Abstract
The retinoblastoma protein (Rb) plays a critical role in cell proliferation, differentiation, and development. To decipher the mechanism of Rb function at the molecular level, we have systematically characterized a number of Rb-interacting proteins, among which is the clone C5 described here, which encodes a protein of 1,978 amino acids with an estimated molecular mass of 230 kDa. The corresponding gene was assigned to chromosome 14q31, the same region where genetic alterations have been associated with several abnormalities of thyroid hormone response. The protein uses two distinct regions to bind Rb and thyroid hormone receptor (TR), respectively, and thus was named Trip230. Trip230 binds to Rb independently of thyroid hormone while it forms a complex with TR in a thyroid hormone-dependent manner. Ectopic expression of the protein Trip230 in cells, but not a mutant form that does not bind to TR, enhances specifically TR-dependent transcriptional activity. Coexpression of wild-type Rb, but not mutant Rb that fails to bind to Trip230, inhibits such activity. These results not only identify a coactivator molecule that modulates TR activity, but also uncover a role for Rb in a pathway that responds to thyroid hormone.
The retinoblastoma gene is a prototypic tumor suppressor (1, 2). Inactivation of this gene has been found in all retinoblastomas and a variety of tumors examined (3). The retinoblastoma gene encodes a nuclear protein of 110 kDa (4). The protein is phosphorylated at serine and threonine residues in a cell cycle-dependent manner, and the modification of the protein by phosphorylation is believed to provide a way to regulate the protein function (5–7). Recently, evidence has been accumulated to suggest that the retinoblastoma protein (Rb) plays a critical role in cell cycle progression, differentiation, and development (8). For example, introduction of Rb into cells early in G1 inhibits progression into S phase (9–10). Overexpression of human Rb in transgenic mice results in dose-dependent growth retardation at the level of the organism (11). Mouse embryos that express no functional Rb fail to survive embryonic stages and die by gestational day 14.5 with defects in erythroid and neuronal developments (12–14). Moreover, Rb has been shown to be involved in differentiation of several cell types, including myoblasts, monocytes, and adipocytes (15–17).
One approach to understanding the mechanism of Rb function in cell growth and differentiation at the molecular level has been to identify associated partners with which Rb interacts. Rb binds a number of proteins, among which is E2F-1, a transcription factor important for the expression of several genes involved in cell cycle progression from G1 to S (18). Rb inhibits E2F-1 activity by blocking its transactivation region (19–21). In contrast, Rb has been shown to have the ability to increase the transactivating activity of the members of the CCAAT/enhancer binding protein (C/EBP) family, and to be required for C/EBPs-dependent adipocyte and monocytes differentiation (16–17). These findings thus suggest that one mechanism of Rb function is to modulate, either negatively or positively, the activity of specific factors that are critical in cellular proliferation, differentiation, and development (22).
To systematically identify factors whose activities are controlled by Rb through binding, we have screened human cDNA libraries for clones that interact with Rb. Using this approach, two dozen proteins were identified in vitro (19) and in yeast (23). Here we describe the characterization of one Rb-associated protein named Trip230 (for thyroid hormone receptor and Rb interaction with a molecular mass of 230 kDa). Interestingly, Trip230 also has the ability to bind to human thyroid hormone receptor β. The thyroid hormone receptor (TR) is a thyroid hormone (T3)-modulated transcription factor with profound effects on growth, development, and homeostasis (24–28). In the presence of T3, Trip230 can enhance the corresponding transcription activity of TR. Rb, through binding to Trip230, inhibits the transactivating activity of TR. The present study therefore not only identifies a regulatory molecule, Trip230, that modulates TR activity, but also uncovers a role of Rb in the thyroid hormone responsive pathway.
MATERIALS AND METHODS
Cloning and Sequence Analysis of Trip230 cDNA.
The 0.9-kb cDNA fragment originally isolated from the yeast two-hybrid screen (23) was labeled with [32P]dATP and used as a probe to screen a human fibroblast cDNA library (generously provided by M.-L. Chu at Thomas Jefferson University, Philadelphia, PA). Several overlapping clones were isolated and cloned into the pBluescript plasmid from the Lambda Zap II vector (Stratagene). The longest clone was sequenced from both directions, and the other clones were partially sequenced to confirm that they are derived from the same transcription unit. DNA sequencing was performed by the dideoxynucleotide termination method, and the sequences were analyzed by the computer program dnastar (Madison, WI).
Chromosomal Localization of the Gene Encoding Trip230.
The originally isolated 0.9-kb Trip230 cDNA fragment was labeled with [32P]dATP and used as a probe in Southern blot analysis of a number of mouse-human and Chinese hamster-human cell hybrids (obtained from the National Institute of General Medical Sciences, Camden, NJ) to localize human chromosomes. The same cDNA fragment also was labeled with [3H]dATP and used as a probe for in situ hybridization. Hybridization to human metaphase chromosomes, posthybridization wash, and emulsion autoradiography were carried out as described previously (29). The chromosomes then were G-banded using Wright’s stain for silver grains on chromosome 14. No other sites were stained above background.
Northern Blot Analysis.
Poly(A)+ RNA was prepared as described (19). The isolated RNA was separated by the glyoxal/dimethyl sulfoxide method and transferred to a piece of nitrocellulose membrane and hybridized with the 0.9-kb cDNA fragment of Trip230 labeled with [32P]dATP. The probe was detected by autoradiography.
In Vitro Transcription and Translation.
35S-methionine- labeled proteins were synthesized using a TNT kit according to the manufacturer’s instructions (Promega).
Immunoprecipitation.
Lysates prepared from 1 × 107 cells were lysed in cold lysis 250 buffer (50 mM Tris⋅HCl, pH 7.4/250 mM NaCl/5 mM EDTA/0.1% Nonidet P-40/50 mM NaF/1 mM phenylmethylsulfonyl fluoride/1 μg/ml leupeptin/1 μg/ml antipain) and centrifuged for 5 min at 16,000 g at 4°C. The supernatant was incubated with the antibody at 4°C for 1 hr before the addition of protein A-Sepharose beads. The beads were collected after another hour of incubation at 4°C. After being washed six times with lysis 250 buffer, the bound antigens then were separated by SDS/PAGE and detected by Western blot analysis or autoradiography. Antibodies specific for Trip230 were produced in mice immunized by subcutaneous injection of glutathione S-transferease (GST)-fusion proteins, GST-C5 (amino acids 1,099–1,372 of Trip230), and GST-C5N (amino acids 78–376 of Trip230). The corresponding antisera were obtained according to standard procedures (4).
In Vitro Binding Assay.
GST-fusion proteins containing different regions of Trip230 were produced in Escherichia coli and purified as described (19). In vitro transcription/translation of TR was performed in the presence of [35S]methionine using the TNT kit according to manufacturer’s instructions (Promega). The assays were carried out at room temperature using glutathione-agarose beads containing 5 μg of GST or GST-fusion proteins in lysis 250 buffer. Bound proteins were washed six times with lysis 150 buffer, separated by SDS/PAGE, and detected by Western blot analysis or autoradiography.
Yeast Two-Hybrid System.
The wild-type N-terminal truncated Rb cDNA and a series of mutant Rb constructs containing a deletion or a point mutation were fused in-frame with the Gal4 transactivating domain in the yeast expression vector pAS1. A fragment of Trip230 corresponding to amino acids 1,099–1,382 was fused with the Gal4 DNA-binding domain in another vector pGAD10, a yeast expression vector derived from pSE1107. Yeast strain Y153 was cotransformed with the above vectors as described (23). Cotransformants were analyzed for β-galactosidase activity by the filter lift method and quantitated by chlorophenyl-red β-d-galactopyranoside (CPRG) assays as described (23).
Transient Transfection Assay.
Expression vectors for Rb, Rb(H209), Trip230, Trip230ΔTR, and Trip230ΔRb were constructed by cloning the corresponding cDNAs downstream of the cytomegalovirus promoter. Expression vectors for TR, retinoic acid receptor (RAR), retinoic X receptor (RXR), progesterone receptor (PR), glucocorticoid receptor (GR), and estrogen receptor (ER) were constructed by cloning the cDNAs of the nuclear receptors downstream of the Rous sarcoma virus promoter (30). Nuclear receptor responsive reporter gene were constructed by cloning the hormone responsive elements, including DR4 (TR), DR2 (RAR), DR5 (RXR), PRE (PR), GRE (GR), ERE (ER), upstream of the thymidine kinase (TK) promoter. The resulting TK promoters then were used to drive the expression of the chloramphenicol acetyltransferase (CAT) reporter gene (30). About 5 × 106 WERI-Rb-27 cells grown in DMEM supplemented with 10% of charcoal-stripped serum for 48 hr were transfected by the calcium phosphate-mediated method as described (16). Plasmids used were: reporter vector (5 μg), nuclear receptor expression vectors (2 μg), Rb and Trip230 expression vectors (1 μg), and β-galactosidase expression vector (1.5 μg). Sixteen hours after transfection, cells were washed twice with PBS and fed with fresh DMEM supplemented with 10% charcoal-stripped serum, either with or without an appropriate nuclear receptor ligand. The concentrations of the ligand used were: 1 μM T3 (TR), 1 μM retinoic acid (RAR), 1 μM 9-cis retinoic acid (RXR), 0.1 μM progesterone (PR), 1 μM dexamethasome (GR), and 0.1 μM 17 β-estadiol (ER). Forty-eight hours after transfection, cells were collected, and lysates were prepared. To monitor transfection efficiency, total DNA of the transfected cells was isolated and amplified in the linear range with PCR using a pair of primers specific for CAT. The primers used were: 5′-CGCCTGATGAATGCTCATCCGG-3′ and 5′-AACTGGTGAAACTCAGGGAGGG-3′. The amplified 265-bp PCR product were separated by agarose gel electrophoresis and subjected to Southern blot analysis using a 32P-dATP-labeled cDNA fragment of the CAT gene as a probe. Alternatively, β-galactosidase activities were used to monitor transfection efficiency. Transcription of the reporter genes was determined by the corresponding CAT activities as assayed by the method described (31).
RESULTS
Molecular Cloning of Trip230 cDNA.
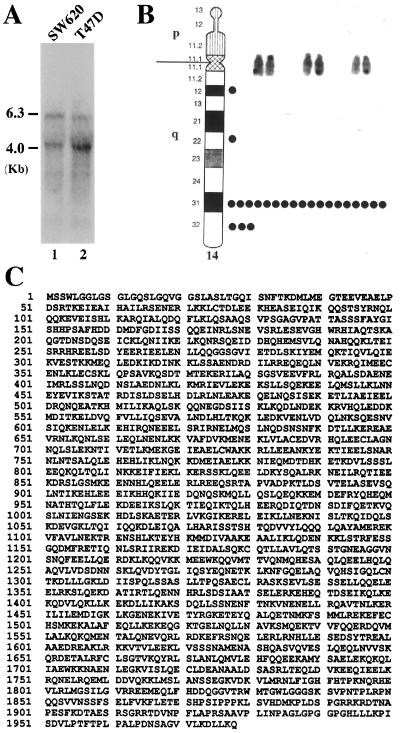
By using the yeast two-hybrid system, we previously have identified several clones that interact specifically with the N-terminal truncated form of Rb, p56RB (23). One of these clones, C5, contains an insert of 0.9 kb. The 0.9-kb fragment of this clone was used as a probe in RNA blot analysis to detect the expression of the corresponding mRNA. Two major transcripts of 6.3 kb and 4 kb were identified in the human cell lines SW620 and T47D (Fig. 1A). To map the chromosomal localization of the corresponding gene, the same probe was used in Southern blot analysis to screen a panel of mouse-human and Chinese hamster-human somatic cell hybrids. Two human PstI fragments (1.25 kb and 0.9 kb) together with a mouse fragment (1.5 kb) and a hamster fragment (1.8 kb) were detected. The two human fragments were present only in hybrids containing human chromosome 14 (data not shown). The 0.9-kb cDNA probe then was used to localize the gene to chromosome 14q31 by in situ hybridization of human metaphase spreads (Fig. 1B). Genetic alterations in the 14q31 region have been found to associate with several abnormalities of thyroid hormone response, including congenital hyperthyroidism, nongoiterous hypothyroidism, Graves disease, and hyperfunctioning thyroid adenoma (32). These results indicated that the protein encoded by this gene may play a role in thyroid hormone (T3) response pathway. To obtain a full-length cDNA, the 0.9-kb probe was used to screen a human fibroblast cDNA library. Several overlapping clones spanning 6.4 kb were isolated. The longest clone contains a single ORF able to encode a protein of 1,978 amino acids with a calculated molecular mass of ≈230 kDa (Fig. 1C). There are multiple stop codons in all three reading frames upstream of the first ATG, indicating that the cDNA clone contains the full-length coding sequence. The putative protein is glutamate-rich (≈13%) and has a pI of ≈5.2. A search of GenBank revealed that the predicted protein does not share significant sequence homology with any known protein except that the very C-terminal region of the protein (amino acids 1,754–1,978) is identical to that of Trip11 (GenBank L40380), a polypeptide fragment interacting with the ligand-binding domain of TR in a T3-dependent manner (33). Together, these results suggest that the isolated cDNA clone encodes a protein, designated as Trip230 (for thyroid hormone receptor and retinoblastoma protein interacting protein with an apparent molecular mass of 230 kDa), which is able to interact with both Rb and TR (see below).
Figure 1.
Cloning of a Rb-interacting protein, Trip230. (A) The corresponding mRNA of Trip230 was detected by using poly(A)+ RNA isolated form human cell lines SW620 (lane 1) and T47D (lane 2) and Northern blot analysis. The 0.9-kb cDNA fragment originally isolated from yeast two-hybrid screen was used as a probe. Two major transcripts of 6.4 kb and 4 kb were identified as indicated. (B) The same 0.9-kb cDNA probe was used to first localize the gene encoding Trip230 to chromosome 14 by Southern blot analysis. Human metaphase spreads then were analyzed by in situ hybridization using the same 0.9-kb cDNA fragment as a probe to map the gene to chromosome 14q31. (C) Amino acid sequence of Trip230 as deduced from the cloned cDNA sequence.
Characterization of Trip230 Protein and its Interaction with Rb and TR.
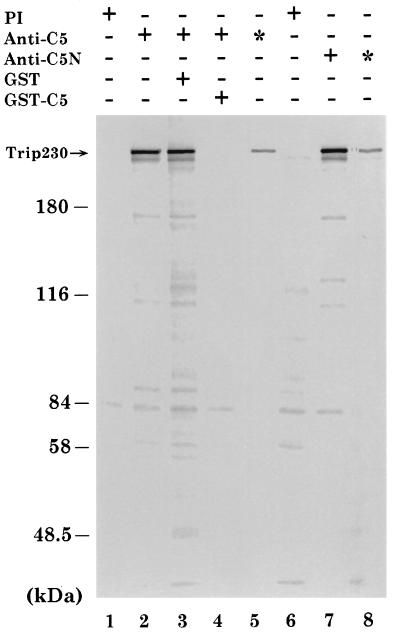
The expression of cellular Trip230 protein was demonstrated by immunoprecipitation experiments using lysates prepared from metabolically labeled T24 cells, a human bladder carcinoma cell line, and two antibodies, anti-C5 and anti-C5N, which were raised against two GST-fusion proteins containing distinct regions of Trip230. As shown in Fig. 2, both antibodies recognized a protein of 230 kDa that was not detected by the preimmune serum (Fig. 2, compare lanes 2 and 7 with lanes 1 and 6). Depletion of the antibodies by pretreatment with the corresponding antigens abolished the specific recognition of the 230-kDa protein (Fig. 2, compare lanes 2 and 4), while incubation with GST alone had no effect (Fig. 2, compare lanes 2 and 3). The specificity of these two antibodies was further confirmed by double immunoprecipitation experiments, which showed the detection of only the 230-kDa protein (Fig. 2, lanes 5 and 8). Similar results were obtained using lysates prepared from WERI-Rb-27 cells, a human retinoblastoma cell line (not shown). These data thus strongly suggest that the detected 230-kDa cellular protein is Trip230. The Trip230-specific antibodies then were used to investigate the tissue distribution pattern of Trip230. Our results indicated that Trip230 is ubiquitously expressed in various amounts in all of the mouse tissues and human cell lines examined (not shown).
Figure 2.
Identification of cellular Trip230 protein by antibodies. Lysates prepared from T24 cells metabolically labeled with [35S]methionine were immunoprecipitated with two antibodies, anti-C5 (lanes 2 to 5) and anti-C5N (lanes 6 to 10), which were raised against two GST-fusion proteins containing amino acids 1,099–1,372 and amino acids 78–376 of Trip230, respectively. These two antibodies recognized a protein of 230 kDa that is not detected by the preimmune serum (PI). Inclusion of the anti-C5 antibody with the GST-C5 fusion protein that contains the corresponding antigen, but not GST alone, resulted in the failure of the antibody to detect the 230-kDa protein, indicating that the antibody is specific. Moreover, results from double immunoprecipitation experiments (∗, lanes 5 and 8) showed the detection of only the 230-kDa protein by both antibodies.
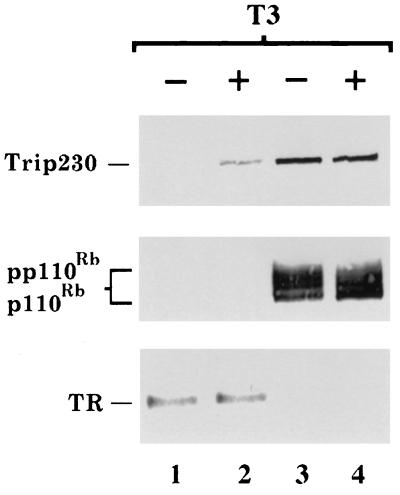
Because Trip230 was independently identified in yeast two-hybrid screen using Rb or TR as a bait, the possibility that it also may have the ability to interact with both proteins in human cells was investigated. The in vivo interactions of Rb and TR with Trip230 were detected by coimmunoprecipitation experiments using lysates prepared from WR2E3 cells, an Rb-reconstituted human retinoblastoma cell line (16). Results from Western blot analysis of such affinity-selected proteins indicated that the formation of Trip230/TR complex only occurs in the presence of T3 (Fig. 3, compare lanes 1 and 2). This result is consistent with the report showing that the interaction between Trip11 and the ligand-binding domain of TR requires T3 (33). In contrast, T3 had no effect on the formation of a complex between Trip230 and Rb (Fig. 3, compare lanes 3 and 4), indicating that the binding of Trip230 to Rb is independent of T3. No Trip230/Rb/TR ternary complex was detected in these experiments, indicating that the binding of Rb and TR to Trip230 may be mutually exclusive. Together, these results suggest that Trip230 is able to specifically interact with Rb and TR in cells.
Figure 3.
In vivo interactions of Trip230 with Rb and TR. Lysates prepared from WR2E3 cells grown in medium without T3 (lanes 1 and 3) or with T3 (lanes 2 and 4) were immunoprecipitated with antibodies specific to TR (lanes 1 and 2) or Rb (lanes 3 and 4). After immunoprecipitation, the antibody/antigen complexes were separated by SDS/PAGE and subjected to Western blot analysis using antibodies specific to Trip230, Rb or TR.
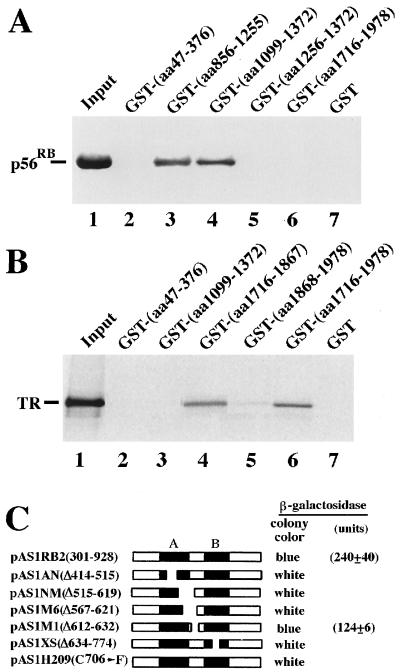
Trip230 Uses Two Distinct Domains to Bind to Rb and TR.
Protein interactions often are mediated by specific domains. To map the regions of Trip230 responsible for its interaction with Rb and TR, a panel of GST-fusion proteins containing different portions of Trip230 were prepared and immobilized on glutathione Sepharose beads. The immobilized proteins were allowed to interact with either purified p56RB or in vitro-translated TR. Results showed that the domains required for the binding of Trip230 to Rb and TR are located at regions containing amino acids 1,099–1,255 (Fig. 4A) and amino acids 1,716–1,867 (Fig. 4B), respectively. Because Trip11 represents a polypeptide containing amino acids 1,754–1,978 of Trip230, the region required for the binding of Trip230 to TR can be further narrowed down to amino acids 1,754–1,867. Taken together, these results suggest that, by using two distinct domains, Trip230 directly interacts with Rb and TR. Next, the yeast two-hybrid assay was carried out to map the domains required for the binding of Rb to Trip230. As shown in Fig. 4C, the wild-type Rb is able to interact with the Rb-binding domain of Trip230. In contrast, Rb with a deletion or a point mutation in the A and B domains, which are required for the binding of the simian virus 40 large T antigen, was unable to interact with Trip230. Accordingly, the binding of Rb to Trip230 depends on the T antigen-binding domain, the same domain required for the interaction of Rb with most of its interacting proteins (8).
Figure 4.
Mapping of interaction domains of Trip230, Rb and TR. (A and B) Two distinct domains mediated the direct binding of Trip230 to Rb and TR. To determine whether Trip230 directly interacts with Rb and TR and which domains are responsible for the interaction, a series of GST-fusion proteins containing different regions of Trip230 were incubated with either purified p56RB (A) or in vitro-translated, 35S-methionine-labeled TR (B). Bound proteins were separated by SDS/PAGE and detected by Western blot analysis (A) or autoradiography (B). (C) The simian virus 40 large T antigen-binding domain mediates the binding of Rb to Trip230. To locate regions required for the binding of Rb to Trip230, the wild-type Rb and a panel of mutant Rb containing a deletion or a point mutation were fused in frame with the Gal4 DNA-binding domain in the expression vector pAS1. The Rb-binding region of Trip230 (amino acids 1,099–1,382) was fused in frame with the Gal4 transactivation domain in another expression vector pGAD10. Yeast cells cotransformed with these vectors as indicated then were assayed for β-galactosidase activity to determine protein interactions.
Trip230 Enhances TR-Regulated Transcription and Rb Represses Trip230-Mediated Activation of TR-Regulated Transcription.
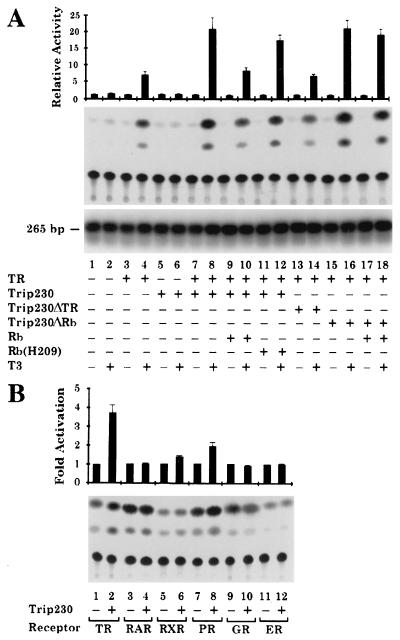
TR belongs to a large superfamily of ligand-activated transcription factors that regulate gene expression by binding to specific DNA elements located in the proximity of the target genes. A number of nuclear receptor-associated proteins have been shown to serve as coactivators in nuclear receptor-mediated transcription (34–45). Because Trip230 specifically interacts with TR in the presence of T3, we therefore investigated whether Trip230 also can function as a coactivator of TR. To address the role of Trip230 in TR-mediated transcription, transient transfection experiments were carried out using a CAT reporter gene driven by a TK promoter that harbors a TR responsive element (30). Results showed that Trip230 alone was unable to stimulate the transcription of the reporter gene either in the absence or the presence of T3 (Fig. 5A, lanes 5 and 6). In contrast, TR activated the transcription of the same reporter gene in the presence of T3 (Fig. 5A, compare lanes 3 and 4). The introduction of exogenous Trip230 by cotransfection further enhanced the TR-mediated transcription by 3-fold (Fig. 5A, compare lanes 4 and 8). The effect of Trip230 on TR-mediated transcription is specific, as a mutant Trip230 that is unable to bind TR had no such effect (Fig. 5A, compare lanes 4 and 14). These results thus indicate that Trip230 is a bona fide coactivator of TR that activates TR-mediated transcription in the presence of T3. The moderate activation by exogenous expression of Trip230 probably is due to a high endogenous level of this protein in the cells. Because a cell line without endogenous Trip230 is yet to be found, the true activating potential of Trip230 on TR-mediated transcription cannot be precisely determined at present.
Figure 5.
The roles of Trip230 and Rb in TR-mediated transcription. (A) Trip230 enhances TR-mediated transcriptional activity, and this enhancement can be abolished by Rb. Trip230, TR, and Rb expression vectors in different combinations as indicated were cotransfected with a CAT reporter gene driven by a TK promoter containing a TR-binding element into WERI-Rb-27 cells grown in the presence or absence of T3. Transfection efficiency was determined by PCR amplification of total DNA isolated from the transfected cells with two primers specific for the CAT gene. The 265-bp amplified fragment was detected by Southern blot analysis. Relative CAT activities then were normalized by the transfection efficiency and plotted as shown. (B) Trip230 is a TR-specific coactivator in WERI-Rb-27 cells. A panel of nuclear receptor-expressing vectors as indicated were transfected with CAT reporter genes driven by a series of TK promoters containing the appropriate nuclear receptor-binding elements. The effect of Trip230 on the nuclear receptor-mediated transcriptional activation was tested by cotransfection of a Trip230 expression vector in the presence of the corresponding hormones. A β-galactosidase expression vector also was included in the transfection experiments to control transfection efficiency. Relative CAT activities were normalized against β-galactosidase activities. To compare the effect of Trip230 on different nuclear receptor-mediated transcription, the reporter activities obtained in the absence of the Trip230 expression vector were set as one as shown.
Rb has been shown to stimulate GR-mediated transcription by direct interaction with the GR coactivator, hBrm (46). To determine whether Rb can regulate TR-mediated transcription through interaction with Trip230, we carried out cotransfection experiments using WERI-Rb-27 cells, which are devoid of endogenous Rb. As shown in Fig. 5A, the enhancement of TR-mediated transcription by Trip230 was abolished by the expression of the wild-type Rb, but not that of Rb(H209), which contains a single point mutation and fails to bind to Trip230 (Fig. 5A, compare lanes 8 and 10, and lanes 8 with 12). Moreover, the effect of Rb was not observed when the wild-type Trip230 was replaced by a mutant form that cannot bind to Rb (Fig. 5A, compare lanes 8 and 18). These data indicate that Rb can down-regulate TR-mediated transcription by direct interaction with Trip230.
To determine whether Trip230 can modulate gene transcription potentiated by other nuclear receptors in WERI-Rb-27 cells, cotransfection experiments were performed using a panel of nuclear receptor expression vectors and reporter genes that are responsive to the corresponding receptors in the presence of the appropriate ligand. As shown in Fig. 5B, Trip230 significantly up-regulated only TR-mediated transcription in the presence of T3, but not other nuclear receptors tested. This result suggests that the stimulation effect of Trip230 is limited to TR-mediated transcription in WERI-Rb-27 cells.
DISCUSSION
The Rb protein controls cell-cycle progression and cellular differentiation by interacting with key regulatory proteins (8). In this report, we describe the cloning and characterization of a Rb-interacting protein Trip230. Significantly, Trip230 also was found to interact with TR in a T3-dependent manner and function as a coactivator of TR-mediated transcription. This latter finding is consistent with a recent report showing that Trip11, a polypeptide corresponding to the amino acid residues 1,754–1,978 of Trip230, was able to interact with rat TRβ in the presence of T3 (33). Furthermore, we showed that, by interacting with Trip230, Rb can down-regulate TR-mediated transcription. Because Rb has been found to potentiate GR-mediated transcription by interacting with hBrm (46), we suggest that Rb may play dual roles in regulating nuclear receptor-mediated transcription, a positive role for GR and a negative role for TR. Interestingly, by using the same T-antigen-binding domain to interact with transcription factors E2F-1 (19) and CCAAT/enhancer binding proteins (16), Rb also has been found to display apparently disparate functions, inhibiting the activity of the former, but activating the activity of the latter.
The opposite influence of GR and TR by Rb may be linked to the different physiological and temporal functions of these two hormone regulatory systems. Thyroid hormone (T3) is normally secreted at a basal level, with little minute-to-minute or hour-to-hour modulation in response to exogenous signals. In contrast, cortisol and other glucocorticoids have a shorter half-life and are modulated more expediently in response to various physiologic conditions. Thus, conceivably, Rb may function to suppress the expression of genes normally activated by thyroid hormone in the basal state, but to enhance the expression of genes that are expressed in response to glucocorticoids. Therefore, in addition to the control of cell cycle progression and cellular differentiation, results obtained from this study suggest that Rb also may play a role in integrating hormonal signals regulating general cellular metabolic pathways. These findings also may help to explain how Rb influences cellular proliferation, differentiation, and development, such as why overexpression of Rb in transgenic mice resulted in dwarfism (11).
To date, a number of nuclear receptor coactivators have been identified (34–45). Results presented here are consistent with the notion that nuclear receptors with their associated factors may form multiprotein complexes to regulate the expression of their target genes. Conceptually, different cell types may have different compositions of the associated factors in such nuclear receptor complexes. Individual factors in a given complex thus may serve as a communicator that integrates distinct physiological signals, as suggested by the finding that the activity of Trip230, a coactivator of TR-mediated transcription, is negatively regulated by Rb, a critical factor for cell cycle progression and cellular differentiation.
How these coactivators activate nuclear receptor-mediated transcription is largely unclear. However, p300/CBP, a coactivator of nuclear receptor-mediated transcription (34), recently was found to possess intrinsic histone acetyltransferase activity (47–48). This finding implies that p300/CBP may contribute directly to transcriptional activation by targeted acetylation of nucleosomes, which would result in the alteration or disruption of the repressive chromatin structure. Because p300/CBP appears to be a global coactivator for many transcriptional activators, whether other specific nuclear receptor coactivators including Trip230, also function in such a way remains to be determined.
Acknowledgments
We thank Diane Jones for the preparation of antibodies against Trip230, Chia-Yu Ku for the construction of GST-fusion protein expression vectors, Dr. Bei Shan for providing the mammalian expression vectors of Rb, and Drs. Andrew A. Farmer and Z. D. Sharp for critical reading of the manuscript. The work was supported by grants from the National Institutes of Health (EY05758 and CA58318 to W.H.L. and HD08188 to B.W.O.) and the Council for Tobacco Research (to W.H.L.).
ABBREVIATIONS
- Rb
retinoblastoma protein
- TR
thyroid hormone receptor
- GST
glutathione S-transferease
- CAT
chloramphenicol acetyltransferase
- GR
glucocorticoid receptor
- TK
thymidine kinase.
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF007217).
References
- 1.Goodrich D W, Lee W H. Biochim Biophys Acta. 1993;1155:43–61. doi: 10.1016/0304-419x(93)90021-4. [DOI] [PubMed] [Google Scholar]
- 2.Riley D J, Lee E Y, Lee W H. Annu Rev Cell Biol. 1994;10:1–29. doi: 10.1146/annurev.cb.10.110194.000245. [DOI] [PubMed] [Google Scholar]
- 3.Bookstein R, Lee W H. Crit Rev Oncog. 1991;2:211–227. [PubMed] [Google Scholar]
- 4.Lee W H, Bookstein R, Hong F, Young L J, Shew J Y, Lee E Y. Science. 1987;235:1394–1399. doi: 10.1126/science.3823889. [DOI] [PubMed] [Google Scholar]
- 5.Buchkovich K, Duffy L A, Harlow E. Cell. 1989;58:1097–1105. doi: 10.1016/0092-8674(89)90508-4. [DOI] [PubMed] [Google Scholar]
- 6.Chen P L, Scully P, Shew J Y, Wang J Y, Lee W H. Cell. 1989;58:1193–1198. doi: 10.1016/0092-8674(89)90517-5. [DOI] [PubMed] [Google Scholar]
- 7.DeCaprio J A, Ludlow J W, Lynch D, Furukawa Y, Griffin J, Piwnica-Worms H, Huang C M, Livingston D M. Cell. 1989;58:1085–1095. doi: 10.1016/0092-8674(89)90507-2. [DOI] [PubMed] [Google Scholar]
- 8.Chen P L, Riley D J, Lee W H. Crit Rev Eukaryotic Gene Expression. 1995;5:79–95. [PubMed] [Google Scholar]
- 9.Goodrich D W, Wang N P, Qian Y W, Lee E Y, Lee W H. Cell. 1991;67:293–302. doi: 10.1016/0092-8674(91)90181-w. [DOI] [PubMed] [Google Scholar]
- 10.Hinds P W, Mittnacht S, Dulic V, Arnold A, Reed S I, Weinberg R A. Cell. 1992;70:993–1006. doi: 10.1016/0092-8674(92)90249-c. [DOI] [PubMed] [Google Scholar]
- 11.Bignon Y J, Chen Y, Chang C Y, Riley D J, Windle J J, Mellon P L, Lee W H. Genes Dev. 1993;7:1654–1662. doi: 10.1101/gad.7.9.1654. [DOI] [PubMed] [Google Scholar]
- 12.Clarke A R, Maandag E R, van Roon M, van der Lugt N M, van der Valk M, Hooper M L, Berns A, Riele H. Nature (London) 1992;359:328–330. doi: 10.1038/359328a0. [DOI] [PubMed] [Google Scholar]
- 13.Jacks T, Fazeli A, Schmitt E M, Bronson R T, Goodell M A, Weinberg R A. Nature (London) 1992;359:295–300. doi: 10.1038/359295a0. [DOI] [PubMed] [Google Scholar]
- 14.Lee E Y, Chang C Y, Hu N, Wang Y C, Lai C C, Herrup K, Lee W H, Bradley A. Nature (London) 1992;359:288–294. doi: 10.1038/359288a0. [DOI] [PubMed] [Google Scholar]
- 15.Gu W, Schneider J W, Condorelli G, Kaushal S, Mahdavi V, Nadal-Ginard B. Cell. 1993;72:309–324. doi: 10.1016/0092-8674(93)90110-c. [DOI] [PubMed] [Google Scholar]
- 16.Chen P L, Riley D J, Chen-Kiang S, Lee W H. Proc Natl Acad Sci USA. 1996;93:465–469. doi: 10.1073/pnas.93.1.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen P L, Riley D J, Chen Y, Lee W H. Genes Dev. 1996;10:2794–2804. doi: 10.1101/gad.10.21.2794. [DOI] [PubMed] [Google Scholar]
- 18.Nevins J R. Science. 1992;258:424–429. doi: 10.1126/science.1411535. [DOI] [PubMed] [Google Scholar]
- 19.Shan B, Zhu X, Chen P L, Durfee T, Yang Y, Sharp D, Lee W H. Mol Cell Biol. 1992;12:5620–5631. doi: 10.1128/mcb.12.12.5620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shirodkar S, Ewen M, DeCaprio J A, Morgan J, Livingston D M, Chittenden T. Cell. 1992;68:157–166. doi: 10.1016/0092-8674(92)90214-w. [DOI] [PubMed] [Google Scholar]
- 21.Helin K, Harlow E, Fattaey A. Mol Cell Biol. 1993;13:6501–6508. doi: 10.1128/mcb.13.10.6501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee W H, Xu Y, Hong F, Durfee T, Mancini M A, Ueng Y C, Chen P L, Riley D. Cold Spring Harbor Symp Quant Biol. 1994;59:97–107. doi: 10.1101/sqb.1994.059.01.013. [DOI] [PubMed] [Google Scholar]
- 23.Durfee T, Becherer K, Chen P L, Yeh S H, Yang Y, Kilburn A E, Lee W H, Elledge S J. Genes Dev. 1993;7:555–569. doi: 10.1101/gad.7.4.555. [DOI] [PubMed] [Google Scholar]
- 24.Kastner P, Mark M, Chambon P. Cell. 1995;83:859–869. doi: 10.1016/0092-8674(95)90202-3. [DOI] [PubMed] [Google Scholar]
- 25.Beato M, Herrlich P, Schutz G. Cell. 1995;83:851–857. doi: 10.1016/0092-8674(95)90201-5. [DOI] [PubMed] [Google Scholar]
- 26.Tsai M J, O’Malley B W. Annu Rev Biochem. 1994;63:451–486. doi: 10.1146/annurev.bi.63.070194.002315. [DOI] [PubMed] [Google Scholar]
- 27.Lazar M A. Endocr Rev. 1993;14:184–193. doi: 10.1210/edrv-14-2-184. [DOI] [PubMed] [Google Scholar]
- 28.Glass C K, Holloway J M. Biochim Biophys Acta. 1990;1032:157–176. doi: 10.1016/0304-419x(90)90002-i. [DOI] [PubMed] [Google Scholar]
- 29.Yang-Feng T L, Floyd-Smith G, Nemer M, Drouin J, Francke U. Am J Hum Genet. 1985;37:1117–1128. [PMC free article] [PubMed] [Google Scholar]
- 30.Leng X, Blanco J, Tsai S Y, Ozato K, O’Malley B W, Tsai M J. J Biol Chem. 1994;269:31436–31442. [PubMed] [Google Scholar]
- 31.Seed B, Sheen J Y. Gene. 1988;67:271–277. doi: 10.1016/0378-1119(88)90403-9. [DOI] [PubMed] [Google Scholar]
- 32.Mckusick V A. Mendelian Inheritance in Man. 11th Ed. Vol. 1. Baltimore: Johns Hopkins Univ. Press; 1994. pp. 2232–2237. [Google Scholar]
- 33.Lee J W, Choi H S, Gyuris J, Brent R, Moore D D. Mol Endocrinol. 1995;9:243–254. doi: 10.1210/mend.9.2.7776974. [DOI] [PubMed] [Google Scholar]
- 34.Chakravarti D, LaMorte V J, Nelson M C, Nakajima T, Schulman I G, Juguilon H, Montminy M, Evans R M. Nature (London) 1996;383:99–103. doi: 10.1038/383099a0. [DOI] [PubMed] [Google Scholar]
- 35.Smith C L, Onate S A, Tsai M J, O’Malley B W. Proc Natl Acad Sci USA. 1996;93:8884–8888. doi: 10.1073/pnas.93.17.8884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fondell J D, Ge H, Roeder R G. Proc Natl Acad Sci USA. 1996;93:8329–8333. doi: 10.1073/pnas.93.16.8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Voegel J J, Heine M J, Zechel C, Chambon P, Gronemeyer H. EMBO J. 1996;15:3667–3675. [PMC free article] [PubMed] [Google Scholar]
- 38.May M, Mengus G, Lavigne A C, Chambon P, Davidson I. EMBO J. 1996;15:3093–3104. [PMC free article] [PubMed] [Google Scholar]
- 39.Sande S, Privalsky M L. Mol Endocrinol. 1994;8:1455–1464. doi: 10.1210/mend.8.11.7877615. [DOI] [PubMed] [Google Scholar]
- 40.Onate S A, Tsai S Y, Tsai M J, O’Malley B W. Science. 1995;270:1354–1357. doi: 10.1126/science.270.5240.1354. [DOI] [PubMed] [Google Scholar]
- 41.Lee J W, Ryan F, Swaffield J C, Johnston S A, Moore D D. Nature (London) 1995;374:91–94. doi: 10.1038/374091a0. [DOI] [PubMed] [Google Scholar]
- 42.Cavailles V, Dauvois S, L’Horset F, Lopez G, Hoare S, Kushner P J, Parker M G. EMBO J. 1995;14:3741–3751. doi: 10.1002/j.1460-2075.1995.tb00044.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cavailles V, Dauvois S, Danielian P S, Parker M G. Proc Natl Acad Sci USA. 1994;91:10009–10013. doi: 10.1073/pnas.91.21.10009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.LeDouarin B, Zechel C, Garnier J M, Lutz Y, Tora L, Pierrat P, Heery D, Gronemeyer H, Chambon P, Losson R. EMBO J. 1995;14:2020–2033. doi: 10.1002/j.1460-2075.1995.tb07194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Halachmi S, Marden E, Martin G, MacKay H, Abbondanza C, Brown M. Science. 1994;264:1455–1458. doi: 10.1126/science.8197458. [DOI] [PubMed] [Google Scholar]
- 46.Singh P, Coe J, Hong W. Nature (London) 1995;374:562–565. doi: 10.1038/374562a0. [DOI] [PubMed] [Google Scholar]
- 47.Ogryzko V V, Schiltz R L, Russanova V, Howard B H, Nakatani Y. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 48.Bannister A J, Kouzarides T. Nature (London) 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]