Abstract
A simple in vitro system that supports chromatin assembly was developed for Saccharomyces cerevisiae. The assembly reaction is ATP-dependent, uses soluble histones and assembly factors, and generates physiologically spaced nucleosomes. We analyze the pathway of histone recruitment into nucleosomes, using this system in combination with genetic methods for the manipulation of yeast. This analysis supports the model of sequential recruitment of H3/H4 tetramers and H2A/H2B dimers into nucleosomes. Using a similar approach, we show that DNA ligase I can play an important role in template repair during assembly. These studies demonstrate the utility of this system for the combined biochemical and genetic analysis of chromatin assembly in yeast.
Keywords: nucleosome assembly, histone, Saccharomyces cerevisiae, DNA repair
The nuclear DNA of eukaryotes is assembled with histones into chromatin, of which the basic building block is the nucleosome (reviewed in refs. 1 and 2). The assembly of chromatin has been intensively studied in a variety of biological systems, using many sophisticated experimental approaches. Studies in Saccharomyces cerevisiae have exploited powerful genetic methods and a broad spectrum of biochemical techniques. However one important approach developed in metazoans, the analysis of chromatin assembly in simple cell extracts, has not been available in yeast. In the present report we describe a system for yeast that supports this approach. This system uses a whole-cell extract previously shown to be competent for transcription by all three nuclear RNA polymerases (3, 4). The assembly reaction occurs under physiological salt conditions, is ATP-dependent, and uses soluble histones and chromatin assembly factors.
Studies of complex biochemical processes such as replication and transcription have benefited from the advantages of combined biochemical and genetic analysis in yeast. We apply this approach to the study of chromatin assembly using the in vitro system described here. Strains expressing selected histones from an inducible promoter (5, 6) are used to characterize the pathway of histone recruitment during chromatin assembly in yeast, and a temperature-sensitive strain with a mutated DNA ligase I gene (7) is used to analyze template repair during assembly. These studies provide evidence for the expected stepwise recruitment of H3/H4 tetramers and H2A/H2B dimers into nucleosomes, and they establish that DNA ligase can function to repair DNA in the context of the chromatin assembly reaction. Our work shows that in combination with available genetic techniques, the yeast in vitro assembly system should provide novel opportunities for analyzing the mechanism and regulation of chromatin assembly.
MATERIALS AND METHODS
Extract Preparation.
S. cerevisiae was grown in 1% yeast extract/2% peptone/2% dextrose (8), usually at 30°C, and harvested at densities of OD600 = 0.5–10. Extract was prepared in the cold room as previously described (3), except for the method of grinding (below) and the omission of chymostatin from the mixture of proteinase inhibitors. Small-scale extracts. One to 3 g of cells is processed in an unmodified home coffee mill (any brand) using dry ice as the coolant. The mill is chilled by covering the blades with nuggets of dry ice and running it until the dry ice is a powder (the lid should not be clamped so tightly as to prevent venting of gas). Enough dry ice powder should be present to just cover both blades. The desired weight of frozen cell “noodles” (yeast paste extruded from a syringe into liquid nitrogen) is added and the mill is then run until the mixture is reduced to a frozen powder (usually 5 min: timing depends on the amount of dry ice and brand of mill). The frozen powder is quickly transferred to a cold beaker; further processing is as previously described (3), although we note that thawing the frozen powder of broken cells at room temperature is not deleterious to the activity of the extract. Large-scale extracts. Five to 50 g of frozen cells are broken efficiently in a motorized mortar and pestle (Retsch mortar grinder, model RMO, hardened chrome steel mortar and pestle; Brinkmann Instruments) cooled with liquid nitrogen. The motor of the grinder is run at room temperature for 5 min, then the mortar and pestle (cooled overnight to −20°C) are mounted and further cooled by adding liquid nitrogen to the mortar while it is spinning. Frozen cells and more liquid nitrogen are then added with the machine running. Grinding is continued until the noodles have been pulverized (5–15 min), with slow addition of liquid nitrogen about every 2 min. The torsion knob for the pestle is adjusted during grinding to ensure that it rotates throughout the run. The mortar is inverted over a cold beaker to collect the frozen powder. Further processing is as described (3). Both methods routinely yield extracts with 15–35 mg of protein per ml. Regarding general maintenance, coffee mills and the steel mortar and pestle should be thoroughly rinsed with distilled water and absolute ethanol after use to minimize rust. Details of all methods are available upon request.
Plasmid Supercoiling Assays.
Nucleosome assembly was monitored by plasmid supercoiling (9) in reactions performed at 30°C in 20 μl final volume. Stock solutions are added as follows: 10× chromatin assembly buffer (75 mM MgCl2/10 mM dithiothreitol/0.5 mM EDTA), 2 μl; ATP (60 mM), 1 μl; creatine kinase (2 μg/ml), 1 μl; creatine phosphate (400 mM), 1 μl; and template (10 ng/μl), 2 μl. The template is either unlabeled closed circular pBluescript II KS (+) (Stratagene) relaxed with calf DNA topoisomerase I (BRL), or the same plasmid uniquely radiolabeled at the HindIII site (10) and mixed 1:4 with purified, unlabeled plasmid relaxed with topoisomerase I. Extract is added in 13 μl, obtained by adding buffer [YDBI in ref. 3; 20 mM Hepes–KOH, pH 7.9/50 mM KCl/5 mM EGTA/0.05 mM EDTA/2.5 mM dithiothreitol/20% (vol/vol) glycerol] to the appropriate volume of extract. Two procedures are suitable for stopping the reactions and isolating the DNA. For rapid screening purposes, reactions are stopped by vigorous mixing with 10 vol of 0.3 M sodium acetate/0.5% SDS/10 mM EDTA, then products are extracted (phenol/chloroform, chloroform) and precipitated. Alternatively, to ensure uniform recovery of the template, reactions in 20 μl are stopped with 5 μl of 2.5 g/100 ml Sarkosyl/100 mM EDTA, then digested at 37°C with 1 mg/ml proteinase K (30 min). After mixing with 200 μl of 0.3 M sodium acetate/0.5% SDS/10 mM EDTA, protein is extracted and the products are precipitated with 10 μg of Escherichia coli tRNA as carrier. The products were resolved by native agarose gel electrophoresis using 0.8 or 1% agarose and Tris/acetate/EDTA running buffer. Two-dimensional gels were performed with 1.5% agarose and chloroquine at 8 and 24 μg/ml in the first and second dimensions, respectively; the running buffer was TPE (see guidelines in ref. 11).
Micrococcal Nuclease Analysis.
Assembly reaction mixtures of 100 μl received 6 μl of 0.1 M CaCl2 and 4.4 μl of micrococcal nuclease (400 units/ml; Sigma) and were digested at 37°C. The amount of micrococcal nuclease was reduced 10-fold for digestions of naked DNA. At various time intervals 20-μl aliquots were withdrawn and the reactions were stopped by adding 1.5 μl of 0.5 M EDTA, and the samples were processed according to the Sarkosyl method of Becker and Wu (12). Specifically, each sample received 5 μl of 2.5 g/100 ml Sarkosyl/100 mM EDTA, and was then digested at 37°C with 200 μg/ml RNase A (15 min) followed by 1 mg/ml proteinase K (30 min). This was followed by routine extraction and precipitation. Carrier RNA was included in control reactions performed without extract. The products were resolved by electrophoresis in 1.5% agarose, with Tris/borate/EDTA or Tris/glycine running buffer. Reaction products were detected by Southern blotting using GeneScreenPlus (NEN) and 32P-labeled pBluescript II KS probe prepared by random-primed synthesis (oligonucleotides were from the DNA Core Facility, Biochemistry Department, University of Alberta). The 123-bp ladder (BRL) used as marker was labeled with 32P according to the supplier’s recommendations. We confirmed the comigration of oligonucleosome fragments generated by micrococcal nuclease digestion of in vitro-assembled chromatin with oligonucleosome fragments from the contaminating cellular chromatin by stripping blots and reprobing with a fragment of the yeast rDNA repeat (not shown).
Depletion of Cellular Chromatin and RNA from the Extract.
Frozen dialyzed extract was thawed, made 2.5 mM MgCl2, and spun in 50-μg aliquots for 10 min at 60,000 × g (4°C; Beckman F2402 rotor) to pellet contaminating chromatin. To digest RNA, 50 μg of extract was treated with 0.5 unit of RNase cross-linked to acrylic beads (Sigma R-7005, resuspended at 0.25 units/μl of YDBI) for 30 min at 37°C (13). The supernatant was recovered after spinning for 3 sec to pellet the beads.
DNA Ligase and Histone Depletion Experiments.
Strains A364A (a ade1 ade2 ura1 his7 lys2 tyr1 gal1 CDC9) and STX435–1-3B (α ade1 ade2 lys2 ura1 his7 leu1 gal1 cdc9–1) (14, 15) were used to analyze the function of DNA ligase I during chromatin assembly. Cultures were grown at 25°C to OD600 = 3 as above, then shifted to the restrictive temperature (37°C) for 3 hr and harvested. Strains MHY102 (5) and UKY403 (6), respectively for H2B and H4 depletion, were grown at 30°C in 1% yeast extract/2% peptone/2% galactose to OD600 = 3, washed twice in water, then resuspended in the original volume of 1% yeast extract/2% peptone/2% dextrose. MHY102 was grown for a further 4 hr and UKY403 was grown for a further 2 hr, at which point microscopic examination revealed the expected cell cycle arrest phenotypes (5, 6). Cells were then harvested according to the standard protocol. Histone depletion was confirmed indirectly by micrococcal nuclease digestion analysis of cellular chromatin from cells used to prepare extracts (not shown).
Histone Purification.
Extract (2.0 ml) was mixed on ice with 8.0 ml of 1 M HCl, then spun for 10 min at 4,200 × g. Perchloric acid (1.1 ml of 70%) was added to the supernatant and the resultant precipitate was collected by 10 min of centrifugation at 4,200 × g. The pellet was washed twice with acetone, dried, and resuspended in 1.5 ml of YDBI. Insoluble material was removed (5 min at full speed in a microcentrifuge) prior to concentrating the supernatant in a Centricon 10 (Amicon). Histones purified from the extract (85–95% pure) migrated in SDS/polyacrylamide gels close to the positions of the corresponding calf thymus histones (Boehringer Mannheim) and the nominal H4 protein in our preparations declined in abundance when the H4 depletion strain (above) was cultured under repressing conditions (not shown). Histones recovered from the extract exist in multiple states of post-translational modification, as determined by Triton/acid/urea gel electrophoresis (not shown).
RESULTS AND DISCUSSION
General Properties of the Assembly Extract.
Extracts were prepared from frozen yeast cells broken open by a simple technique that preserves specific transcription by all nuclear RNA polymerases (3). We have improved this method by adapting motorized grinders for breaking frozen cells, either a kitchen coffee/spice mill or a Retsch mortar grinder. Thawing and extraction of the broken cell powder is followed by centrifugation to obtain a 100,000 × g supernatant (S100) that is dialyzed and then used as the assembly extract. The S100 obtained by all grinding methods is competent for chromatin assembly and for transcription by RNA polymerases I, II, and III (not shown). It contains most cellular proteins as well as a variable amount of nucleic acid, including mature ribosomal RNAs, tRNAs, and double-stranded DNA (up to 3.5 μg of DNA per ml of S100). Analysis by micrococcal nuclease digestion revealed that a significant amount of the DNA is in the form of chromatin (not shown), which can be easily removed if necessary (see below).
Characterization of the Assembly Reaction.
Nucleosome deposition onto duplex covalently closed circular DNA molecules, in the presence of topoisomerase activity, results in the appearance of one negative supercoil per nucleosome after removal of proteins from the DNA. Therefore, the level of negative supercoiling of the deproteinized template is a measure of the number of nucleosomes originally assembled onto the template (9).
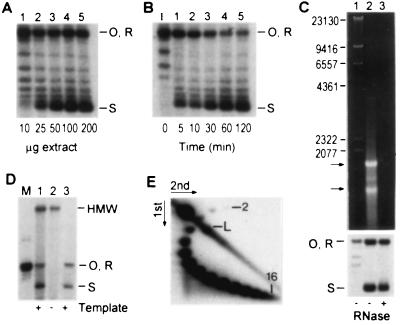
Using supercoiling as an assay, standard conditions for assembly were established on the basis of our experience with over 400 independent extracts, made from 10 standard lab strains and 120 uncharacterized temperature-sensitive mutants derived from strain A364A (16). From this survey it was clear that it is not advantageous to use protease-deficient strains. The reaction has a protein optimum of 100 μg with 20 ng of template (Fig. 1A), a capacity similar to crude chromatin assembly extracts from mammalian somatic cells (17, 18). At the temperature optimum of 30°C (not shown) the reaction is usually complete in 60 min (Fig. 1B); in more active preparations supercoiling with 50 μg of extract is complete in 5–10 min, consistent with the time required for nucleosome assembly in vivo (19–22). Potassium chloride inhibits the reaction (30–50 mM KCl is optimal; not shown), whereas ATP is essential for high levels of supercoiling (see below). The reaction is relatively insensitive to MgCl2 concentration (not shown). While RNA alone can facilitate nucleosome deposition (23), the supercoiling capacity of yeast S100 is only marginally affected when contaminating RNA is completely digested with RNase (Fig. 1C). Depleting the extract of cellular chromatin did not significantly affect the efficiency of assembly onto exogenous DNA (Fig. 1D), and, on the basis of comparisons of otherwise similar extracts, there was no correlation between the activity of an extract and its content of cellular chromatin (not shown). Since supercoiling is not significantly impaired by removal of RNA or DNA, we conclude that the extract contains soluble protein factors that are responsible for supercoiling. Under standard conditions in a typical extract, the level of supercoiling determined by two-dimensional agarose gel electrophoresis corresponds to the deposition of up to 15 nucleosomes per 2,961-bp plasmid (Fig. 1E). This density of packing of physiologically spaced nucleosomes is close to the theoretical maximum of 17 that could be accommodated by this plasmid.
Figure 1.
Characterization of the chromatin assembly reaction in yeast whole-cell extract by DNA supercoiling assays. Open circular (O), relaxed (R), and highly supercoiled (S) products are resolved by agarose gel electrophoresis. (A) Protein titration. Reactions were performed at 30°C for 30 min. (B) Time course of supercoiling with 50 μg of yeast whole-cell extract. The gradual disappearance of open circular/relaxed DNA species is accompanied by the progressive accumulation of highly supercoiled products. (C) RNA in the extract does not play a role in de novo assembly. S100 was treated with RNase linked to acrylic beads to digest the RNA (Upper). Untreated (lane 2) and treated (lane 3) extracts have similar capacities to assemble the template (Lower). Arrows mark the positions of the most prominent RNA species. The size (bp) of DNA markers (lane 1) is indicated on the left. (D) Cellular chromatin in the extract does not play a role in de novo assembly. S100 (lane 1) contains high molecular weight (HMW) DNA that can be removed by low-speed centrifugation (lane 2, DNA pellet) without significantly affecting assembly (lane 3). The template (relaxed pBluescript) was labeled with [32P]dCTP by repair synthesis during assembly. (E) Analysis of template linking number by two-dimensional agarose gel electrophoresis. Up to 16 topoisomers can be resolved (numbered). The faintest spot (number one) was visible only upon prolonged exposure. The intense spot in the upper left corner is open circular template.
Three experiments were performed to demonstrate that the supercoiling reaction is due to chromatin assembly. We first determined that the deproteinized template DNA is efficiently untwisted by E. coli DNA topoisomerase I (not shown), which relaxes negative but not positive supercoils. Therefore the template DNA was positively supercoiled prior to protein removal, as expected when supercoiling is driven by chromatin assembly.
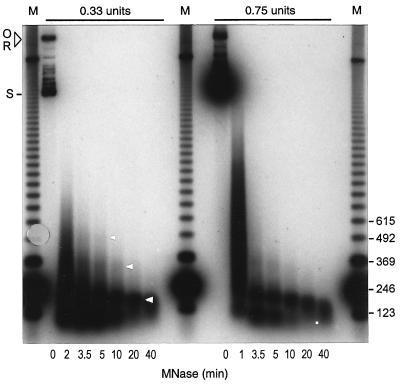
We performed micrococcal nuclease digestion experiments as the second step in demonstrating that supercoiling in whole S100 (Fig. 2), and in S100 depleted of contaminating chromatin (not shown), is due to nucleosome assembly. In both cases nuclease digestion generated a product of approximately 165 bp that resisted prolonged enzyme treatment. Since this size of product corresponds to the DNA repeat protected by a yeast nucleosome in vivo (1), we conclude that the in vitro system contains all the components necessary for the assembly of a correctly organized and stable yeast nucleosome under physiological conditions. Products smaller than the monosome are also generated by micrococcal nuclease digestion. These products are less stable than the monosome, having largely disappeared after 20 min of digestion, when the repeat-length fragment is prominent.
Figure 2.
Characterization of the chromatin assembly reaction in yeast whole-cell extract by micrococcal nuclease digestion. Reactions were treated with micrococcal nuclease (0.33 or 0.75 unit/20-μl aliquot) for the indicated times and the reaction products were detected by Southern blotting. This representative experiment shows a prominent mononucleosome, a disome, and a faint trinucleosome (arrowheads). The dot indicates the position of a sub-nucleosomal fragment (1- to 10-min time points) that is less stable than the monosome. Nucleosome-sized products were never observed in digestions performed without added yeast extract (Fig. 3D Left). Open circular (O), relaxed (R), and highly supercoiled (S) products are indicated. The DNA marker (M) is a 123-bp ladder.
Depending upon the extract used, we observe, in addition to a monosome-sized product, bands that migrate at the positions expected for di-, tri-, and tetranucleosomes assembled in vivo (a disome and faint trisome are evident in Fig. 2). Therefore physiological spacing of nucleosomes is supported in the extract. It is noteworthy, however, that we typically obtain a background smear of digestion products between bands corresponding to the nucleosomes in higher-order arrays (Fig. 2). Consequently, except in some rare extracts, it is difficult to detect tetrasomes and higher-order arrays. The failure to detect extensive arrays persists in reactions supplemented with partially purified yeast histones (not shown). These properties are in contrast to the results obtained with extracts from metazoan cells, in which extensive nucleosome arrays can be generated (2).
Since a monosome is readily detected in our assays, a defect in assembly itself is unlikely to account for the failure to observe extended nucleosomal arrays in the yeast system. The poor resolution of oligonucleosomes may reflect the presence of templates which at the time of digestion were not fully packed with nucleosomes. On the other hand, the digestion pattern we observe is remarkably reminiscent of the pattern obtained when nucleosomes are assembled on linear DNA in a Xenopus extract (24). In the frog system, this pattern was attributed to nucleosome instability on linear as compared with closed circular DNA. Extrapolating from the Xenopus work, we speculate that yeast whole-cell extract may contain a nucleosome-destabilizing activity and that nucleosomes become more susceptible to this activity when the DNA is linearized during micrococcal nuclease digestion. Consistent with the presence of a nucleosome-destabilizing factor in the yeast system, we recently detected an ATP-dependent activity in some extracts that causes pre-assembled plasmids to lose supercoiling (M.C.S. and P. Thiara, unpublished work). Since poly(l-glutamate) stabilizes nucleosome arrays in HeLa cell assembly extract apparently by counteracting a destabilizing activity (25), we tested if oligonucleosomes are more readily detected when assembly is performed in the yeast system in the presence of poly(l-glutamate). Under conditions where poly(l-glutamate) on its own cannot assemble nucleosomes in the presence of topoisomerase I and purified histones, this indeed is the case (W.L.G., D.J.H., and M.C.S., unpublished work; ref. 26). These preliminary results indicate that a soluble factor in yeast extract can destabilize nucleosomes, and they lend support to the proposal that chromatin reconstituted in biological systems may be in a highly dynamic state (27). Ongoing efforts to purify and clone the nucleosome destabilizing activity will provide avenues for the detailed analysis of its physiological relevance.
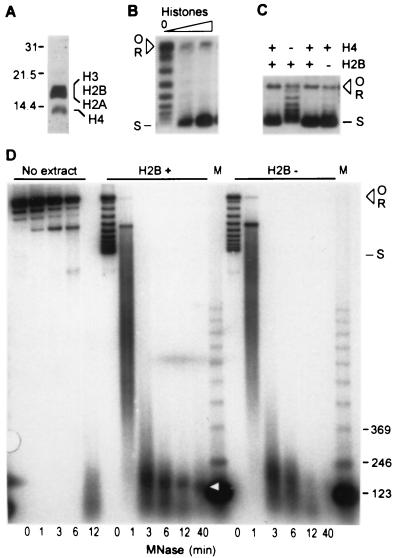
The third experiment we performed to demonstrate that supercoiling is due to chromatin assembly was to show directly that the reaction depends upon soluble histones. For this experiment we assayed supercoiling in limiting amounts of extract supplemented with yeast histones prepared from the assembly extract by acid extraction (Fig. 3A). As shown in Fig. 3B, added histones strongly stimulated DNA supercoiling. Furthermore, assembly in an extract depleted of H4 (see below) was stimulated by exogenous, purified yeast H4 (B.A.A., S. Patel, and M.C.S., unpublished results). We conclude that the assembly reaction in the yeast extract uses soluble histones.
Figure 3.
(A) Analysis of histones purified from yeast chromatin assembly extract. Proteins were resolved by SDS/polyacrylamide gel electrophoresis (15%) and stained with silver (Bio-Rad). The migration of protein markers (kDa) is indicated on the left. (B) Purified yeast histones (A) stimulate plasmid supercoiling in a limiting amount of yeast assembly extract (10 μg). (C) Histone requirements for assembly-driven supercoiling. Extracts were prepared from histone H4 (lanes 1 and 2) and histone H2B (lanes 3 and 4) depletion strains before and after shifting cells to conditions that result in histone depletion. Supercoiling reactions were performed under standard conditions and the products were resolved by native agarose gel electrophoresis. (D) The nucleosomal pattern of micrococcal nuclease (MNase) protection depends upon the presence of H2B. Reactions were performed as in Fig. 2, using extracts from the H2B depletion strain prepared before (H2B+, Center) and after (H2B−, Right) histone depletion. The arrowhead shows the position of the mononucleosome-sized product in H2B+ extract. This product was barely detectable in reactions using H2B− extract and was not observed after digestion of naked template (No extract).
In summary, we have shown that yeast whole cell extract supports a histone-dependent reaction that introduces positive supercoils into plasmid DNA and assembles nucleoprotein complexes that protect DNA from micrococcal nuclease digestion in a pattern corresponding to that generated by yeast nucleosomes assembled in vivo. We conclude that this in vitro reaction corresponds to the de novo assembly of nucleosomes independent of DNA replication. In a series of preliminary experiments using plasmid supercoiling and micrococcal nuclease digestion analysis, we detected assembly during second strand synthesis of single-stranded circular DNA primed with a 17-residue oligonucleotide (D.J.H. and M.C.S., unpublished results). Therefore nucleosome assembly coupled to replication is also supported in this extract. Ongoing experiments are aimed at improving the efficiency of the replication-coupled assembly reaction.
Histone Recruitment into Nucleosomes.
A combined biochemical and genetic approach was used to examine the pathway of histone recruitment into nucleosomes in the extract. Previous work has established that chromatin assembly is a step-wise process involving the association of a tetramer of histone H3/H4 with the DNA followed by the incorporation of H2A/H2B dimers to form the nucleosome (reviewed in refs. 1, 2, 28, and 29). The H3/H4 tetramer itself (but not H2A/H2B dimers) is able to form a nucleosome-like structure whose assembly in the presence of a topoisomerase can be detected as supercoiling of closed-circular DNA (30–32). On the basis of these observations, selective depletion of H3 or H4 in an assembly extract is expected to interfere with supercoiling. Since H3/H4 tetramers can associate with the DNA in the absence of H2A/H2B dimers, supercoiling may not be disrupted when either H2A or H2B is depleted. We tested these predictions using strains in which a histone under control of an inducible promoter can be depleted in vivo by shifting cells to culture conditions that repress the promoter [kindly provided by M. Grunstein (5, 6)]. Assembly was analyzed in extracts prepared from histone depletion strains before and after depletion. Depletion of histone H4 severely impairs supercoiling (Fig. 3C, lanes 1 and 2), indicating that a critical step in supercoiling is driven by a reaction involving association of H4 [likely in the form of an H3/H4 tetramer (1, 28–32)] with the DNA. Supercoiling was only slightly affected by depletion of H2B (Fig. 3C, lanes 3 and 4), as expected when the critical step in supercoiling is the association of H3/H4 with the DNA. When H2B is absent, however, complete nucleosomes should not be formed. Indeed, a stable monosome is not observed upon micrococcal nuclease digestion of DNA assembled in the H2B-depleted extract (Fig. 3D Right). We conclude that in yeast whole-cell extract H3/H4 deposition drives supercoiling and is followed by the recruitment of H2A/H2B to form a complete nucleosome.
Using the strategy outlined above, we have begun a structure–function analysis of the role of the highly conserved amino-terminal tails of the histones in chromatin assembly. This analysis has revealed that assembly in vitro is sensitive to the overall structure of the histone tails (33). Furthermore, when specific lysines in the tails of H3 and H4 that are acetylated in vivo are changed to glycines, assembly is disrupted (X.-J. Ma, B.A.A., M.C.S., and M. Grunstein, unpublished results). This sensitivity is consistent with assembly by an activity that is homologous to the mammalian chromatin assembly complex (CAC), which is preferentially associated with tail-acetylated isoforms of H3 and H4 (34). It follows that Hat2p, a yeast protein that associates in vitro with the histone acetyltransferase Hat1p (35) and is homologous to the p48 subunit of the human CAC (34, 35), may function as a chromatin assembly factor in the yeast extract. Small acidic proteins such as Nap1p (28, 36–39) and high mobility group proteins 1 and 2 (40) are able to assemble nucleosomes, and could also contribute to the reconstitution reaction. However it is unlikely that the yeast reaction is solely dependent upon Nap1p, since supercoiling is not impaired in extracts from a nap1 null mutant (not shown; strain kindly provided by P. Lewis).
ATP Requirement.
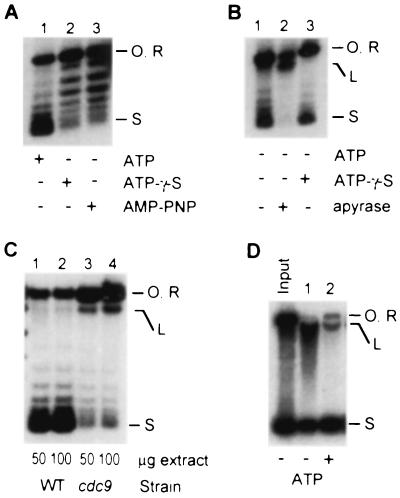
The properties of the assembly reaction in yeast typify physiologically relevant reconstitution reactions in other biological systems (12, 17, 27, 41–48) except that ATP is required for efficient supercoiling in the yeast extract (Fig. 4). γ-Thio-ATP and β,γ-imido-ATP, nonhydrolyzable analogs of ATP, do not support supercoiling (Fig. 4A). Some extracts contain enough endogenous ATP to support supercoiling in the absence of added ATP. Supercoiling in such extracts is strongly inhibited by ATP depletion with native apyrase (Fig. 4B), whereas heat-denatured apyrase (90°C, 10 min) has no effect (not shown). We conclude that ATP is required as a positive cofactor for chromatin assembly, most likely because of its participation in phosphoryl-transfer reactions.
Figure 4.
Role of ATP during assembly-driven supercoiling in yeast. (A) An extract dependent on added ATP for supercoiling (lane 1) was supplemented with adenosine 5′-[γ-thio]triphosphate (ATP-γ-S; lane 2) or adenosine 5′-[β,γ-amido]triphosphate (AMP-PNP; lane 3) instead of ATP. (B) In extracts with enough endogenous ATP to support supercoiling (lane 1), the reaction is not affected by added ATP-γ-S (lane 3) but is strongly inhibited by 3 units of apyrase (lane 2). The reactions in A and B received 100 μg of extract and were stopped after 30 min. (C) DNA ligase I (Cdc9p) functions efficiently in the context of the chromatin assembly machinery. Reactions were performed at 37°C using extracts from wild-type (lane 1) and cdc9 (lane 2) cells. Extracts were heated for 10 min at 37°C before other reaction components were added. The accumulation of nicked template in the cdc9 extract was confirmed by electrophoresis in the presence of ethidium bromide (not shown). (D) Accumulation of full-length linear DNA during assembly in the absence of ATP. The products of a standard assembly reaction are resolved by agarose gel electrophoresis in the presence of 0.25 μg/ml ethidium bromide. Open circular (O), relaxed (R), linear (L), and highly supercoiled (S) products are indicated.
An important role of ATP could be as a cofactor for an enzyme that repairs nicked DNA and therefore preserves the closed-circular configuration of plasmid templates (a prerequisite for detecting supercoiling). One ATP-dependent enzyme that repairs nicks and double-strand breaks is DNA ligase. We therefore tested if Cdc9p, the ATP-dependent DNA ligase I of yeast (7, 49), plays a role in template repair during assembly. Extracts were prepared from a temperature-sensitive cdc9 strain and a wild-type strain (7, 15) and compared for assembly using the plasmid supercoiling assay. Fig. 4C shows that linear and open circular/relaxed reaction products preferentially accumulate in the cdc9 reaction. Therefore, Cdc9p does function in wild-type extract to reseal DNA during assembly. Consistent with this result, we observed that full-length linear DNA accumulates in reactions performed in the absence of ATP (Fig. 4B, lane 2, and 4D). We conclude that the ATP dependence of the supercoiling reaction partly reflects the involvement of DNA ligase in template repair during the chromatin assembly reaction. Interestingly, in reactions performed without ATP, the DNA that remains covalently closed is not efficiently supercoiled (compare the topoisomer distribution in lanes 1 and 2 of Fig. 4A). This observation suggests a requirement for ATP distinct from template repair. This distinct requirement could reflect the involvement of an ATP-dependent topoisomerase in assembly, with the topoisomerase functioning to relieve torsional strain resulting from nucleosome deposition on the closed-circular template. According to this scenario, the most abundant ATP-dependent topoisomerase in yeast, DNA topoisomerase II (50), would be the likely ATP-dependent activity required for chromatin assembly. However, our recent work shows that assembly-driven supercoiling remains ATP-dependent when topoisomerase II activity is eliminated (26). Therefore it is unlikely that assembly is ATP-dependent because of a requirement for topoisomerase II. The function of ATP that is independent of DNA repair and topoisomerase II activity remains under investigation.
Summary and Perspectives.
We have established a simple yeast system that supports the ATP-dependent assembly of physiologically spaced nucleosomes in vitro. This system should be useful for combined biochemical and genetic studies of the molecular mechanism and regulation of genome packaging in eukaryotes. The interaction of the chromatin assembly apparatus with biochemical machineries that use the chromatin substrate will also be amenable to analysis by this approach.
Acknowledgments
M.C.S. thanks Ron Reeder for initial support and encouragement and Michael Grunstein, Susan Rosenberg, and Charlotte Spencer for comments on the manuscript. Many colleagues in the chromatin field made useful technical suggestions. Jean Finnegan is thanked for suggesting that we try coffee mills; Ata Ghavidel kindly helped in developing their use. Histone depletion strains were provided by M. Grunstein, and STX435–1-3 was from the Yeast Genetic Stock Center. J. Wang is thanked for E. coli DNA topoisomerase I. This work was supported by grants from the Alberta Heritage Foundation for Medical Research, the Central Research Fund (University of Alberta), and the Medical Research Council. M.C.S. is a Scholar of the Medical Research Council; T.A.A.H. is a Postdoctoral Fellow of the Natural Sciences and Engineering Research Council; and W.I.G. has been supported by a University of Alberta Ph.D. Scholarship and a Scholarship from the Alberta Heritage Foundation for Medical Research.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.van Holde K E. Chromatin. New York: Springer; 1988. [Google Scholar]
- 2.Wolffe A. Chromatin. Structure and Function. London: Academic; 1992. [Google Scholar]
- 3.Schultz M C, Choe S Y, Reeder R H. Proc Natl Acad Sci USA. 1991;88:1004–1008. doi: 10.1073/pnas.88.3.1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schultz M C, Reeder R H, Hahn S. Cell. 1991;69:697–702. doi: 10.1016/0092-8674(92)90233-3. [DOI] [PubMed] [Google Scholar]
- 5.Han M, Chang M, Kim U-J, Grunstein M. Cell. 1987;48:589–597. doi: 10.1016/0092-8674(87)90237-6. [DOI] [PubMed] [Google Scholar]
- 6.Kim U-J, Han M, Kayne P, Grunstein M. EMBO J. 1988;7:2211–2219. doi: 10.1002/j.1460-2075.1988.tb03060.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnston L H, Nasmyth K A. Nature (London) 1978;274:891–893. doi: 10.1038/274891a0. [DOI] [PubMed] [Google Scholar]
- 8.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current Protocols in Molecular Biology. New York: Wiley; 1995. pp. 13.0.1–13.14.17. [Google Scholar]
- 9.Germond J E, Hirt B, Oudet P, Gross-Bellard M, Chambon P. Proc Natl Acad Sci USA. 1975;72:1843–1847. doi: 10.1073/pnas.72.5.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Razvi F, Gargiulo G, Worcel A. Gene. 1983;23:175–183. doi: 10.1016/0378-1119(83)90049-5. [DOI] [PubMed] [Google Scholar]
- 11.Pruss G J. J Mol Biol. 1985;185:51–63. doi: 10.1016/0022-2836(85)90182-2. [DOI] [PubMed] [Google Scholar]
- 12.Becker P B, Wu C. Mol Cell Biol. 1992;12:2241–2249. doi: 10.1128/mcb.12.5.2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Halmar L, Gruss C. Nucleic Acids Res. 1995;23:773–778. doi: 10.1093/nar/23.5.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Culotti J, Hartwell L H. Exp Cell Res. 1971;67:389–401. doi: 10.1016/0014-4827(71)90424-1. [DOI] [PubMed] [Google Scholar]
- 15.Hartwell L H, Mortimer R K, Culotti J, Culotti M. Genetics. 1973;74:267–286. doi: 10.1093/genetics/74.2.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hartwell L H. J Bacteriol. 1967;93:1662–1670. doi: 10.1128/jb.93.5.1662-1670.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith S, Stillman B. Cell. 1988;58:15–25. doi: 10.1016/0092-8674(89)90398-x. [DOI] [PubMed] [Google Scholar]
- 18.Gruss C, Gutierrez C, Burhans W C, DePamphilis M L, Koller T, Sogo J M. EMBO J. 1990;9:2911–2922. doi: 10.1002/j.1460-2075.1990.tb07482.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Worcel A, Han S, Wong M L. Cell. 1978;15:969–977. doi: 10.1016/0092-8674(78)90280-5. [DOI] [PubMed] [Google Scholar]
- 20.Ryoji M, Worcel A. Cell. 1984;37:21–32. doi: 10.1016/0092-8674(84)90297-6. [DOI] [PubMed] [Google Scholar]
- 21.Cusick M E, Lee K-S, DePamphilis M L, Wasarman P M. Biochemistry. 1983;22:3873–3884. doi: 10.1021/bi00285a024. [DOI] [PubMed] [Google Scholar]
- 22.Smith P A, Jackson V, Chalkey R. Biochemistry. 1984;23:1576–1581. doi: 10.1021/bi00302a036. [DOI] [PubMed] [Google Scholar]
- 23.Nelson T, Wiegand R, Brutlag D. Biochemistry. 1981;20:2594–2601. doi: 10.1021/bi00512a035. [DOI] [PubMed] [Google Scholar]
- 24.Almouzni G, Méchali M. EMBO J. 1988;7:4355–4365. doi: 10.1002/j.1460-2075.1988.tb03334.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Banerjee S, Cantor C R. Mol Cell Biol. 1990;10:2863–2873. doi: 10.1128/mcb.10.6.2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garinther W I, Schultz M C. Mol Cell Biol. 1997;17:3520–3526. doi: 10.1128/mcb.17.7.3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Varga-Weisz P D, Blank T A, Becker P B. EMBO J. 1995;14:2209–2216. doi: 10.1002/j.1460-2075.1995.tb07215.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaufman P D, Botchan M R. Curr Opin Genet Dev. 1994;4:229–235. doi: 10.1016/s0959-437x(05)80049-8. [DOI] [PubMed] [Google Scholar]
- 29.Krude T. Curr Biol. 1995;5:1232–1234. doi: 10.1016/s0960-9822(95)00245-4. [DOI] [PubMed] [Google Scholar]
- 30.Camerini-Otero R D, Felsenfeld G. Nucleic Acids Res. 1977;4:1159–1181. doi: 10.1093/nar/4.5.1159-a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oudet P, Germond J E, Sures M, Gallwitz D, Bellard M, Chambon P. Cold Spring Harbor Symp Quant Biol. 1977;42:301–312. doi: 10.1101/sqb.1978.042.01.031. [DOI] [PubMed] [Google Scholar]
- 32.Bina-Stein M. J Biol Chem. 1978;253:5213–5219. [PubMed] [Google Scholar]
- 33.Ling X, Harkness T A A, Schultz M C, Fisher-Adams G, Grunstein M. Genes Dev. 1996;10:686–699. doi: 10.1101/gad.10.6.686. [DOI] [PubMed] [Google Scholar]
- 34.Verreault A, Kaufman P D, Kobayashi R, Stillman B. Cell. 1996;87:95–104. doi: 10.1016/s0092-8674(00)81326-4. [DOI] [PubMed] [Google Scholar]
- 35.Parthun M R, Widom J, Gottschling D E. Cell. 1996;87:85–94. doi: 10.1016/s0092-8674(00)81325-2. [DOI] [PubMed] [Google Scholar]
- 36.Fujii-Nakata T, Ishimi Y, Okuda A, Kikuchi A. J Biol Chem. 1992;267:20980–20986. [PubMed] [Google Scholar]
- 37.McQuibban A, Lewis P N. Can Fed Biol Soc Proc. 1994;37:153. (abstr.). [Google Scholar]
- 38.Simon H-U, Mills G B, Kozlowski M, Hogg D, Branch D, Ishimi Y, Siminovitch K A. Biochem J. 1994;297:389–397. doi: 10.1042/bj2970389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ito T, Bulger M, Kobayashi R, Kadonaga J T. Mol Cell Biol. 1996;16:3112–3124. doi: 10.1128/mcb.16.6.3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bonne-Andrea C, Harper F, Sobczak J, DeRecondo M. EMBO J. 1984;3:1193–1199. doi: 10.1002/j.1460-2075.1984.tb01950.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dilworth S M, Black S J, Laskey R A. Cell. 1987;51:1009–1018. doi: 10.1016/0092-8674(87)90587-3. [DOI] [PubMed] [Google Scholar]
- 42.Glikin G C, Ruberti I, Worcel A. Cell. 1984;37:33–41. doi: 10.1016/0092-8674(84)90298-8. [DOI] [PubMed] [Google Scholar]
- 43.Kamakaka R T, Bulger M, Kadonaga J T. Genes Dev. 1993;7:1779–1795. doi: 10.1101/gad.7.9.1779. [DOI] [PubMed] [Google Scholar]
- 44.Kamakaka R T, Kaufman P D, Stillman B, Mitsis P G, Kadonaga J T. Mol Cell Biol. 1994;14:5114–5122. doi: 10.1128/mcb.14.8.5114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kaufman P D, Kobayashi R, Kessler N, Stillman B. Cell. 1995;81:1105–1114. doi: 10.1016/s0092-8674(05)80015-7. [DOI] [PubMed] [Google Scholar]
- 46.Kleinschmidt J A, Fortkamp E, Krohne G, Zentgraf H, Franke W W. J Biol Chem. 1985;260:1166–1176. [PubMed] [Google Scholar]
- 47.Krude T, Knippers R. J Biol Chem. 1993;268:14432–14442. [PubMed] [Google Scholar]
- 48.Laskey R A, Mills A D, Morris N R. Cell. 1977;10:237–243. doi: 10.1016/0092-8674(77)90217-3. [DOI] [PubMed] [Google Scholar]
- 49.Kornberg A, Baker T A. DNA Replication. 2nd Ed. New York: Freeman; 1992. [Google Scholar]
- 50.Wang J C. J Biol Chem. 1991;266:6659–6662. [PubMed] [Google Scholar]