Abstract
Exposure of cells to protein tyrosine phosphatase (PTP) inhibitors causes an increase in the phosphotyrosine content of many cellular proteins. However, the level at which the primary signaling event is affected is still unclear. We show that Jaks are activated by tyrosine phosphorylation in cells that are briefly exposed to the PTP inhibitor pervanadate (PV), resulting in tyrosine phosphorylation and functional activation of Stat6 (in addition to other Stats). Mutant cell lines that lack Jak1 activity fail to support PV-mediated [or interleukin 4 (IL-4)-dependent] activation of Stat6 but can be rescued by complementation with functional Jak1. The docking sites for both Jak1 and Stat6 reside in the cytoplasmic domain of the IL-4 receptor α-chain (IL-4Rα). The glioblastoma-derived cell lines T98G, GRE, and M007, which do not express the IL-4Rα chain, fail to support Stat6 activation in response to either IL-4 or PV. Complementation of T98G cells with the IL-4Rα restores both PV-mediated and IL-4-dependent Stat6 activation. Murine L929 cells, which do not express the γ common chain of the IL-4 receptor, support PV-mediated but not IL-4-dependent Stat6 activation. Thus, Stat6 activation by PV is an IL-4Rα-mediated, Jak1-dependent event that is independent of receptor dimerization. We propose that receptor-associated constitutive PTP activity functions to down-regulate persistent, receptor-linked kinase activity. Inhibition or deletion of PTP activity results in constitutive activation of cytokine signaling pathways.
Keywords: signal transducer and activator of transcription, Janus kinase, pervanadate, protein tyrosine kinase, interleukin 4 receptor
Cytokines transmit their signals through transmembrane receptors that are physically associated with members of the Jak family of protein tyrosine kinases (PTK) (1–8). Aggregation of receptors, resulting in the association of Jaks in a receptor complex and a conformational change in the kinase domain, is an early step in cytokine receptor activation (2–11). Trans-phosphorylation of the conserved tyrosine residues in the kinase activation segments promotes kinase activity (11). The activated Jaks then phosphorylate the receptor-associated and downstream signaling molecules, including Stat proteins recruited to the receptor complex (1–8, 12–15).
Protein tyrosine phosphatases (PTP) are associated with many cytokine receptors and are implicated in the down-regulation of ligand-induced signaling through dephosphorylation of the activated Jaks and the receptors (15–20). Dephosphorylation of activated Stat proteins is catalyzed by different PTP activities in the nucleus (21–23). We and others have found that cytokine receptor-associated PTP activities not only down-regulate cytokine-induced signals but also suppress the constitutive activities of Jak proteins (21, 24–26). To understand better the role of PTP activity in the negative regulation of receptor-linked constitutive Jak activity, we used the interleukin 4 (IL-4)-activated Jak-Stat signaling pathway as a paradigm.
IL-4 initiates transmembrane signaling through two types of receptors. The type I receptor comprises two subunits, IL-4Rα (the high-affinity binding chain for IL-4) and the γ common-chain (γc), which functions as an accessory signal transducer molecule and is shared by other cytokines, including IL-2, -7, -9, and -15 (2–6, 27–29). IL-4Rα is also used by IL-13 for signaling, providing the molecular basis for the overlapping actions of these two cytokines (27–29). Furthermore, IL-4Rα in association with IL-13Rα, a low-affinity binding chain for IL-13, is known to function as the type II receptor for IL-4 (28, 29). A homodimer of IL-4Rα has also been proposed to function as the type II receptor for IL-4 (28, 30, 31). Jak1 is found associated with IL-4Rα, whereas γc is coupled to Jak3 (3–5, 27–29, 32, 33). The cytoplasmic domain of IL-4Rα contains docking sites for Stat6, which can be activated by both IL-4 and IL-13 (13, 14). Initially, it was thought that Stat6 was unique to the IL-4/IL-13 system. However, recently leptin, platelet-derived growth factor, and anti-IgM-induced DNA-binding complexes in specific cell types have been shown to contain Stat6 (34–36). The targeted disruption of the Stat6 gene in mice shows that it is required for the IL-4-dependent activation of the CD23, major histocompatibility complex II, Iɛ, and IL-4Rα genes (37–39). An insulin receptor substrate-docking site is also located in the cytoplasmic domain of IL-4Rα (40), and stimulation of growth by IL-4 involves the activation of insulin receptor substrate proteins and probably their downstream partners, including GRB2, the p85 subunit of PI3Kinase, and SHP-2 (27, 28, 41, 42).
To examine the requirement for specific Jaks in the ligand-independent activation of different Stat proteins, we used mutant cell lines lacking individual Jaks. We show that the activation of Stat6, but not Stat1 or Stat3, through the inhibition of PTP requires Jak1 activity. This Jak1-dependent activation of Stat6 by PTP inhibition also requires IL-4Rα. Furthermore, we demonstrate that inhibition of PTP activity can bypass the receptor dimerization step, leading to kinase activation and signal transduction. These data show that PTP activity functions to down-regulate receptor-linked constitutive kinase activity.
MATERIALS AND METHODS
Cells, Reagents, Electrophoretic Mobility-Shift Assay (EMSA), Immunoprecipitation, and Immunoblot Analysis.
Daudi cell lines, 2fTGH, 2C4, and the mutant cell lines derived from them, were grown as described previously (21). T98G, GRE, M007, COS-7, and L929 cells were grown in DMEM containing 10% fetal calf serum and 50 units/ml of both penicillin and streptomycin. Pervanadate (PV) solution (20 mM) was prepared as described (21).
The duplex oligonucleotide probes are as follows: (a) acute phase responsive element, 5′-TCGACCTTCCCGGAATTC-3′ (43); (b) N6-GAS, 5′-GATCGCTCTTCTTCCCAGGAACTCAATG-3′ (13, 14), were used in EMSA as described (21). Human IL-4 was obtained from Schering-Plough. Murine IL-4 and murine IL-13 were purchased from R & D Systems. Polyclonal antibodies to Stat6 and Jaks were purchased from Santa Cruz Biotechnology; anti-phosphotyrosine monoclonal antibodies 4G10 and P-Y20 were purchased from Upstate Biotechnology (Lake Placid, NY) and Signal Transduction Laboratories (Lexington, KY), respectively. Immunoprecipitation and Western blot analyses using anti-phosphotyrosine antibody were performed as described (44).
Cell Transfections.
Expression plasmids containing the complete coding sequence of the human IL-4Rα chain (pKCR-4α) (30, 45) or truncated IL-4Rα containing the N-terminal 404 amino acids (lacking the Stat 6 docking sites) in the same vector were cotransfected with a neomycin marker plasmid. Stable clones, isolated by G-418 selection, were examined for expression of transgenes by RT–PCR. Transient transfections of COS-7 and L-929 cells with an IL-4-responsive luciferase construct were performed by using lipofectamine (21, 46). Twelve hours after transfection, the cells were trypsinized and grown in duplicate plates for a further 12 hr and then treated with PV for the indicated time period or left untreated. Luciferase activity was measured as described previously (21).
RESULTS
The Activation of Stats Mediated by PTP Inhibition Requires Jaks.
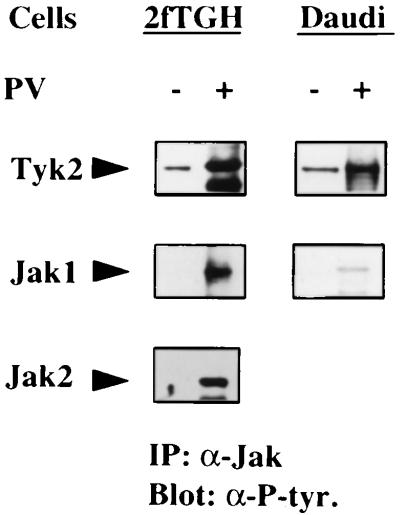
In resting cells, the phosphotyrosine content is maintained at levels of less than 1% of the total of phosphoamino acids through the constitutive activities of a large number of PTPs (47). Inhibition of constitutive PTP activities results in an increase in phosphotyrosine content of many PTKs, which in turn phosphorylate their cellular substrates. To determine if tyrosine-phosphorylated Jaks can be detected in cells exposed briefly to PV, a potent inhibitor of PTP, the human fibrosarcoma cell line 2fTGH and the human lymphoblastoid cell line Daudi were treated with PV and the immunoprecipitated Jak1, Jak2, and Tyk2 proteins were analyzed in Western blot transfers by using anti-phosphotyrosine antibodies (Fig. 1). Each Jak exhibited an increase in phosphotyrosine content in both cell lines, with the exception of Jak2, which is not expressed in Daudi cells (ref. 21; unpublished observations).
Figure 1.
PV treatment causes tyrosine phosphorylation of Tyk2, Jak1, and Jak2. 2fTGH and Daudi cells were treated with 200 μM of PV for 20 min or left untreated and lysates were immunoprecipitated with the appropriate anti-Jak antibody. The protein complexes were resolved in a SDS/7.5% polyacryamide gel and immunoblotted with a mixture of anti-phosphotyrosine monoclonal antibodies.
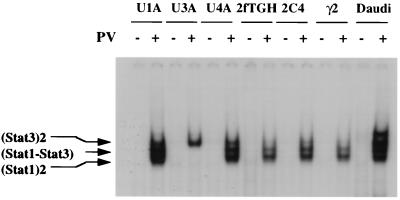
Different Stat proteins are phosphorylated by the activated Jaks through the engagement of various cytokine receptors on the cell surface (1–8, 10, 12–15). The treatment of cells with PTP inhibitors also results in the activation of different Stat proteins in a ligand-independent manner (21, 24–26). When 2fTGH, 2C4, or mutant cell lines derived from them are exposed to PV, Stat1 and Stat3 become activated and bind to the cognate GAS elements (ref. 43; Fig. 2). The Tyk2-minus cell line U1A (48), the Jak1-minus cell line U4A (49), the Jak2-minus cell lines γ2 (50), and Daudi all support PV-mediated activation of both Stat1 and Stat3 (ref 43; Fig. 2). Clearly, the lack of any single Jak does not prevent the activation of Stat1 and Stat3 in PV-treated cells.
Figure 2.
Activation of Stat1 and Stat3 by PV treatment of cells. EMSA was performed using an acute phase responsive element probe and whole cell extract (WCE) prepared from U1A (lacking Tyk2), U3A (lacking Stat1), U4A (lacking Jak1), 2fTGH (parental control for U1A, U3A and U4A), γ2 (lacking Jak2), 2C4 (parental control for γ2), or Daudi cells treated with 200 μM of PV for 30 min or left untreated. The DNA–protein complexes were resolved in a 6% native polyacryamide gel. The U3A cell extract was used as a positive control for Stat3 activation by PV.
Stats 1, 3, and 5 are activated by a number of cytokines through cognate receptors that associate with Jaks (1–7). In contrast, the activation of Stats 2, 4, and 6 has been shown to be more ligand specific (1, 3, 5, 8, 29). In most cell types that express IL-4Rα and Jak1, Stat6 is activated by IL-4 and IL-13 (27–29). Stat6, which binds specifically to the N6-GAS element (13, 14, 46), is activated in the absence of any added cytokine in PV-treated cells (Fig. 3A). Like ligand-dependent Stat6 activation, PV-mediated activation of Stat6 does not require Tyk2 or Jak2 activity (data not shown). However, PV treatment of the mutant Daudi cell line Daudi-100K (unpublished observations), which does not express functional Jak1 protein, fails to activate Stat6 protein (Fig. 3B). The requirement of Jak1 activity for the PV-mediated and ligand-dependent activation of Stat6 was confirmed in another Jak1-mutant cell line U4A (49). U4A cells fail to support either ligand-dependent or PV-mediated phosphorylation and activation of Stat6. However, when U4A cells are complemented with functional Jak1, but not kinase-inactive Jak1, activation of Stat6 by PV or IL-4 is restored (Fig. 3C). These results indicate that Jak1 activity is necessary for the activation of Stat6 in IL-4- or PV-treated cells.
Figure 3.
(A) Activation of Stat6 in PV-treated Daudi cells. The cells were treated with 200 μM of PV for 30 min, 20 ng/ml of human IL-4 for 30 min, or were left untreated. WCE was prepared. An N6-GAS oligonucleotide, end-labeled with 32P, was used for EMSA and an unlabeled N6-GAS probe was used at a 50-fold molar excess in the competition assay (lane 4), showing the specificity of the DNA-protein complex. A supershift was performed with anti-Stat6 antibody (lane 3) to verify the presence of Stat6 in the DNA-protein complex. (B) Stat6 activation by PV requires Jak1 activity. Wild-type and Jak1 mutant Daudi-100K cells were treated with 200 μM of PV for 30 min or left untreated; WCEs prepared were used in EMSA with the N6-GAS probe. (C) Restoration of Stat6 activation by PV or IL-4 in complemented Jak1 mutant cells. U4A cells lacking functional Jak1 activity and U4A cells complemented with either a functional Jak1 or a kinase-inactive Jak1 were treated with 200 μM of PV or 20 ng/ml of human IL-4 for 30 min or left untreated. WCE and the N6-GAS probe were used for EMSA.
IL-4Rα Is Required for Stat6 Activation by PTP Inhibition.
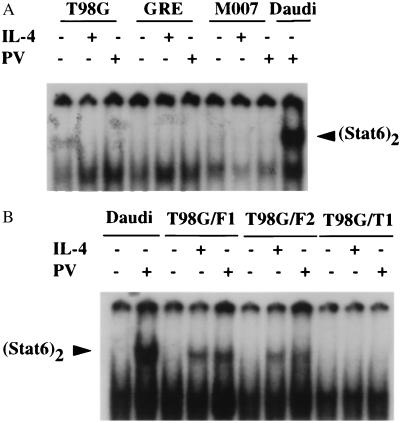
Stat1 overexpressed in Cos cells is constitutively phosphorylated, possibly independent of receptor activation (51). However, ligand-dependent activation of the Jak-Stat pathway is mediated through cytokine receptors, where the specificity of the pathway relies on specific protein–protein interactions (1–8, 12–15). To determine if PV activation of the Jak–Stat pathway depends on cytokine receptor activation, we examined the IL-4Rα chain, which contains docking sites for both Jak1 and Stat6. The human glioblastoma-derived cell lines T98G, GRE, and M007, which do not express detectable levels of IL-4Rα, express comparable levels of Stat6 and IRS-1 (data not shown). These cells respond to both IFN-α and IFN-γ, indicating that they express functional Jak1 protein (52) and they support PV-mediated activation of Stat1 and Stat3 (data not shown). However, when treated with IL-4, they do not activate Stat6 or support PV-mediated Stat6 phosphorylation (Fig. 4A). When T98G cells are stably transfected with full-length human IL-4Rα, both PV-mediated and IL-4-dependent activation of Stat6 occurs, whereas cells expressing a truncated IL-4 Rα, which retains the Jak1 binding site but lacks the two Stat6 binding sites, tyrosine 576 and tyrosine 606 (13, 14), fail to support either IL-4-dependent or PV-mediated Stat6 activation (Fig. 4B). These data indicate that Stat6 activation in PV-treated cells is mediated through an interaction with the IL-4Rα chain.
Figure 4.
(A) Lack of Stat6 activation in human glioblastoma cell lines. T98G, GRE, and M007 cells were treated with human IL-4 (50 ng/ml) or PV (500 μM) for 30 min or left untreated. WCE prepared from these cells were subjected to EMSA with the N6-GAS probe. WCE from PV-treated Daudi cells was used as positive control for activated Stat6. (B) Stat6 activation by PV or IL-4 in T98G cells expressing full-length human IL-4Rα protein. Two representative clones of T98G cells that stably express the full-length IL-4Rα (T98G/F1 and T98G/F2) and one stable clone (T98G/T1) expressing a truncated IL-4Rα (the N-terminal 404 amino acids lacking the Stat6 docking sites), all under the simian virus 40 promoter, were treated with human IL-4 (20 ng/ml) or PV (200 μM) for 30 min or left untreated. WCE prepared from these cells were used in EMSA with an N6-GAS probe.
Receptor Dimerization-Independent Activation of the IL-4 Signaling Pathway via PTP Inhibition.
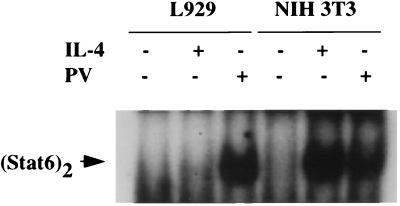
PV-mediated activation of cytokine signaling results from irreversible inhibition of PTP activities associated with cytokine receptors (53). Since simultaneous treatment of cells with the strong kinase inhibitor staurosporine blocks PV-mediated as well as cytokine-dependent activation of the Jak–Stat pathway (data not shown), inhibition of PTP results in constitutive activation of Jaks (Fig. 1). In the presence of constitutive PTP activity, cytokine-dependent signaling requires receptor aggregation, leading to enhanced activation of receptor-linked kinases. Accordingly, dimerization of IL-4Rα and γc is required for IL-4 signaling through the type I receptor (27–29). To determine if heterodimer formation is necessary for PV-mediated Jak1 activation and the subsequent activation of Stat6, we tested murine L929 cells, which do not express the γc chain and are not responsive to IL-4 through its type I receptor (54). A robust activation of Stat6 was observed with PV (Fig. 5). Thus, heterodimerization of the IL-4 type I receptor is not required for PV-mediated activation of Stat6.
Figure 5.
Receptor dimerization-independent activation of Stat6 in PV-treated L929 cells. L929 cells (lacking the γc chain) and NIH 3T3 cells (positive control) were treated with murine IL-4 (20 ng/ml) or PV (200 μM) for 30 min or left untreated. WCE derived from these cells were subjected to EMSA using the N6-GAS probe.
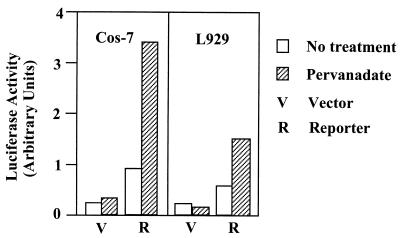
To determine if PV-mediated Stat6 phosphorylation leads to the transcriptional activation of IL-4 responsive genes, we transfected L929 or COS-7 cells, which express the IL-4Rα chain (data not shown), with a reporter construct encoding the luciferase gene driven by a minimal thymidine kinase promoter linked to three copies of a Stat6-binding GAS element derived from the human IL-1rα gene (46). Short-term treatment of the transfected cells with PV results in a 2.5- to 3.5-fold induction of luciferase activity, indicating that PV-induced signaling is functional at the transcription level (Fig. 6). Taken together, these results indicate that inhibition of receptor-associated PTP activities can lead to dimerization-independent activation of the IL-4 signaling pathway.
Figure 6.
Transcriptional activation of a heterologous promoter-linked N6-GAS element in PV-treated COS-7 and L929 cells. Cells were transfected with a reporter construct encoding luciferase under control of a minimal thymidine kinase (tk) promoter linked to 3 copies of the N6-GAS element (45). Twenty-four hours after transfection, the cells were split into two plates and grown for 20–24 hr before treatment with 50 μM of PV. L929 cells and COS-7 cells were similarly treated for 2 hr and 4 hr, respectively.
DISCUSSION
The cytokine-dependent activation of the Jak–Stat signal transduction pathway results in the phosphorylation of Jaks, cytokine receptors, and Stat proteins on tyrosine residues. We have previously proposed that the dephosphorylation of these proteins is catalyzed by at least two classes of PTP activities, PTP-X and PTP-Y (21). PTP-X is postulated to dephosphorylate activated Jaks and cytokine receptors, whereas PTP-Y dephosphorylates activated Stat proteins, possibly in the nucleus (21, 23). Here, we have addressed the role of PTPs in modulating the constant exposure of cells to tyrosine kinase-derived signal inputs. Negative regulation of constitutive signal inputs is of prime importance in executing cellular functions in an orderly manner. Protein phosphorylation is reversible. In mammalian cells, 1000 genes are estimated to encode PTKs and 500 to encode PTPs (55). PTPs have 10 to 1000 times higher specific activity than PTKs (56), a differential that probably prevails intracellularly. Therefore, an increase in kinase activity is required to drive tyrosine phosphorylation in a forward direction to generate and transmit a cytokine signal.
An alignment of the amino acids of known kinase domains reveals that all tyrosine kinases and many serine/threonine kinases have an arginine residue immediately preceding the catalytic aspartic acid, hence the term “RD” kinases (11). Many RD kinases, including the Jaks, are activated by phosphorylation of the conserved tyrosine residue located in their activation segments (Fig. 7). Structural features suggest that RD kinases require charge neutralization of a cluster of basic amino acids, including this arginine, by the phosphotyrosine of the activation segment (11). The structures of the insulin receptor kinase (IRK) and fibroblast growth factor receptor 1 kinase (FGFR1K) reveal that, despite having quite different conformations of the activation loops, both kinase domains are auto-inhibited and require trans-phosphorylation to stimulate kinase activity. This process is mediated by receptor aggregation (11, 57, 58), normally through ligand binding. However, patho-physiological conditions may also cause ligand-independent receptor aggregation, resulting in the induction of constitutive signals. Examples include mutations in the extracellular or transmembrane domains, gene amplification, or gene rearrangements of receptor tyrosine kinases (10).
Figure 7.
Activation segments of three classes of RD (arginine aspartic acid) kinases. Insulin receptor kinase (IRK), fibroblast growth factor receptor 1 (FGFR1), platelet-derived growth factor receptor-β (PDGFRβ), and colony-stimulating factor 1 receptor (CSF1R) represent the receptor tyrosine kinases. Janus kinases (Jaks) and cyclic-AMP-dependent protein kinase (cAPK) represent non-receptor tyrosine kinases and serine/threonine kinase respectively. These kinases are activated by autophosphorylation of conserved tyrosine/threonine residues (underlined) (11). This segment is located between the highly conserved triplets DFG of kinase subdomain VII and APE of kinase subdomain VIII. The activation segment of IRK which causes autoinhibition has five extra amino acids and adopts a different conformation of the segment in cAPK (11, 58).
PV, which directly inhibits cellular PTP activities by an irreversible oxidation of the catalytic cysteine residue of the PTP catalytic domain, causes an increase in the tyrosine phosphorylation levels of many cellular proteins (21, 24–26, 53). We found that PV treatment of cells results in the activation of Jaks and subsequent activation of Stats (Figs. 1 and 3). IL-4- or IL-13-dependent phosphorylation of Stat6 is also catalyzed by Jak1 and is mediated through the IL-4Rα (27–29). Surprisingly, we found that ligand-independent PV activation of Stat6 also requires the IL-4Rα chain (Fig. 4). In mice, intraperitoneal administration of PV causes tyrosine phosphorylation and activation of the receptors for epidermal growth factor, insulin, and hepatocyte growth factor in liver and kidney (26). Possibly the insulin-mimetic effect of vanadium oxides and peroxides also results from receptor activation (53). With the exception of members of the insulin receptor family, which are heterotetramers in the absence of ligand, other receptor tyrosine kinases and kinase-linked cytokine receptors exist as monomers in resting cells (9, 10). PV-mediated activation of tyrosine kinase-linked receptors and subsequent activation of the pathways leading to transcriptional activation of the cognate genes suggest that PTP inhibition causes dimerization-independent receptor activation (Fig. 5). This is shown in L929 cells in which IL-4Rα is activated in the absence of its dimerization partner γc (ref. 54; Fig. 5). Thus, in the absence of an associated PTP activity, the basal activity of receptor linked-kinases seems to be sufficient to catalyze the receptor activation reaction and subsequent signal transduction (Fig. 6).
Auto-inhibition is one mechanism though which kinase activity is regulated in cells (11, 57, 58). Receptors whose kinase domains are always in close proximity may require stronger auto-inhibition than other types. Indeed, IRK exhibits stronger auto-inhibition than does FGFR1K (58). Our results suggest that the constitutive PTP activities associated with cytokine receptor complexes provide a second level of negative regulation of receptor-linked kinase activities. A further understanding of the role of PTPs in the regulation of transmembrane signaling through tyrosine kinase-linked receptors requires the identification of the PTPs that constitutively down-regulate the endogenous signal input. The IL-4 signaling pathway should serve as a paradigm for identifying specific PTPs through biochemical and genetic approaches.
Acknowledgments
We thank Drs. James Ihle and Warren Leonard for cell lines, Drs. Yoshihiro Ohmori and Thomas Hamilton for the luciferase reporter construct, and the Schering–Plough Research Institute for the recombinant human IL-4. This work was supported by Grant No. PO1-CA62220 from the National Cancer Institute of the National Institutes of Health. W.K. and K.F. acknowledge support by the Deutsche Forschungsgemeinschaft through Sonderforschungsbereich 176.
ABBREVIATIONS
- PTP
protein tyrosine phosphatase
- PV
pervanadate
- IL
interleukin
- PTK
protein tyrosine kinase
- WCE
whole cell extract
References
- 1.Darnell J E, Jr, Kerr I M, Stark G R. Science. 1994;264:1415–1421. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- 2.Kishimoto T, Taga T, Akira S. Cell. 1994;76:253–262. doi: 10.1016/0092-8674(94)90333-6. [DOI] [PubMed] [Google Scholar]
- 3.Schindler C, Darnell J E., Jr Annu Rev Biochem. 1995;64:621–651. doi: 10.1146/annurev.bi.64.070195.003201. [DOI] [PubMed] [Google Scholar]
- 4.Taniguchi T. Science. 1995;268:251–255. doi: 10.1126/science.7716517. [DOI] [PubMed] [Google Scholar]
- 5.Ihle J N, Witthuhn B A, Quelle F W, Yamamoto K, Silvennoinen O. Annu Rev Immunol. 1995;13:369–398. doi: 10.1146/annurev.iy.13.040195.002101. [DOI] [PubMed] [Google Scholar]
- 6.Ihle J N. Nature (London) 1995;377:591–594. doi: 10.1038/377591a0. [DOI] [PubMed] [Google Scholar]
- 7.Ihle J N. Cell. 1996;84:331–334. doi: 10.1016/s0092-8674(00)81277-5. [DOI] [PubMed] [Google Scholar]
- 8.Leaman D W, Leung S, Li X, Stark G R. FASEB J. 1996;10:1578–1588. [PubMed] [Google Scholar]
- 9.Lemon M A, Schlessinger J. Trends Biol Sci. 1994;19:459–463. doi: 10.1016/0968-0004(94)90130-9. [DOI] [PubMed] [Google Scholar]
- 10.Heldin C-H. Cell. 1995;80:213–223. doi: 10.1016/0092-8674(95)90404-2. [DOI] [PubMed] [Google Scholar]
- 11.Johnson L N, Noble M E, Owen D J. Cell. 1996;85:149–158. doi: 10.1016/s0092-8674(00)81092-2. [DOI] [PubMed] [Google Scholar]
- 12.Greenlund A C, Farrar M A, Viviano B L, Screiber R D. EMBO J. 1994;13:1591–1600. doi: 10.1002/j.1460-2075.1994.tb06422.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hou J, Schindler U, Henzel W J, Ho T C, Brasseur M, McKnight S L. Science. 1994;265:1701–1706. doi: 10.1126/science.8085155. [DOI] [PubMed] [Google Scholar]
- 14.Schindler U, Wu P, Rothe M, Brasseur M, McKnight S L. Immunity. 1995;2:689–697. doi: 10.1016/1074-7613(95)90013-6. [DOI] [PubMed] [Google Scholar]
- 15.Stahl N, Farruggella T J, Boulton T G, Zhong D, Darnell J E, Jr, Yancopoulos G D. Science. 1995;267:1349–1353. doi: 10.1126/science.7871433. [DOI] [PubMed] [Google Scholar]
- 16.Yi T, Ihle J N. Mol Cell Biol. 1993;13:3350–3358. doi: 10.1128/mcb.13.6.3350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yi T, Mui A A-F, Krystal G, Ihle J N. Mol Cell Biol. 1993;13:7577–7586. doi: 10.1128/mcb.13.12.7577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kingmüller U, Lorenz U, Cantley L C, Neel B G, Lodish H. Cell. 1995;80:729–738. doi: 10.1016/0092-8674(95)90351-8. [DOI] [PubMed] [Google Scholar]
- 19.David M, Chen H E, Geolz S, Larner A C, Neel B G. Mol Cell Biol. 1995;15:7050–7058. doi: 10.1128/mcb.15.12.7050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Petricoin E, III, David M, Igarashi K, Benjamin C, Ling L, Goelz S, Finbloom D S, Larner A C. Mol Cell Biol. 1996;16:1419–1424. doi: 10.1128/mcb.16.4.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haque S J, Flati V, Deb A, Williams B R G. J Biol Chem. 1995;270:25709–25714. doi: 10.1074/jbc.270.43.25709. [DOI] [PubMed] [Google Scholar]
- 22.Shuai K, Liao J, Song M M. Mol Cell Biol. 1996;16:4932–4941. doi: 10.1128/mcb.16.9.4932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haspei R L, Salditt-Georgieff M, Darnell J E., Jr EMBO J. 1996;15:6262–6268. [PMC free article] [PubMed] [Google Scholar]
- 24.Igarashi K, David M, Larner A C, Finbloom D S. Mol Cell Biol. 1993;13:3984–3989. doi: 10.1128/mcb.13.7.3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Igarashi K, Garotta G, Ozmen L, Ziemiecki A, Wilks A F, Harpur A G, Larner A C, Finbloom D S. J Biol Chem. 1994;269:14333–14336. [PubMed] [Google Scholar]
- 26.Ruff S J, Chen C, Cohen S. J Biol Chem. 1997;272:1263–1267. doi: 10.1074/jbc.272.2.1263. [DOI] [PubMed] [Google Scholar]
- 27.Lin J-X, Migone T-S, Tsang M, Friemann M, Weatherbee J A, Zhou L, Yamauchi A, Bloom E T, Mietz J, John S, Leonard W J. Immunity. 1995;2:331–339. doi: 10.1016/1074-7613(95)90141-8. [DOI] [PubMed] [Google Scholar]
- 28.Keegan A D, Ryan J J, Paul W E. Immunologist. 1996;4:194–198. [Google Scholar]
- 29.Callard R E, Matthews D J, Hibbert L. Immunol Today. 1996;17:108–110. doi: 10.1016/0167-5699(96)80600-1. [DOI] [PubMed] [Google Scholar]
- 30.Kammer W, Lischke A, Moriggl R, Groner B, Ziemiecki A, Gurniak C B, Berg L J, Friedrich K. J Biol Chem. 1996;271:23634–23637. doi: 10.1074/jbc.271.39.23634. [DOI] [PubMed] [Google Scholar]
- 31.Lai S Y, Molden J, Liu K D, Puck J M, White M D, Goldsmith M A. EMBO J. 1996;15:4506–4514. [PMC free article] [PubMed] [Google Scholar]
- 32.Yin T, Tsang M S-L, Yang Y-C. J Biol Chem. 1994;269:26614–26617. [PubMed] [Google Scholar]
- 33.Miyajaki T, Kawahara A, Fujii H, Nakagawa Y, Minami Y, Lui Z-J, Ioshi I, Silvennoinen O, Whitthuhn B A, Ihle J N. Science. 1994;266:1045–1047. doi: 10.1126/science.7973659. [DOI] [PubMed] [Google Scholar]
- 34.Ghilardi N, Ziegler S, Wiestner A, Stoffel R, Heim M H, Skoda R C. Proc Natl Acad Sci USA. 1996;93:6231–6235. doi: 10.1073/pnas.93.13.6231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Patel B K R, Wang L-M, Lee C-C, Taylor W G, Pierce J H, LaRochelle W J. J Biol Chem. 1996;271:22175–22182. doi: 10.1074/jbc.271.36.22175. [DOI] [PubMed] [Google Scholar]
- 36.Karras J G, Wang Z, Coniglio S J, Frank D A, Rothstein T L. J Immunol. 1996;157:39–47. [PubMed] [Google Scholar]
- 37.Kaplan M H, Schindler U, Smiley S T, Grusby M J. Immunity. 1996;4:313–319. doi: 10.1016/s1074-7613(00)80439-2. [DOI] [PubMed] [Google Scholar]
- 38.Takeda K, Tanaka T, Shi W, Matsumoto M, Minami M, Kashiwamura S-I, Nakanishi K, Yoshida N, Kishimoto T, Akira S. Nature (London) 1996;380:627–629. doi: 10.1038/380627a0. [DOI] [PubMed] [Google Scholar]
- 39.Shimoda K, Deursen J, Sangster M Y, Sarawar S R, Carson R T, Tripp R A, Chu C, Quelle F W, Nosaka T, Vignali D A A, Doherty P C, Grosveld G, Paul W E, Ihle J N. Nature (London) 1996;380:630–633. doi: 10.1038/380630a0. [DOI] [PubMed] [Google Scholar]
- 40.Keegan A D, Nelms K, White M, Wang L-M, Pierce J H, Paul W E. Cell. 1994;76:811–820. doi: 10.1016/0092-8674(94)90356-5. [DOI] [PubMed] [Google Scholar]
- 41.Sun X-J, Wang L-M, Zhang Y, Yanush L, Myers M G, Glasheen E, Lane W S, Pierce J H, White M F. Nature (London) 1995;377:173–177. doi: 10.1038/377173a0. [DOI] [PubMed] [Google Scholar]
- 42.White M F, Kahn C R. J Biol Chem. 1994;269:1–4. [PubMed] [Google Scholar]
- 43.Akira S, Nishio Y, Inoue M, Wang X-J, Wei S, Matsusaka T, Yoshida K, Sudo T, Naruto M, Kishimoto T. Cell. 1994;77:63–71. doi: 10.1016/0092-8674(94)90235-6. [DOI] [PubMed] [Google Scholar]
- 44.Flati V, Haque S J, Williams B R G. EMBO J. 1996;15:1566–1571. [PMC free article] [PubMed] [Google Scholar]
- 45.Lischke A, Kammer W, Friedrich K. Eur J Biochem. 1995;234:100–107. doi: 10.1111/j.1432-1033.1995.100_c.x. [DOI] [PubMed] [Google Scholar]
- 46.Ohmori Y, Smith M F, Jr, Hamilton T A. J Immunol. 1996;157:2058–2065. [PubMed] [Google Scholar]
- 47.Hunter T, Sefton B M. Proc Natl Acad Sci USA. 1980;77:1311–1317. doi: 10.1073/pnas.77.3.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Velazquez L, Fellous M, Stark G R, Pellegrini S. Cell. 1992;70:313–322. [PubMed] [Google Scholar]
- 49.Muller M, Briscoe J, Laxton C, Guschin D, Ziemiecki A, Silvennoinen O, Harpur A G, Barbieri G, Witthuhn B A, Schindler C, Pellegrini S, Wilks A F, Ihle J N, Stark G R, Kerr I M. Nature (London) 1993;366:129–135. doi: 10.1038/366129a0. [DOI] [PubMed] [Google Scholar]
- 50.Watling D, Guschin D, Muller M, Silvennoinen O, Witthuhn B A, Quelle F W, Rogers N C, Schindler C, Stark G R, Ihle J N, Kerr I M. Nature (London) 1993;366:166–170. doi: 10.1038/366166a0. [DOI] [PubMed] [Google Scholar]
- 51.Heim M H, Kerr I M, Stark G R, Darnell J E., Jr Science. 1995;267:1347–1349. doi: 10.1126/science.7871432. [DOI] [PubMed] [Google Scholar]
- 52.Hannigan G, Williams B R G. EMBO J. 1986;5:1607–1613. doi: 10.1002/j.1460-2075.1986.tb04403.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huyer G, Liu S, Kelly J, Moffat J, Payette P, Kennedy B, Tsaprailis G, Gresser M J, Ramachandran C. J Biol Chem. 1997;272:843–851. doi: 10.1074/jbc.272.2.843. [DOI] [PubMed] [Google Scholar]
- 54.Russell S M, Keegan A D, Harada N, Nakamura Y, Noguchi M, Leland P, Friedmann M C, Miyajima A, Puri R K, Paul W E, Leonard W J. Science. 1993;262:1880–1883. doi: 10.1126/science.8266078. [DOI] [PubMed] [Google Scholar]
- 55.Hunter T. Cell. 1995;80:225–236. doi: 10.1016/0092-8674(95)90405-0. [DOI] [PubMed] [Google Scholar]
- 56.Fischer E H, Charbonneau H, Tonks N K. Science. 1991;253:401–406. doi: 10.1126/science.1650499. [DOI] [PubMed] [Google Scholar]
- 57.Mahammadi M, Schlessinger J, Hubbard S R. Cell. 1996;86:577–587. doi: 10.1016/s0092-8674(00)80131-2. [DOI] [PubMed] [Google Scholar]
- 58.Hubbard S R, Wei L, Ellis L, Hendrickson W A. Nature (London) 1994;372:746–753. doi: 10.1038/372746a0. [DOI] [PubMed] [Google Scholar]