Abstract
Retinoic acid receptors (RARs) are hormone-regulated transcription factors that control key aspects of normal differentiation. Aberrant RAR activity may be a causal factor in neoplasia. Human acute promyelocytic leukemia, for example, is tightly linked to chromosomal translocations that fuse novel amino acid sequences (denoted PML, PLZF, and NPM) to the DNA-binding and hormone-binding domains of RARα. The resulting chimeric receptors have unique transcriptional properties that may contribute to leukemogenesis. Normal RARs repress gene transcription by associating with ancillary factors denoted corepressors (also referred to as SMRT, N-CoR, TRAC, or RIP13). We report here that the PML-RARα and PLZF-RARα oncoproteins retain the ability of RARα to associate with corepressors, and that this corepressor association correlates with certain aspects of the leukemic phenotype. Unexpectedly, the PLZF moiety itself can interact with SMRT corepressor. This interaction with corepressor is mediated, in part, by a POZ motif within PLZF. Given the presence of POZ motifs in a number of known transcriptional repressors, similar interactions with SMRT may play a role in transcriptional silencing by a variety of both receptor and nonreceptor transcription factors.
Retinoids regulate many aspects of vertebrate cell proliferation and differentiation through receptors that function as hormone-regulated transcription factors (1–6). Two major classes of retinoid receptors have been identified: retinoic acid receptors (RARs) and retinoid X receptors (RXRs) (3–6). Both RARs and RXRs bind to specific sites on the DNA, and they can activate or repress expression of adjacent target genes (1–6). Aberrant RARs appear to play a crucial role in human acute promyelocytic leukemia (APL) (7–15). In over 95% of all APL patients, specific chromosomal translocations create abnormal RARs by replacing the N terminus of RARα with novel ORFs. The breakpoint in RARα is identical in all cases, whereas the nature of the novel N-terminal sequences can vary. In the common t(15;17) translocation, the RARα N terminus is replaced with an ORF denoted PML (for promyelocytic leukemia) (7–12). Alternatively, t(11;17) and t(5;17) translocations result in chimeric proteins denoted PLZF (promyelocytic leukemia zinc finger)-RARα and NPM (nucleophosmin)-RARα, respectively (13–15). Although possessing a number of recognizable structural motifs, including several prevalent in transcription factors, the PML, PLZF, and NPM-derived sequences exhibit little structural interrelatedness, and the functions of these proteins in the normal cell are not fully understood (reviewed in refs. 16–18).
The near-invariant association of acute promyelocytic leukemia with the expression of chimeric RARα proteins, and the ability of retinoids to drive leukemias bearing the PML-RARα translocation into differentiation and clinical remission (16–18), suggest an active role for these chimeras in conferring the leukemogenic phenotype. Furthermore, ectopic expression of PML-RARα in murine or avian systems can lead to leukemia and hepatocellular carcinoma (e.g., see refs. 19–22). However, the molecular basis by which these chimeric RARα proteins contribute to neoplasia is unclear. PML-RARα and PLZF-RARα bind to many of the same target DNA sites recognized by RARα (14, 23–25). Therefore, the chimeric RARα proteins may function in leukemias by leading to aberrant regulation of genes that are targets for RARα in normal cells. Alternatively, if PML and PLZF are themselves involved in transcriptional regulation, the chimeras may aberrantly regulate genes normally under PML or PLZF control.
In the absence of hormone, RARs (and the closely related thyroid hormone receptors, T3Rs) act as transcriptional repressors, a process mediated, in part, by the physical association of these receptors with ancillary proteins termed corepressors (26–33). Addition of hormone converts these receptors into transcriptional activators, a process that correlates with dissociation of the corepressors from the receptor and a corresponding recruitment of a novel set of coactivator proteins (26–34). In vertebrates, corepressors are encoded by two known, interrelated loci, denoted SMRT (or TRAC) and N-CoR (or RIP13), with each locus expressed as a variety of alternatively spliced mRNAs (e.g., TRAC-1, TRAC-2, RIP13A; refs. 26–33). Mutant T3Rs exhibiting aberrant interactions with corepressors display aberrant transcriptional regulatory properties (26–29, 33). Therefore we asked if the PML-RARα and PLZF-RARα chimeras exhibited similar aberrant interactions with corepressors that might contribute to their oncogenic proclivities. The results of these studies are reported below.
MATERIALS AND METHODS
In Vitro Receptor/Corepressor Binding Assays.
Glutathione S-transferase (GST), and GST-SMRT fusion constructions representing codons 71–769 of TRAC-1 were previously described (29, 33). Similar GST fusion constructs restricted to SMRT(TRAC-1) codons 71–394, 406–769, or 611–769 were created by subcloning of appropriate restriction fragments into pGEX-KG (33). GST-PLZF fusion proteins representing PLZF codons 1–456, 1–120, or 121–456 were generated by use of synthetic primers and PCR to introduce EcoRI and XhoI sites flanking the regions of interest and by insertion of the resulting DNA fragments into the EcoRI and XhoI sites of pGEX-KG (35, 36). GST-PLZF fusion proteins representing codons 1–137, 365–456, or 435–456 were obtained by cleavage of a PLZF insert in pCR2.1 (Invitrogen) with BamHI–NcoI, NcoI–XhoI, or SacI–HindIII, respectively, and insertion of the resulting fragments into the corresponding restriction sites in pGEX-KG. GST fusion proteins were expressed in Escherichia coli and were immobilized on glutathione-agarose as previously described (29, 33).
35S-radiolabeled PLZF, PLZFΔc, PML, RARα, PML-RARα, PLZF-RARα, and SMRT(TRAC-1) were synthesized by a coupled in vitro transcription and translation system (TnT kit, Promega), using appropriate pBluescript- or pSG5-based plasmids. SMRT-association-defective (“AHT”) mutants of RARα, PML-RARα, or PLZF-RARα were generated as previously described for T3R and RAR (27, 33). The radiolabeled proteins were incubated with the immobilized GST fusion proteins in HEMG buffer (29), the agarose matrix was extensively washed, and the bound proteins were eluted with glutathione and analyzed by SDS/PAGE (29, 33). The electrophoretograms were visualized by autoradiography and were quantified either by phosphoimager analysis or by scanning densitometry.
Mammalian Two-Hybrid Assay.
The HindIII fragment of pACT2 (CLONTECH) was inserted between a linker-modified EcoRI and BamHI site of pSG5 (Stratagene) to create the pSG5-GAL4AD vector (AD, activation domain). PLZF-activation domain fusions were first created in pACT2 by subcloning of suitable portions of PLZF (codons 1–456, 1–120, or 121–456) by using synthetic primers and PCR, then transferring these subclones as MluI to XhoI fragments into the pSG5-GAL4AD vector (36). pSG5-GAL4AD-T3Rα was constructed by inserting the HindIII fragment from pACT2-T3Rα into the modified pSG5 HindIII site (29). The pSG5-GAL4DBD vector (DBD, DNA-binding domain) was created by transferring the HindIII to BamHI portion of pGBT9 (CLONTECH) as a blunt-end fragment into the similarly treated EcoRI and BamHI sites of pSG5. The SMRT (TRAC-1) codon fusion was constructed by inserting the EcoRI fragment of pGBT9-SMRT(TRAC-1) into the pSG5-GAL4DBD vector (29). pSG5-GAL4DBD vectors bearing subdomains of SMRT(TRAC-1) were created by use of appropriate restriction sites and subcloning.
Transient transfections of CV-1 cells were performed by a calcium phosphate coprecipitation method. Each 60-mm plate, representing approximately 3 × 105 cells, was transfected with 500 ng of a pGAL-(17-mer)-Luc reporter (31), 125 ng of the pSG5-GAL4DBD vector, 500 ng of the pSG5-GAL4AD vector, 500 ng of a pCH110 vector (Pharmacia) as an internal β-galactosidase control, and sufficient pUC19 to normalize the total DNA to 10 μg. Luciferase activity was determined after 46 hr using a luciferase assay kit (Promega) and a TD 20/20 luminometer (Turner Design, Sunnyvale, CA). The relative light units were normalized to the β-galactosidase activity.
The 5XGAL-β-globin-CAT reporter contains five GAL4(17-mer) binding sites inserted upstream of a β-globin promoter/chloramphenicol acetyltransferase construct and was kindly provided by P. Chambon (Institut Pasteur). GAL4DBD fusions to full-length PLZF, or to human HP1α were expressed under the control of the simian virus 40 promoter. Human Chang liver cells (CCL13), maintained in DMEM with 10% calf serum, were seeded and transfected using a calcium phosphate procedure similar to that described for the CV-1 cells. Each plate received 0.5 μg of pCH110, 1.5 μg of the 5XGAL-β-globin-CAT reporter plasmid, the appropriate GAL4DBD-fusion plasmid as indicated, and sufficient pSG5 plasmid to bring the total DNA to 7.5 μg. Cell lysates were prepared 48 hr later by freeze–thaw in 250 mM Tris⋅HCl (pH 7.8). CAT activity, relative to β-galactosidase, was determined.
RESULTS
PML-RARα and PLZF-RARα Retain the Ability to Bind to SMRT Corepressor in Vitro.
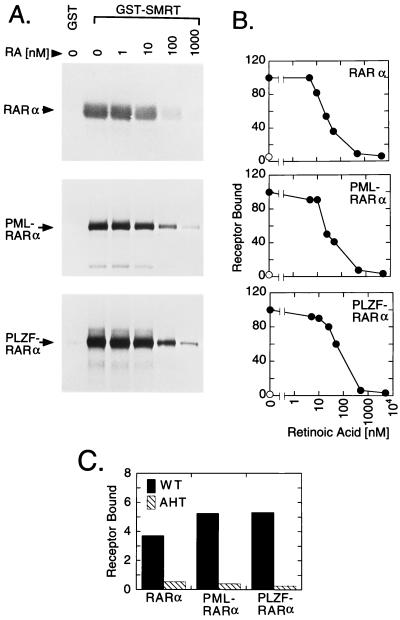
The chromosomal breakpoints that generate the PML-RARα and PLZF-RARα chimeras preserve intact the receptor-derived D and E domains that mediate binding of SMRT and N-CoR corepressors to the wild-type RARs (Fig. 1). To test if the chimeric RAR oncoproteins retained the ability to interact with corepressors, we employed an in vitro binding assay (29, 33). An immobilized GST derivative of SMRT (GST-SMRT) was incubated with 35S-radiolabeled receptor. Any receptor binding to the GST-SMRT matrix was subsequently eluted and analyzed by SDS/PAGE and autoradiography. Consistent with previous observations for RARβ (29), RARα readily bound to GST-SMRT in the absence of hormone, but not to a nonrecombinant GST employed as a negative control (Fig. 2A). Also consistent with previous work (26–33), addition of all-trans-retinoic acid to the binding assay led to a progressive release of RARα from the GST-SMRT matrix, with a 50% release observed at approximately 25 nM all-trans-retinoic acid (Fig. 2A, quantified in Fig. 2B). In contrast, no significant release of RARα from GST-SMRT was observed with ethanol carrier alone (Fig. 2B) or with comparable concentrations of retinoids that are not high-affinity ligands for RARα, such as all-trans-retinal, 9-cis-retinal, or all-trans-retinol (data not shown).
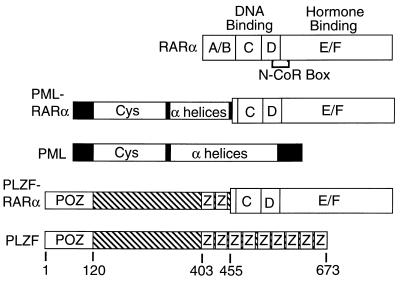
Figure 1.
Schematic representations of the RARα, PML, PML-RARα, PLZF, and PLZF-RARα proteins. Proteins are depicted from N to C terminus. Sequences derived from RARα are subdivided into domains A to F, as previously described, with the receptor regions involved in DNA and hormone binding indicated above the schematic, and the receptor N-CoR box, necessary for corepressor association, indicated below (1–6). Putative structural motifs identified within PML or PLZF-derived sequences include a cysteine-rich RING finger/B-box motif (Cys), an α-helical/coiled-coil domain (α-helices), a POZ domain (37), and zinc-finger motifs (Z). Codon positions within PLZF are numbered from the N terminus.
Figure 2.
Binding of RARα, PML-RARα, and PLZF-RARα to GST-SMRT corepressor. (A) Binding of RARα derivatives to corepressor in vitro. Radiolabeled receptors, synthesized in vitro, were incubated with immobilized GST, or with immobilized GST-SMRT(TRAC-1), over a range of all-trans-retinoic acid concentrations, or with ethanol carrier alone, as indicated above the lanes. Receptors bound to the immobilized GST derivatives were eluted and were subsequently analyzed by SDS/PAGE and autoradiography. (B) Quantitation of the SMRT in vitro binding assay. The amount of each receptor derivative bound to GST (⊙) or to GST-SMRT(TRAC-1) (•) under different all-trans-retinoic acid concentrations was analyzed as in A and quantified. Receptor binding in the absence of hormone was defined as 100%. (C) Comparison of SMRT binding by wild-type receptor versus mutant receptors bearing defects in corepressor association. Receptor binding to GST-SMRT(TRAC-1) was determined as in A and was quantified as the percentage of the input radiolabel. All determinations were in the absence of hormone. Binding of wild-type RARα, PML-RARα, and PLZF-RARα to corepressor (solid bars) was compared with that of equivalent receptors bearing N-CoR box mutations (AHT mutants; hatched bars).
In common with RARα, both PML-RARα and PLZF-RARα also strongly bind to GST-SMRT, but not to nonrecombinant GST, and the binding of the chimeric RARs to corepressor was destabilized by all-trans-retinoic acid, but not by ethanol alone (Fig. 2 A and B) or by noncognate ligands (data not shown). An “N-CoR box” sequence motif in the receptor has been implicated in the ability of RARs to interact with corepressors (Fig. 1 and refs. 26–33). Indeed, mutation of the conserved N-CoR box sequence (AHT mutants) severely disrupted the ability of RARα, PML-RARα, and PLZF-RARα to bind to SMRT in vitro (Fig. 2C). We conclude that both PML-RARα and PLZF-RARα retain the ability to bind SMRT corepressor, and that hormone destabilizes this interaction for the chimeric receptors with dynamics comparable to, although not identical with, dynamics for RARα. Furthermore, the principal SMRT/receptor interaction domain in the chimeric receptors, as determined by in vitro binding assay, appears to be the same as that previously elucidated for RARα.
The PLZF Domain Possesses an Autonomous Ability to Bind to SMRT Corepressor in Vitro.
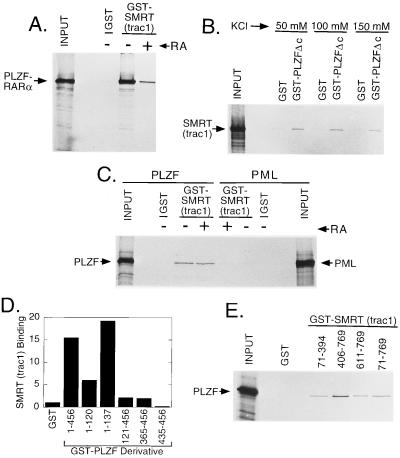
The primary interaction between SMRT and the chimeric RARs in vitro appeared to be through the same receptor-derived, D and E domain determinants as in the wild-type RARα. However, close inspection revealed a residual binding to the GST-SMRT matrix that was not abrogated by even very high concentrations of hormone (Figs. 2A and 3A). Unexpectedly, one component of this residual, hormone-resistant interaction between SMRT and PLZF-RARα mapped to the PLZF portion of the chimera (GST-PLZF-Δc in Fig. 3B) and was also observed for the full-length (i.e., nontranslocated) PLZF protein (Fig. 3C). These SMRT/PLZF interactions were detectable by using either the GST-SMRT construct and a radiolabeled PLZF derivative (Fig. 3C), or reciprocally, a GST-PLZF construct and a radiolabeled SMRT derivative (Fig. 3B and data not shown). Although the in vitro interaction between PLZF and SMRT was relatively weak compared with that observed between RARα and SMRT, this binding of PLZF to SMRT was nonetheless highly reproducible, was detectable over a variety of buffer conditions, and was not observed for nonrecombinant GST constructs employed as negative controls (Fig. 3 B and C). Also of note, the PML gene product exhibited little or no binding to SMRT in parallel assays (Fig. 3C). We conclude that PLZF-RARα possesses at least two distinct corepressor binding domains, a C-terminal, hormone-labile, SMRT-interaction domain shared with RARα and PML-RARα and a separate, hormone-insensitive, SMRT interaction domain mapping to the PLZF domain itself.
Figure 3.
Binding of PLZF and PLZF derivatives to SMRT corepressor. (A) Residual SMRT binding by PLZF-RARα at high hormone concentrations. The ability of radiolabeled PLZF-RARα to bind to GST or to GST-SMRT(TRAC-1) was determined as in Fig. 2A, but in the absence (−) or presence (+) of 40 μM all-trans-retinoic acid, as indicated above the lanes. An aliquot of the in vitro translation products used in the binding assay was analyzed in the input lane. (B) Binding of the PLZF moiety by SMRT corepressor. Radiolabeled SMRT(TRAC-1) was incubated with immobilized GST, or with GST-PLZFΔc (codons 1–456 of PLZF), using HEMG buffer containing the KCl concentrations indicated. Radiolabeled SMRT(TRAC-1) remaining bound to the matrix after extensive washing was eluted and analyzed by SDS/PAGE and autoradiography as in Fig. 2A. (C) Binding of full-length PLZF, but not PML, to SMRT corepressor. Radiolabeled full-length PLZF or PML was incubated with immobilized GST or with immobilized GST-SMRT(TRAC-1), in HEMG buffer containing 100 mM KCl and lacking (−) or containing (+) 5 μM all-trans-retinoic acid, as indicated. Proteins bound to the immobilized matrices were subsequently eluted and analyzed by SDS/PAGE and autoradiography. (D) Mapping of domains of PLZF able to confer SMRT binding in vitro. The ability of radiolabeled SMRT(TRAC-1) to bind to GST fusions containing different portions of PLZF was tested using the protocol described for B; the results were quantified by phosphoimager analysis and are expressed as arbitrary units. (E) Mapping of domains of SMRT(TRAC-1) capable of binding to PLZF. The same protocol as in C was employed, but using GST fusions bearing the different domains of SMRT(TRAC-1) indicated. Codons are indicated using the TRAC-1 numbering system (codon 769 of TRAC-1 is equivalent to codon 1495 of SMRT(TRAC-2)).
The POZ Domain in PLZF Is Sufficient to Mediate an Interaction with SMRT in Vitro.
A number of amino acid motifs have been identified within the Krüppel-like PLZF polypeptide that mediate protein–protein or protein–DNA interactions in other protein contexts. These include the N-terminal POZ domain and two internal zinc-finger motifs (Fig. 1 and refs. 37–39). PLZF constructs encompassing just the POZ domain, expressed as either codons 1–120 or codons 1–137, were sufficient to confer interaction with SMRT in vitro (Fig. 3D). In contrast, constructs encompassing the remainder of the PLZF sequence in PLZF-RARα (codons 121–456) or subsets of this sequence representing one or both zinc-finger motifs (codons 365–456, 435–456) interacted much more weakly, or not at all, with SMRT in vitro (Fig. 3D). We suggest that the POZ domain is likely the principal mediator of the PLZF/SMRT interaction observed in vitro, although additional PLZF domains may play contributory roles.
Conversely, at least two distinct domains within SMRT appeared to mediate the corepressor’s interaction with PLZF in vitro (Fig. 3E). One PLZF-interaction domain mapped to the same SMRT C-terminal domain involved in binding SMRT to nuclear hormone receptors (29), as seen with constructs representing SMRT(TRAC-1) codons 406–769 or 611–769 (Fig. 3E). In addition, however, codons 71–394 of SMRT (TRAC-1) were also able to bind to PLZF in vitro (Fig. 3E); these more N-terminal SMRT determinants do not bind nuclear hormone receptors (29), suggesting that PLZF binding by SMRT may involve multiple contacts, including novel SMRT determinants not involved in interactions with the nuclear hormone receptors.
A Strong PLZF Interaction with SMRT Is Observed in Vivo.
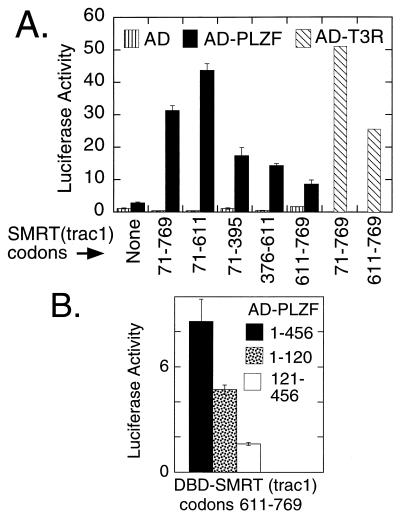
We next tested if the PLZF/SMRT interaction could be detected in vivo. Employing a Saccharomyces cerevisiae two-hybrid system (29), we could detect a consistent, but low-level, activation of a Gal4-responsive β-galactosidase reporter when a GAL4DBD fusion of SMRT was coexpressed with a GAL4AD fusion of PLZF (data not shown). Notably, however, the interaction between SMRT and PLZF in the yeast two-hybrid assay, as in vitro, was extremely weak. We reasoned that these very modest interactions between PLZF and SMRT might be enhanced if assayed in a more physiological context. Therefore we next tested the ability of PLZF and SMRT to interact in a mammalian two-hybrid system. Indeed, a very strong activation of a luciferase reporter containing GAL4 (17-mer) binding sites was observed when a GAL4DBD-SMRT fusion was coexpressed in CV-1 monkey cells with a PLZF-AD fusion (Fig. 4A). This SMRT/PLZF interaction in the mammalian two-hybrid assay was now comparable in magnitude to the interaction observed between SMRT and T3Rs when tested in the same context (Fig. 4A). In contrast, negative controls, representing DBD or AD vectors lacking SMRT or PLZF sequences, conferred little or no stimulation of the luciferase reporter (Fig. 4A).
Figure 4.
Mammalian two-hybrid analysis of the PLZF interaction with SMRT. (A) Two-hybrid interaction mediated by PLZFΔc. Different portions of SMRT(TRAC-1) fused to a GAL4DBD, or the GAL4DBD alone, were employed as indicated below the bars, using the numbering system in Fig. 3E. The GAL4AD alone (AD; vertical stripes), the AD fused to PLZF codons 1–456 (AD-PLZF; filled bars), or the AD fused to the D to F domains of avian T3Rα (AD-T3R; hatched bars) was then coexpressed with the DBD derivatives by transfections of CV-1 cells. The activity of a cointroduced GAL4(17-mer)/tk-luciferase reporter was determined, relative to a cytomegalovirus (CMV) lacZ construct employed as an internal control. (B) Two-hybrid interaction mediated by subdomains of PLZF. Activation domain fusions of PLZF codons 1–456 (filled bars), of PLZF codons 1–120 (stippled bars), or PLZF codons 121–456 (empty bars) were tested in the same two-hybrid protocol as in A, using the DBD-SMRT construct indicated.
Paralleling our in vitro binding data, both the N-terminal (e.g., codons 71–395) and the C-terminal (e.g., codons 611–769) domains of SMRT(TRAC-1) could independently interact with PLZF in the mammalian two-hybrid assay, with a SMRT(TRAC-1) construct containing both N- and C-terminal domains (codons 71–769) exhibiting an additive response (Fig. 4A). Also consistent with our in vitro data, a portion of PLZF limited to the POZ domain (codons 1–120) displayed a detectable interaction with certain of the SMRT constructs in the mammalian two-hybrid assay (Fig. 4B). Notably, however, this POZ-domain/SMRT interaction in the mammalian two-hybrid assay was weak, and it was only one component of the much stronger interaction observed with SMRT when a codon 1–456 version of PLZF was tested (Fig. 4A). We suggest that the PLZF/SMRT interaction in vitro is mediated primarily by POZ/SMRT contacts, but this interaction in vivo may be further enhanced by additional proteins binding to the non-POZ domains of PLZF.
PLZF Can Function as a Transcriptional Repressor.
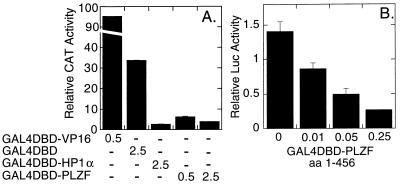
Is the ability of PLZF to interact with corepressor manifested as an ability to repress transcription? No authentic DNA-binding sites have been isolated so far for PLZF, so we created a GAL4DBD-PLZF fusion, and tested the ability of this fusion protein to repress basal transcription from a promoter containing five GAL4(17-mer) binding sites and driving a reporter gene. The GAL4DBD-PLZF construct functioned as a potent dose-dependent repressor under these conditions, equivalent in strength to a GAL4DBD construct of HP-1α, a known transcriptional repressor (40) (Fig. 5A). In contrast, an empty GAL4DBD construct employed as a negative control had no effect on reporter gene expression (Fig. 5A). Notably, a GAL4 fusion limited to PLZF amino acids 1–456 (the domain found in PLZF-RARα) also exhibited strong transcriptional silencing properties (Fig. 5B), indicating that the PLZF sequences within PLZF-RARα are sufficient to mediate transcriptional repression. Repression by the GAL4DBD-PLZF constructs was detectable in a variety of experimental contexts, employing a range of promoters, reporter genes, and cell types (Fig. 5 and data not shown).
Figure 5.
Transcriptional repression by GAL4-PLZF fusion proteins. (A) Repression by GAL4DBD-PLZF. Chang liver cells were transfected with a 5XGAL-β-globin-CAT reporter, a pCH110 internal standard, and 0.5 or 2.5 μg per plate of the GAL4DBD fusion vectors indicated. CAT activity was determined as percentage of chloramphenicol converted to acetylated form, normalized to the internal β-galactosidase standard. Values represent the averages of two independent transfections; no significant differences were observed between duplicate experiments. (B) Repression by a GAL4DBD-PLZF construct restricted to PLZF amino acids 1–456. From 0 to 0.25 μg per plate of a GAL4DBD fusion vector representing amino acids 1–456 of PLZF (GAL4DBD-PLZFΔc) was introduced into CV-1 cells, together with 1 μg of the pGAL4(17-mer)-Luc reporter. The resulting luciferase activity, relative to a β-galactosidase internal control, was determined as described for the two-hybrid system; the average and range obtained from duplicate experiments are presented.
DISCUSSION
Both PML-RARα and PLZF-RARα Retain the Ability of RARα to Associate with the SMRT/TRAC Family of Corepressors.
The transcriptional properties of nuclear hormone receptors are mediated, in part, by the ability of these receptors to physically associate with corepressors and coactivators. Corepressors physically bind to T3Rs and RARs in the absence of hormone, a context in which these receptors typically repress. Addition of hormone leads to release of the corepressor, the association of coactivators, and conversion of the receptor to a positive enhancer of transcription (26–34). Given the central role of corepressors in the actions of the normal RARs, we examined the role of these cofactors in the actions of the PML-RARα and PLZF-RARα oncoproteins. Indeed, both of these chimeric proteins retained the ability of the wild-type RARα to bind to the SMRT corepressor in vitro. Also in common with the wild-type RARα, the association of PML-RARα and PLZF-RARα with corepressor in vitro was disrupted by addition of all-trans-retinoic acid. The hormone concentrations necessary for 50% inhibition of SMRT binding were very similar for all three receptor derivatives, consistent with the near equal hormone-binding affinities previously reported for RARα, PML-RARα, and PLZF-RARα (37). This hormone-labile association of PML-RARα and PLZF-RARα with SMRT appeared to be mediated through the same receptor determinants as previously mapped in RARα, and it was similarly impaired by mutation of a conserved AH--T amino acid motif in the RARα N-CoR box.
PLZF Possesses an Autonomous and Hormone-Independent Ability to Interact with SMRT Corepressor.
In addition to the hormone-labile interaction domain within the RARα moiety, the PLZF-RARα chimera also possessed an unanticipated SMRT-association domain mapping within the PLZF sequences. Notably, the full-length PLZF gene product (encoded by a nontranslocated chromosome 11) also interacts with SMRT corepressor, whereas the full-length PML gene product exhibits little or no SMRT interaction. The PLZF interaction with SMRT observed in vitro was only a fraction of the much stronger SMRT interaction mediated by the RARα determinants under similar conditions. However, the interaction of PLZF with SMRT was greatly enhanced when assayed by a mammalian two-hybrid method, reaching a level comparable to the interaction detected between nuclear hormone receptors and SMRT in the same two-hybrid system. Notably, the SMRT protein itself is just one component of a larger multiprotein corepressor complex that includes mSin3, histone deacetylases, and other polypeptides (refs. 41 and 42 and references therein). Therefore, we suggest that, in vertebrate cells, PLZF makes multiple contacts with other members of this corepressor complex in addition to SMRT, and only a small part of this overall interaction is recapitulated when purified SMRT protein is used in vitro.
What role might corepressor association play in the function of the untranslocated PLZF protein in the normal cell? Unfortunately, the precise role of PLZF remains unclear. PLZF is a Krüppel-like nuclear protein which is likely involved in central nervous system development (13, 14, 25, 37, 39). Although no physiologically relevant DNA-binding sites or target genes have yet been reported for PLZF, many features of PLZF are shared with known transcription factors; it is therefore possible that PLZF is indeed a transcription factor and can regulate gene expression by tethering directly or indirectly (through an intermediary protein) to DNA. Consistent with this proposal, a GAL4DBD fusion of PLZF was a strong transcriptional repressor when tested on promoters containing GAL4 binding sites.
It is provocative that the POZ domain in PLZF is capable of mediating at least some component of the SMRT interaction. Consistent with the studies we present here, deletion of the POZ domain significantly inhibits the dominant-negative transcriptional properties of the PLZF-RARα chimera (37). Furthermore, POZ domains are found in a variety of known transcription factors that can function as repressors, such as the Bcl-6 oncoprotein (43–45). The POZ domain plays an important role in Bcl-6 repression (44, 45). It is tempting to speculate that gene silencing by Bcl-6, like that of RAR, is mediated by interaction with the SMRT/N-CoR family of corepressors, and that SMRT and N-CoR, presumably complexed with additional proteins, serve to mediate repression by a broad variety of nonreceptor transcription factors (refs. 41 and 42 and references therein).
Corepressor Association by PML-RARα and PLZF-RARα Correlates with the Hormone Responsiveness Displayed by the Corresponding Leukemias.
Leukemic cells bearing the t(15;17) PML-RARα translocation differentiate into mature granulocytes when treated with all-trans-retinoic acid, an important basis of the clinical management of this disease (16–18). In contrast, leukemic cells bearing the t(11;15) PLZF-RARα translocation are resistant to all-trans-retinoic acid treatment, remaining in an immature proliferative state and refractory to hormone therapy (46). It is therefore intriguing that PML-RARα exhibits a hormone-labile interaction with corepressor, whereas the PLZF-RARα interaction with SMRT is apparently further stabilized by the additional, hormone-refractory domain in PLZF. This correlation between SMRT association and leukemogenesis may be purely coincidental. Alternatively, PML-RARα and PLZF-RARα may indeed contribute to leukemogenesis by associating with SMRT and repressing the expression of specific genes. These target genes may include those normally regulated by RARα, or, due to changes in the DNA-recognition properties of PML-RARα and PLZF-RARα, the targets of PML-RARα or PLZF-RARα repression may be distinct from normal RARα target genes. More speculatively, if PLZF and PML are transcriptional regulators in their own right, then the RAR chimeric forms of these proteins may act by repressing transcription of PLZF or PML target genes.
It is clear that the molecular biology of acute promyelocytic leukemia is extremely complex and that the chimeric RARs may play more than one role. These actions may include subversion or mimicry of the actions of the untranslocated PML/PLZF proteins, and interference with the actions of RARs, retinoid X receptors, or nonretinoid nuclear hormone receptors present in the same cells. Corepressors may potentially have a function in any or all of these aspects. Clearly, additional work will be necessary to address the potential roles of SMRT in acute promyelocytic leukemia, including use of PML-RARα and PLZF-RARα constructs with altered corepressor association properties in animal models of human acute promyelocytic leukemia.
Acknowledgments
We are indebted to P. Chambon, C. Transy, J. M. Bishop, S. Kogan, M. Lazar, S. Lee-Bond, and S. Elledge for the generous gift of molecular clones and vectors employed in these experiments. This work was supported by National Institutes of Health Grant R37 CA-53394 and by a University of California Faculty Research Grant.
Note Added in Proof
It is worth noting that PLZF also interacts with the TRAC-2 related N-CoR protein (A. Zelent and F. Guidez, personal communication).
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: RAR, retinoic acid receptor; PML, promyelocytic leukemia; PLZF, PML zinc finger; AD, activation domain; DBD, DNA-binding domain; T3R, thyroid hormone receptor; GST, glutathione S-transferase; CAT, chloramphenicol acetyltransferase.
References
- 1.Lazar M A. Endocrinol Rev. 1993;14:184–193. doi: 10.1210/edrv-14-2-184. [DOI] [PubMed] [Google Scholar]
- 2.Tsai M J, O’Malley B W. Annu Rev Biochem. 1994;63:451–486. doi: 10.1146/annurev.bi.63.070194.002315. [DOI] [PubMed] [Google Scholar]
- 3.Mangelsdorf D J, Thummel C, Beato M, Herrlich P, Schutz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, Evans R M. Cell. 1995;83:835–840. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mangelsdorf D J, Evans R M. Cell. 1995;83:841–850. doi: 10.1016/0092-8674(95)90200-7. [DOI] [PubMed] [Google Scholar]
- 5.Beato M, Herrlich P, Schutz G. Cell. 1995;83:851–858. doi: 10.1016/0092-8674(95)90201-5. [DOI] [PubMed] [Google Scholar]
- 6.Kastner P, Mark M, Chambon P. Cell. 1995;83:859–870. doi: 10.1016/0092-8674(95)90202-3. [DOI] [PubMed] [Google Scholar]
- 7.de The H, Chomienne C, Lanotte M, Degos L, Dejean A. Nature (London) 1990;347:558–561. doi: 10.1038/347558a0. [DOI] [PubMed] [Google Scholar]
- 8.Borrow J, Goddard A D, Sheer D, Solomon E. Science. 1990;249:1577–1580. doi: 10.1126/science.2218500. [DOI] [PubMed] [Google Scholar]
- 9.Alcalay M D, Zangrilli D, Paolo P, Mencarelli A, Longo L, Giacomucci A, Rocchi M, Biondi A, Rambalsi A, LoCoco F, Diverio D, Donti E, Grignani F, Pelicci P G. Proc Natl Acad Sci USA. 1991;88:1977–1981. doi: 10.1073/pnas.88.5.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de The H, Lavau C, Marchio A, Chomienne C, Degos L, Dejean A. Cell. 1991;66:675–684. doi: 10.1016/0092-8674(91)90113-d. [DOI] [PubMed] [Google Scholar]
- 11.Kakizuka A, Miller W H, Jr, Umesono K, Warrell R P, Frankel S R, Murty V V S, Dmitrovsky E, Evans R M. Cell. 1991;66:663–674. doi: 10.1016/0092-8674(91)90112-c. [DOI] [PubMed] [Google Scholar]
- 12.Kastner P, Perez A, Lutz Y, Rochette-Egly C, Gaub M-P, Durand B, Lanotte M, Berger R, Chambon P. EMBO J. 1992;11:629–642. doi: 10.1002/j.1460-2075.1992.tb05095.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen Z, Brand N J, Chen A, Chen S J, Tong J H, Wang Z Y, Waxman S, Zelent A. EMBO J. 1993;12:1161–1167. doi: 10.1002/j.1460-2075.1993.tb05757.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Licht J D, Shaknovich R, English M A, Melnick A, Li J Y, Reddy J C, Dong S, Chen S J, Zelent A, Waxman S. Oncogene. 1996;12:323–336. [PubMed] [Google Scholar]
- 15.Redner R L, Rush E A, Faas S, Rudert W A, Corey S J. Blood. 1996;87:882–886. [PubMed] [Google Scholar]
- 16.Warrell R P, Jr, de The H, Wang Z Y, Degos L. N Engl J Med. 1993;329:177–189. doi: 10.1056/NEJM199307153290307. [DOI] [PubMed] [Google Scholar]
- 17.Lavau C, Dejean A. Leukemia. 1994;8:1615–1621. [PubMed] [Google Scholar]
- 18.Grignani F, Fagioli M, Alcalay M, Longo L, Pandolfi P P, Donti E, Biondi A, Lo Coco F, Grignani F, Pelicci P G. Blood. 1994;83:10–25. [PubMed] [Google Scholar]
- 19.Altabef M, Garcia M, Lavau C, Bae S C, Dejean A, Samarut J. EMBO J. 1996;15:2707–2716. [PMC free article] [PubMed] [Google Scholar]
- 20.Grisolano J L, Wesselschmidt R L, Pelicci P G, Ley T J. Blood. 1997;89:376–387. [PubMed] [Google Scholar]
- 21.Brown D, Kogan S, Lagasse E, Weissman I, Alcalay M, Pelicci P G, Atwater S, Bishop J M. Proc Natl Acad Sci USA. 1997;94:2551–2556. doi: 10.1073/pnas.94.6.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.David G, Terris B, Marchio A, Lavau C, Dejean A. Oncogene. 1997;46:1547–1556. doi: 10.1038/sj.onc.1200989. [DOI] [PubMed] [Google Scholar]
- 23.Perez A, Kastner P, Sethi S, Lutz Y, Reibel C, Chambon P. EMBO J. 1993;12:3171–3182. doi: 10.1002/j.1460-2075.1993.tb05986.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jansen J H, Mahfoudi A, Rambaud S, Lavau C, Wahli W, Dejean A. Proc Natl Acad Sci USA. 1995;92:7401–7405. doi: 10.1073/pnas.92.16.7401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen Z, Guidez F, Rousselot P, Agadir A, Chen S J, Wang Z Y, Degos L, Zelent A, Waxman S, Chomienne C. Proc Natl Acad Sci USA. 1994;91:1178–1182. doi: 10.1073/pnas.91.3.1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen J D, Evans R M. Nature (London) 1995;377:454–457. doi: 10.1038/377454a0. [DOI] [PubMed] [Google Scholar]
- 27.Horlein A J, Naar A M, Heinzel T, Torchia J, Gloss B, Kurokawa R, Ryan A, Kamel Y, Soderstrom M, Glass C K, Rosenfeld M G. Nature (London) 1995;377:397–404. doi: 10.1038/377397a0. [DOI] [PubMed] [Google Scholar]
- 28.Kurokawa R, Soderstrom M, Horlein A, Halachmi S, Brown M, Rosenfeld M G, Glass C K. Nature (London) 1995;377:451–454. doi: 10.1038/377451a0. [DOI] [PubMed] [Google Scholar]
- 29.Sande S, Privalsky M L. Mol Endocrinol. 1996;10:813–825. doi: 10.1210/mend.10.7.8813722. [DOI] [PubMed] [Google Scholar]
- 30.Chen J D, Umesono K, Evans R M. Proc Natl Acad Sci USA. 1996;93:7567–7571. doi: 10.1073/pnas.93.15.7567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zamir I, Harding H P, Atkins G B, Horlein A, Glass C K, Rosenfeld M G, Lazar M A. Mol Cell Biol. 1996;16:5458–5465. doi: 10.1128/mcb.16.10.5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seol W, Mahon M J, Lee Y K, Moore D D. Mol Endocrinol. 1996;10:1646–1655. doi: 10.1210/mend.10.12.8961273. [DOI] [PubMed] [Google Scholar]
- 33.Yoh S, Chatterjee V K K, Privalsky M L. Mol Endocrinol. 1997;11:470–480. doi: 10.1210/mend.11.4.9914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Horwitz K B, Jackson T A, Bain D L, Richer J K, Takimoto G S, Tung L. Mol Endocrinol. 1996;10:1167–1177. doi: 10.1210/mend.10.10.9121485. [DOI] [PubMed] [Google Scholar]
- 35.Guan K L, Dixon J E. Anal Biochem. 1991;192:262–267. doi: 10.1016/0003-2697(91)90534-z. [DOI] [PubMed] [Google Scholar]
- 36.Ausubel F M, Brent R, Kinston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current Protocols in Molecular Biology. New York: Wiley; 1997. pp. 8.5.7–8.5.9. [Google Scholar]
- 37.Dong S, Zhu J, Reid A, Strutt P, Guidez F, Zhong H-J, Wang Z-Y, Licht J, Waxman S, Chomienne C, Chen Z, Zelent A, Chen S-J. Proc Natl Acad Sci USA. 1996;93:3624–3629. doi: 10.1073/pnas.93.8.3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bardwell V J, Treisman R. Genes Dev. 1994;8:1664–1677. doi: 10.1101/gad.8.14.1664. [DOI] [PubMed] [Google Scholar]
- 39.Cook M, Gould A, Brand N, Davies J, Strutt P, Shaknovich R, Licht J, Waxman S, Chen Z, Gluecksohn-Waelsch S, Krumlauf R, Zelent A. Proc Natl Acad Sci USA. 1995;92:2249–2253. doi: 10.1073/pnas.92.6.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Le Douarin B, Nielsen A L, Garnier J M, Ichinose H, Jeanmougin F, Losson R, Chambon P. EMBO J. 1996;15:6701–6715. [PMC free article] [PubMed] [Google Scholar]
- 41.Wolffe A P. Nature (London) 1997;387:16–17. doi: 10.1038/387016a0. [DOI] [PubMed] [Google Scholar]
- 42.Pazin M J, Kadonaga J T. Cell. 1997;89:325–328. doi: 10.1016/s0092-8674(00)80211-1. [DOI] [PubMed] [Google Scholar]
- 43.Deweindt C, Albagli O, Bernardin F, Dhordain P, Quief S, Lantoine D, Kerckaert J P, Leprince D. Cell Growth Differ. 1995;6:1495–1503. [PubMed] [Google Scholar]
- 44.Chang C C, Ye B H, Chaganti R S, Dalla-Favera R. Proc Natl Acad Sci USA. 1996;93:6947–6952. doi: 10.1073/pnas.93.14.6947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Seyfert V L, Allman D, He Y, Staudt L M. Oncogene. 1996;12:2331–2342. [PubMed] [Google Scholar]
- 46.Guidez F, Huang W, Tong J H, Dubois C, Balitrand N, et al. Leukemia. 1994;8:312–317. [PubMed] [Google Scholar]