to repress the translation of tra-2 which allows for the specification of male cell fates. Previously, we found that mis-expression of GLD-1 in the soma resulted in masculinization of somatic tissues in hermaphrodite animals (9). We reasoned that if QKI-6 is behaving in a similar manner, expression of QKI-6 in the hermaphrodite soma should also result in the development of male cell fates. To examine this, we heat-shocked wild-type hermaphrodites carrying hsp∷QKI-6 throughout development and assayed for masculinization. We found that 28% (n = 122) of the XX hermaphrodite animals expressing QKI-6 produced truncated tails, indicative of masculinization (Fig. 3 C and D); however, truncated tails were not detected in animals not expressing QKI-6 (n = 200; Fig. 3A). These data indicate that QKI-6 can act similarly to GLD-1 to repress tra-2 translation in vivo.
Figure 3.
Expression of QKI-6 causes masculinization of somatic tissues. (A) Adult wild-type XX hermaphrodite and (B) XO male tails (C and D). Transgenic XX hermaphrodite animals carrying the hsp∷QKI-6 transgene were heat shocked once a day from embryogenesis to adults. The truncated tail is indicative of somatic masculinization.
Because QKI-6 only weakly masculinized wild-type hermaphrodites, we used a second in vivo assay to ask whether QKI-6 could masculinize animals that were intersexual. To do this, we heat-shocked tra-3(lf) animals carrying hsp∷QKI-6 throughout development and assayed for masculinization. tra-3, like tra-2, is required for female development (10) and genetically appears to act upstream of laf-1 to regulate tra-2 translation (28). tra-3(lf) animals are incomplete males. As expected, less than 0.5% of tra-3(e1107lf) animals expressing QKI-6 produced vulva (n = 256), indicative of male development, compared with 10% of the tra-3(lf) animals not expressing QKI-6 (n = 193). The further masculinization of tra-3(lf) animals after overexpression of QKI-6 is consistent with our initial conclusion that QKI-6 can repress tra-2 translation to inhibit female development. We also found that expression of GLD-1 and QKI-6 in the soma resulted in some embryonic lethality (data not shown), possibly indicating that these proteins are disrupting the activity of an essential gene(s).
To test whether the observed masculinization after expression of QKI-6 was acting on tra-2, QKI-6 was expressed in the soma of fem-3(lf) animals by using the hsp∷QKI-6 construct. fem-3 activity is required for male somatic development and tra-2 is thought to promote female development by directly inhibiting fem-3 activity (for review, see ref. 33). QKI-6 was expressed in XX fem-3(lf) animals by using the hsp∷QKI-6 construct. In contrast to wild-type animals, fem-3(lf) animals expressing QKI-6 did not develop truncated tails (n = 192), consistent with QKI-6 acting upstream of fem-3 in a genetic hierarchy. In addition, to determine whether QKI-6 was regulating tra-2 directly through the TGEs, we expressed QKI-6 in tra-2(e2020 gf) animals. The tra-2(e2020 gf) mutation completely removes the TGEs from the tra-2 3′UTR. We observed that in contrast to wild-type and tra-3(lf) animals, all tra-2(gf) expressing QKI-6 had wild-type tails and developed vulva (n = 132). These findings suggest QKI-6 was acting through the TGEs to repress tra-2 translation.
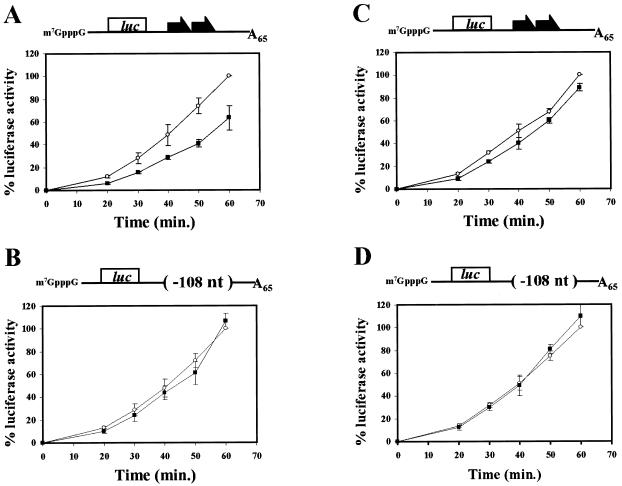
Previously, GLD-1 was shown to repress translation of reporter RNAs that carried TGEs in vitro (9). We examined whether QKI-6 could inhibit translation in a TGE-dependent manner in vitro by using yeast translation extracts and assaying the translation of reporter RNAs containing different 3′UTRs. The reporter RNAs encoded luciferase and carried a wild-type tra-2 3′UTR [tra-2(+)] or a mutant tra-2 3′UTR [tra-2(−108)] in which both TGEs as well as some flanking sequence was deleted. The RNAs were capped and contained a poly(A) tail of 65 adenosine residues. The reporter RNAs were added to the yeast extract with or without a 10-fold molar excess of purified His-QKI-6 protein, and the luciferase activity was quantitated at specific times. The addition of His-QKI-6 protein to reactions containing RNAs carrying tra-2(+) 3′UTR resulted in an approximately 40–50% decrease in luciferase activity (Fig. 4A, closed squares); however, incubation of His-QKI-6 protein with RNAs carrying tra-2(−108) 3′UTR did not significantly affect luciferase activity (Fig. 4B, closed squares). Northern analysis showed that the differences in luciferase activity were not the result of changes in RNA levels (data not shown). Neither GLD-1 nor QKI-6 fully represses translation in the yeast in vitro translation assay. Similar to QKI-6, GLD-1 represses translation by about 50%. It is possible that full repression by QKI-6 and GLD-1 requires other factors not present in the yeast extract. Alternatively, it is possible that 50% repression is sufficient for normal development, because many processes are dosage sensitive. We next examined whether QKkt4 could repress translation in the yeast in vitro assay. Although QKkt4 still binds RNA (see above), it was not able to repress translation of a reporter RNA that contained the wild-type tra-2(+) 3′UTR [Fig. 4C; compare a tenfold molar excess of QKkt4 (open circles) with no protein (closed squares)]. These data indicate it is possible to separate RNA binding from translational control. Perhaps the disruption of the ability of QKI-6 to interact with itself or another factor by the qkkt4 mutation within the QUA1 domain significantly reduces its ability to repress translation.
Figure 4.
QKI-6 represses translation through the TGEs in vitro, whereas QKkt4 containing a glutamic acid to glycine mutation at position 48 does not repress translation. Translation of reporter RNAs were assayed in yeast extracts. (A) A reporter luciferase RNA (6 nM) carrying the tra-2(+) 3′UTR or (B) the tra-2(−108) 3′UTR lacking the TGEs was incubated with 60 nM His-QKI-6 protein (closed squares) and compared with incubations in which protein was not added (open circles). (C) A reporter luciferase RNA (6 nM) carrying the tra-2(+) 3′UTR or (D) the tra-2(−108) 3′UTR was incubated with 60 nM His-QKkt4 protein (closed squares) and compared with incubations in which protein was not added (open circles). The 3′UTRs present within the reporter RNAs are depicted above each graph. Aliquots of the reaction mixture were taken at the indicated time points. Data are plotted as a percent of total luciferase activity at the t = 60 min when only the reporter RNAs were present. All presented data are averages and standard deviations of at least three independent experiments. The absolute luciferase values were similar for each of the reporter RNAs in the absence of His-QKI-6 or His-QKkt4 proteins.
In conclusion, the mouse QKI-6, like the C. elegans GLD-1, can repress translation both in vivo and in vitro, and this repression is dependent on TGEs within the 3′UTR. QKI-6 protein is required for proper myelination in the mouse. How QKI-6 controls development and the nature of its RNA target(s) is unclear. Our findings raise the possibility that QKI-6 regulates development in the mouse by controlling the translation of TGE-containing RNA(s). Immunocytochemistry indicates that QKI-6 is mostly cytoplasmic (16); this is consistent with it acting as a translational regulator. It is also not known how QKI-5 and QKI-7 control gene activity. QKI-5 is predominantly nuclear and shuttles between the nucleus and cytoplasm, suggesting it may act by a different mechanism to control gene expression (16, 17). Perhaps QKI-5 is involved in splicing like the STAR family member SF1/BBP (34, 35). QKI-7 has the same tissue distribution as QKI-6 and is also cytoplasmic (16), and may operate in the same manner as QKI-6. Because STAR proteins are thought to homo- as well as heterodimerize (6, 36), one speculative model is that QKI-7 and QKI-6 interact, and in so doing alter their activity and/or RNA specificity.
The TGE regulation and the STAR proteins are found in both invertebrates and vertebrates (1, 9). Our analysis of C. elegans GLD-1 and the mouse QKI-6 proteins suggests that at least these two STAR family members control gene expression by acting through TGEs to repress translation. Interestingly, sequence analysis of the STAR proteins indicates that they fall into subfamilies (1). For example, GLD-1 and QKI-6 are more similar to the Drosophila WHO/HOW, Xenopus XQUA, and the human HQUA proteins than the mammalian SF1/BPP and SAM-68. It is possible that STAR protein subfamilies function similarly and those proteins most similar to GLD-1 and QKI-6 may also control development by acting as TGE translational regulators.
Acknowledgments
We thank Aurelia Haller and Peter Sarnow for assistance in the yeast in vitro assay and Laimonis Laimins for use of the luminometer. We thank Peter Candido and Colette Witkowski for constructs. We thank Laura Graves, Scott Segal, Sejal Shah, Jackie Yuen, and Young Yoo for valuable discussions. We acknowledge the Caenorhabditis Genetics Center for worm strains. This work was supported by National Institutes of Health Grant GM 51836-01 (to E.B.G. and L.S.), National Institutes of Health Grants HD 10668 and HD 30658 (to K.A.), a Cancer Biology Training Grant (to C.L.), and a National Institutes of Health Carcinogenesis Training Grant (to E.J.).
Abbreviations
- TGE
tra-2 and GLI element
- β-gal
β-galactosidase
- 3′UTR
3′ untranslated region
- STAR
signal transduction and activation of RNA
- GLD-1
germline defective-1
- tra-2
transformer
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Vernet C, Artzt K. Trends Genet. 1997;13:479–484. doi: 10.1016/s0168-9525(97)01269-9. [DOI] [PubMed] [Google Scholar]
- 2.Ebersole T A, Chen Q, Justice M J, Artzt K. Nat Genet. 1996;12:260–265. doi: 10.1038/ng0396-260. [DOI] [PubMed] [Google Scholar]
- 3.Justice M J, Bode V C. Genet Res. 1988;51:95–102. doi: 10.1017/s0016672300024101. [DOI] [PubMed] [Google Scholar]
- 4.Baehrecke E. Development (Cambridge, UK) 1997;124:1323–1332. doi: 10.1242/dev.124.7.1323. [DOI] [PubMed] [Google Scholar]
- 5.Zaffran S, Astier M, Gratecos D, Semeriva M. Development (Cambridge, UK) 1997;124:2087–2098. doi: 10.1242/dev.124.10.2087. [DOI] [PubMed] [Google Scholar]
- 6.Zorn A M, Krieg P A. Genes Dev. 1997;11:2176–2190. doi: 10.1101/gad.11.17.2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Francis R, Barton M K, Kimble J, Schedl T. Genetics. 1995;139:579–606. doi: 10.1093/genetics/139.2.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Francis R, Maine E, Schedl T. Genetics. 1995;139:607–630. doi: 10.1093/genetics/139.2.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jan E, Motzny C K, Graves L E, Goodwin E B. EMBO J. 1999;18:258–269. doi: 10.1093/emboj/18.1.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hodgkin J A, Brenner S. Genetics. 1977;86:275–287. [PMC free article] [PubMed] [Google Scholar]
- 11.Kuwabara P E, Kimble J. Trends Genet. 1992;8:164–168. doi: 10.1016/0168-9525(92)90218-s. [DOI] [PubMed] [Google Scholar]
- 12.Hodgkin J. Genetics. 1980;96:649–664. doi: 10.1093/genetics/96.3.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goodwin E B, Okkema P G, Evans T C, Kimble J. Cell. 1993;75:329–339. doi: 10.1016/0092-8674(93)80074-o. [DOI] [PubMed] [Google Scholar]
- 14.Jan E, Yoon J W, Walterhouse D, Iannaccone P, Goodwin E B. EMBO J. 1997;16:6301–6313. doi: 10.1093/emboj/16.20.6301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doniach T. Genetics. 1986;114:53–76. doi: 10.1093/genetics/114.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hardy R J, Loushin C L, Friederich V L, Jr, Chen Q, Ebersosle T A, Lazzarine R A, Artzt K. J Neurosci. 1996;16:7941–7949. doi: 10.1523/JNEUROSCI.16-24-07941.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu J, Zhou L, Tonissen K, Tee R, Artzt K. J Biol Chem. 1999;274:199–201. doi: 10.1074/jbc.274.41.29202. [DOI] [PubMed] [Google Scholar]
- 18.Sidman R L, Dickie M M, Apllel S H. Science. 1964;144:309–311. doi: 10.1126/science.144.3616.309. [DOI] [PubMed] [Google Scholar]
- 19.Friedrich V L., Jr Brain Res. 1974;82:168–172. doi: 10.1016/0006-8993(74)90902-0. [DOI] [PubMed] [Google Scholar]
- 20.Bode V C. Genetics. 1984;108:457–470. doi: 10.1093/genetics/108.2.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Justice M J, Bode V C. Genet Res. 1986;47:187–192. doi: 10.1017/s0016672300023119. [DOI] [PubMed] [Google Scholar]
- 22.Shedlovsky A, King T R, Dove W F. Proc Natl Acad Sci USA. 1988;85:180–184. doi: 10.1073/pnas.85.1.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brenner S. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hodgkin J. Genetics. 1986;114:15–52. doi: 10.1093/genetics/114.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Edgley M L, Baillie D L, Riddle D L, Rose A M. In: Methods in Cell Biology. Epstein H, Shakes D C, editors. New York: Academic; 1995. pp. 147–184. [PubMed] [Google Scholar]
- 26.Ausubel F M, Brent R, Kingston R E, Moore D, Seidman J G, Smith J A, Struhl K. Current Protocols in Molecular Biology. 1, Suppl. 31. New York: Greene & Wiley; 1995. pp. 3.8.1–3.8.4. [Google Scholar]
- 27.Milligan J F, Uhlenbeck O C. Methods Enzymol. 1989;180:51–62. doi: 10.1016/0076-6879(89)80091-6. [DOI] [PubMed] [Google Scholar]
- 28.Goodwin E B, Hofstra K, Hurney C A, Mango S, Kimble J. Development (Cambridge, UK) 1997;124:749–758. doi: 10.1242/dev.124.3.749. [DOI] [PubMed] [Google Scholar]
- 29.Mello C C, Kramer J M, Stinchcomb D, Ambros V. EMBO J. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iizuka N, Najita L, Franzusoff A, Sarnow P. Mol Cell Biol. 1994;14:7322–7330. doi: 10.1128/mcb.14.11.7322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Preiss T, Hentze M W. Nature (London) 1998;392:516–520. doi: 10.1038/33192. [DOI] [PubMed] [Google Scholar]
- 32.Chen T, Richard S. Mol Cell Biol. 1998;18:4863–4871. doi: 10.1128/mcb.18.8.4863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meyer B J. In: C. elegans II. 2nd Ed. Riddle D L, Blumenthal T J, Meyer B J, Priess J R, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1997. pp. 209–240. [PubMed] [Google Scholar]
- 34.Abovich N, Rosbash M. Cell. 1997;89:403–412. doi: 10.1016/s0092-8674(00)80221-4. [DOI] [PubMed] [Google Scholar]
- 35.Berglund J A, Chua K, Abovich N, Reed R, Rosbash M. Cell. 1997;89:781–787. doi: 10.1016/s0092-8674(00)80261-5. [DOI] [PubMed] [Google Scholar]
- 36.Chen T, Damaj B B, Herrera C, Lasko P, Richard S. Mol Cell Biol. 1997;17:5707–5718. doi: 10.1128/mcb.17.10.5707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stringham E G, Dixon D K, Jones D, Candido E P. Mol Cell Biol. 1992;3:221–233. doi: 10.1091/mbc.3.2.221. [DOI] [PMC free article] [PubMed] [Google Scholar]