Abstract
Using clathrin-minus Dictyostelium cells, we identified a novel requirement for clathrin during cytokinesis. In suspension culture, clathrin-minus cells failed to divide and became large and multinucleate. This cytokinesis deficiency was not attributable to a pleiotropic effect on the actomyosin cytoskeleton, since other cellular events driven by myosin II (e.g., cortical contraction and capping of concanavalin A receptors) remained intact in clathrin-minus cells. Examination of cells expressing myosin II tagged with green fluorescent protein showed that clathrin-minus cells failed to assemble myosin II into a functional contractile ring. This inability to localize myosin II to a particular location was specific for cytokinesis, since clathrin-minus cells moving across a substrate localized myosin II properly to their posterior cortexes. These results demonstrate that clathrin is essential for construction of a functional contractile ring during cell division.
Cytokinesis, which follows nuclear division in a coordinated fashion, is the process by which one cell divides into two (1). During late anaphase/early telophase, as condensed chromosomes move toward opposite poles of a cell, a contractile ring assembles in the center of the cell. As the contractile ring pinches the cell in half, two cells are formed. While many of the cellular properties involved in cytokinesis have been studied extensively, central questions remain. Especially lacking is information about how the plasma membrane and associated proteins integrate with the contractile ring to form a cleavage furrow during cytokinesis.
The best characterized proteins of the contractile ring are actin and myosin II. Actin filaments form the scaffold for the contractile ring, whereas myosin II is the motor that drives constriction of the ring. Additional proteins, both membrane-bound and cytosolic, are certainly involved in contractile ring formation and function. Some proteins identified recently include racE, a small GTPase required for cytokinesis in Dictyostelium (2), and septins, identified first in Saccharomyces cerevisiae (3) and then in Drosophila (4). In addition, the actin-binding protein anillin (5, 6) and the formin-like protein diaphanous (7) are required for cytokinesis in Drosophila. In Dictyostelium, the actin-binding protein cortexillin (8) and the cytoskeletal protein coronin (9) both function in cytokinesis. Finally, the actin-binding protein radixin (10) has been localized to the contractile ring in dividing culture cells. Many other molecules may be involved, but their identities as well as potential interactions with the contractile ring and the plasma membrane remain unknown.
Because functional connections between the actomyosin cytoskeleton and membrane remodeling have been proposed (11, 12), we explored the possible requirement of clathrin-mediated membrane traffic during cytokinesis. Clathrin coated vesicles convey membrane traffic between specific cellular compartments (13, 14). Clathrin heavy and light chains assemble into a characteristic polygonal lattice on membranes. This protein coat, initially a planar lattice on the intracellular surface of the plasma membrane, invaginates to bud coated vesicles into the cytoplasm. Subsequently, vesicles shed their clathrin coat and join the endolysosomal system. In a similar fashion, clathrin coated vesicles bud from the trans-Golgi network to join the endolysosomal system. Clathrin is important for the internalization of receptors from the plasma membrane and the trafficking of vacuolar and lysosomal hydrolases from the trans-Golgi network (13, 15, 16). Conceivably, clathrin-mediated membrane traffic could internalize receptors, signals, and plasma membrane required for effective cytokinesis. Another possible role for clathrin-mediated membrane traffic is in the trafficking and processing of proteins needed for cytokinesis. To examine a possible requirement for clathrin during cytokinesis, we studied this process in clathrin-minus Dictyostelium cells (15).
MATERIALS AND METHODS
Growth Curves and 4,6-Diamidino-2-phenylindole (DAPI) Staining.
Clathrin-minus Dictyostelium cell lines have been engineered by gene replacement (15, 17). For control cells, we used a cell line from the original transformation with nonhomologous integration of the plasmid and wild-type levels of clathrin protein, whose doubling time during growth on plates matched clathrin-minus cells. Alternatively, we examined the parental wild-type cell line Ax2 as a control. Cells were seeded at 1 × 105 cells per ml in HL5 medium (17) and grown at 19°C in either a flask shaken at 220 rpm or on a plastic Petri dish. The number of cells was counted daily and plotted versus time. At 72 hr, a sample of each cell line was allowed to attach to a coverslip and fixed in 1% formaldehyde in methanol at −15°C for 5 min. Cells were stained with 0.1 μg/ml DAPI (2). The cells were visualized using a Zeiss Axiophot microscope equipped with a cooled CCD camera.
Azide Contraction.
Cells were allowed to attach to a substrate in HL5 medium. After 10 min, the medium was replaced with HL5 containing 10 mM sodium azide. Cells were photographed both before and 5–10 min after azide treatment.
Capping.
Cells were allowed to attach to a coverslip for 15 min, washed for 5 min in phosphate buffer (PB) (14.6 mM KH2PO4/2 mM Na2HPO4, pH 6.1), pulsed for 2 min with 0.5 mg/ml fluorescein isothiocyanate–concanavalin A (Con A) (Molecular Probes) in PB, and incubated at room temperature for 5–15 min, before fixing for 5 min at −20°C in 1% formaldehyde in methanol (2).
Myosin Localization.
Clathrin-minus or control cells were transformed as described (15) with the green fluorescent protein (gfp)–myosin heavy chain construct generously supplied by Sheri Moores and Jim Spudich (18). Transformants were selected for growth in 10 μg/ml G418 (GIBCO/BRL). Cells were grown to confluency on a Petri dish, diluted into fresh HL5 medium, and allowed to attach to a coverslip. At 5-min intervals, cells were fixed for 1 min in 3.7% formaldehyde and 0.05% Triton X-100 in PBS, then for 1 hr in 3.7% formaldehyde in PBS. The cells were washed twice in PBS, incubated for 10 min in 50 mM NH4Cl and 0.1 mg/ml DAPI in PBS, washed twice in PBS, and mounted on a slide in 50% glycerol in PBS containing 100 mg/ml diazabicylo[2.2.2]octane (Sigma). For immunofluorescence microscopy, we followed published methods (2) using anti-myosin polyclonal antiserum provided by Arturo De Lozanne (Duke University Medical Center).
RESULTS AND DISCUSSION
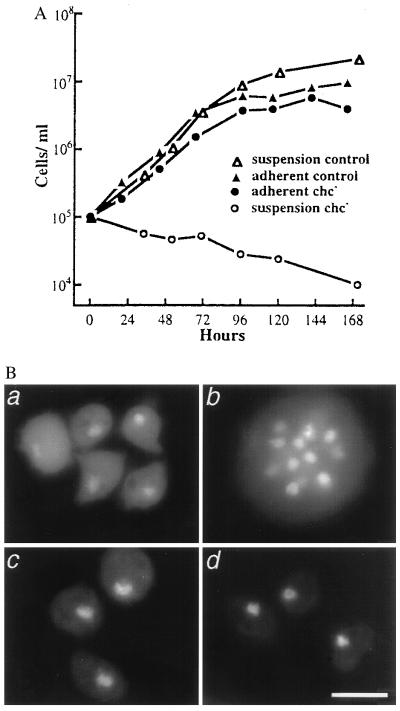
We recently engineered Dictyostelium cell lines that are devoid of clathrin heavy chain and identified defects in specific membrane traffic events such as endocytosis and regulated secretion (15, 19). Clathrin-minus cells also were blocked in development (17). In addition to these defects, we saw that the clathrin-minus cells grew similarly to a matched wild-type control strain when attached to a substrate, but failed to grow in suspension culture (Fig. 1A). This phenotype is reminiscent of the conditional growth defect of cytokinesis mutants such as Dictyostelium myosin heavy chain null cells (20–22). We therefore tested the possibility that clathrin-minus cells harbored a similar defect in cytokinesis. We stained clathrin-minus and control cells, grown in adherent or suspension culture for 72 hr, with the DNA-binding dye DAPI. Clathrin-minus cells grown on a substrate generally contained a single nucleus, as did control cells grown under all conditions (Fig. 1B). Conversely, we found that clathrin-minus cells grown in suspension culture were large and multinucleate, containing as many as 20 nuclei per cell (Fig. 1B). Since the doubling time of this clathrin-minus cell line in adherent culture is normally 18 hr, in 72 hr we would expect to see 16 cells arising from each original cell. However, instead of increasing in titer in 72 hr, the clathrin-minus cells exhibited a small decrease in cell titer accompanied by a 10-fold increase in the number of nuclei per cell (data not shown). Because the number of nuclei but not the cell titer correlated well with the normal doubling time in adherent culture, we inferred that clathrin-minus cells underwent karyokinesis but not cytokinesis in suspension culture. Once clathrin-minus cells accumulated many nuclei, the cells lysed, accounting for the drop in titer over 72 hr. These results suggested that clathrin was essential for cytokinesis.
Figure 1.
Clathrin-minus cells fail to undergo cytokinesis. (A) Clathrin-minus and wild-type control cells were grown in adherent or suspension cultures. (B) After 72 hr, samples were fixed and stained with DAPI, revealing multiple nuclei in clathrin-minus cells grown in suspension cultures. (a and b) Clathrin-minus cells. (c and d) Control cells. (a and c) Adherent cultures. (b and d) Suspension cultures. (Scale bar = 10 μ.)
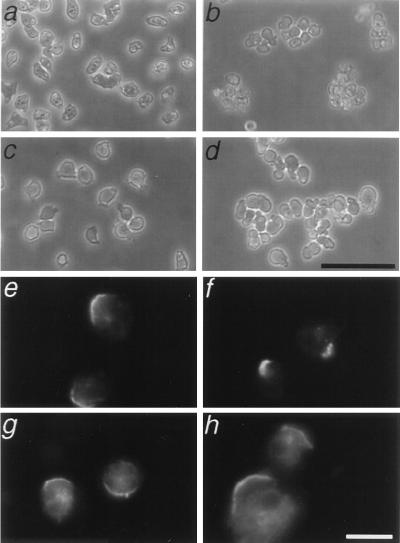
Cytokinesis requires functional myosin II and the actin cytoskeleton (20–22). We examined the possibility that clathrin-minus cells harbored defects in other cellular processes driven by the cytoskeleton. To test the functional integrity of myosin II, we tested the contractile response of cells exposed to azide (23). As a metabolic poison that rapidly depletes intracellular ATP, azide causes an irreversible attachment of myosin II to actin. This rigor attachment results in contraction of the cellular cortex and subsequent detachment of the cell from the substrate. When exposed to azide, clathrin-minus cells behaved the same as wild-type cells (Fig. 2 a–d): both cell types contracted their cortices, rounded up and detached from the plate. As a negative control for our assay, we verified that myosin II null cells failed to contract their cortex and remained flattened and adherent (data not shown). The wild-type response of clathrin-minus cells to azide demonstrated that myosin II can function normally in clathrin-minus cells.
Figure 2.
Clathrin-minus cells contract their cortex and cap Con A receptors. Wild-type (a) or clathrin-minus (c) cells adhere to a substrate, but upon treatment with 10 mM azide, both control (b) and clathrin-minus (d) cells contract, form clumps of cells, and float off the substrate. Wild-type (e and f) and clathrin-minus (g and h) cells cap cell surface receptors for the lectin Con A. (Scale bar = 10 μ.)
As a second measure of a process that requires myosin II, we also tested the ability of clathrin-minus cells to cap cell surface receptors (23). When incubated with the lectin Con A, wild-type cells rapidly gathered the cross-linked Con A receptors into a polar cap (Fig. 2 e and f). Similarly, clathrin-minus cells exposed to Con A also formed a cap at one end of the cell (Fig. 2 g and h). This is in contrast with the complete absence of caps seen in identical experiments with myosin–II null cells (ref. 24 and data not shown). We found that clathrin-minus cells were not as efficient at capping (≈5% of mutant cells formed caps vs. ≈50% of wild-type cells), possibly due to swelling of the mutants in the hypotonic buffer used in the capping assay (19). Nonetheless, the ability of clathrin-minus cells to form Con A caps contrasted markedly with the complete failure of myosin II–null mutants to form caps.
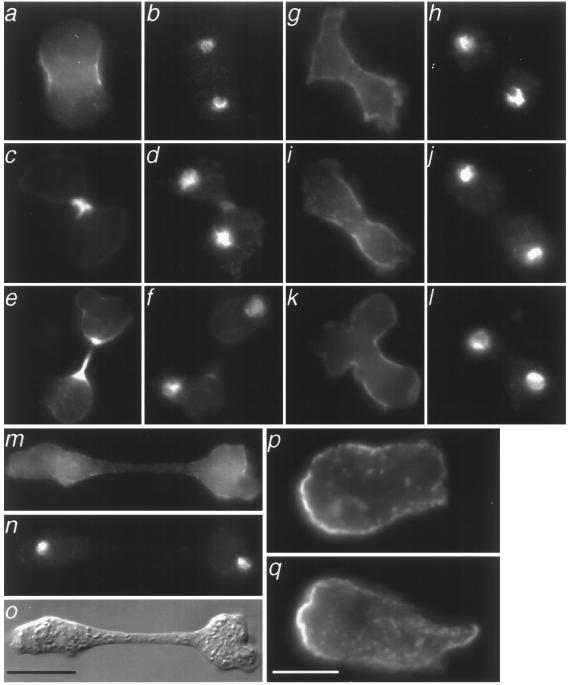
Clathrin-minus cells can contract their cortex and cap Con A receptors, but fail to complete cytokinesis in suspension culture. Therefore, while not required for general actomyosin function, clathrin is required specifically for cytokinesis. To dissect the mechanism of the cytokinesis defect, we examined the possibility that the phenotype arose from a failure to organize a functional contractile ring. We looked for the presence of a contractile ring in clathrin-minus cells dividing on a substrate, a condition permissive for cell division and growth. As a marker for the contractile ring, we examined cells that expressed myosin II tagged with gfp (gfp–myosin) (18). In wild-type cells, we found many examples of gfp–myosin concentrated in a contractile ring within the constricting cleavage furrow (Fig. 3 a–f). Indeed, every example of wild-type cells containing condensed chromosomes typical of late anaphase or telophase had gfp–myosin localized in a contractile ring (Fig. 3 a, c, and e). However, in clathrin-minus cells, we were unable to find a single example of gfp–myosin concentrated in a contractile ring (Fig. 3 g–o). Although we found many examples of clathrin-minus cells with chromosomes in late anaphase or telophase (Fig. 3 h and j), none of these had gfp–myosin assembled into a contractile ring. Instead, we found gfp–myosin associated with the plasma membrane in binucleated clathrin-minus cells. This cortical distribution for myosin is normally found in an interphase cell, not in a dividing cell. The inability of clathrin-minus cells to assemble myosin in a contractile ring was specific for this structure, because during migration, clathrin-minus cells localized myosin II properly to their posterior cortex (Fig. 3 p and q).
Figure 3.
Clathrin-minus cells do not form a functional contractile ring like wild-type cells. Using cells expressing gfp–myosin, we examined cell division on a substrate. We showed by DAPI staining that dividing cells were binucleate (b, d, f, h, j, l, and n). Three examples of wild-type cells in cytokinesis are shown (a–e). Dividing wild-type cells concentrated gfp–myosin (a, c, and e) in a cleavage furrow that was bright in subsequent stages of cytokinesis. Shown are four examples of clathrin-minus cells that had a binucleate, elongate morphology suggestive of cell division (g–o). Clathrin-minus cells showed a cortical distribution of gfp–myosin (g, i, and k), but did not concentrate gfp–myosin between the nuclei of dividing cells. Also shown is a clathrin-minus cell that appears to be undergoing cell division by traction-mediated cytofission on a substratum (m–o). Immunofluorescence of control (p) and clathrin-minus (q) cells without the gfp–myosin cassette are shown. Myosin II, visualized with an anti-myosin II antibody, localized to the posterior cortex of both cells as they migrated to the right. (Scale bar = 10 μ.)
The dramatic absence of contractile rings in clathrin-minus cells raised the question of how clathrin-minus cells divide on a substrate. Since their ability to divide requires attachment to a substrate, one possibility is that clathrin-minus cells divide when opposite ends of a binucleate cell crawl away from each other and break the cell in two (Fig. 3 m–o). This adhesion-dependent mechanism, used by myosin II–null cells, is called traction-mediated cytofission (24). Alternatively, clathrin-minus cells may divide by “attachment assisted mitotic cleavage,” a mechanism of cell division that also does not require contractile ring formation (25). This method of cell division, described recently in myosin II–null cells, is closely synchronized with mitosis and is characterized by cleavage furrow formation in the absence of a contractile ring. It is thought that both adhesion-dependent processes contribute to normal cytokinesis in Dictyostelium cells.
Although the plasma membrane is an obvious cellular participant in cytokinesis, these results are the first identification of a membrane-associated protein required for this process. Because the contractile ring fails to form in clathrin-minus mutants, the cellular requirement for clathrin must precede contractile ring assembly. At this point, we can envision several possible models for the specific role of clathrin in dividing cells. Clathrin at the plasma membrane could serve to remodel the membrane locally, either to create a membrane zone permissive for contractile ring attachment or assembly, or perhaps even to endocytose and thereby remove a membrane protein inhibitory for contractile ring formation. Conceivably, clathrin lattices on the plasma membrane could interact directly with the actomyosin cytoskeleton. Whereas this kind of interaction is relatively unprecedented, recent studies uncovered high-affinity binding between clathrin and a domain of the cytoskeletal protein ankyrin (26). An additional model for the role of clathrin in cytokinesis invokes its trafficking potential. By analogy with the importance of clathrin in trafficking α-mannosidase in Dictyostelium (15), clathrin could also be required for the processing of membrane proteins involved in contractile ring assembly. Ultimately, more experimental studies are needed to further substantiate any of these scenarios. Dictyostelium clathrin-minus cells provide an excellent resource for defining the molecular mechanism of the cellular role of clathrin in cytokinesis.
Acknowledgments
We thank Danny Lew and members of the O’Halloran and De Lozanne labs for helpful comments on this manuscript. This research is supported by National Institutes of Health Grant GM48624 to T.J.O.
ABBREVIATIONS
- Con A
concanavalin A
- DAPI
4,6-diamidino-2-phenylindole
- gfp
green fluorescent protein
References
- 1.Rappaport R. Cytokinesis in Animal Cells. Cambridge, U.K.: Cambridge Univ. Press; 1996. [Google Scholar]
- 2.Larochelle D A, Vithalani K, De Lozanne A. J Cell Biol. 1996;133:1–9. doi: 10.1083/jcb.133.6.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Longtine M S, DeMarini D J, Valencik M L, Al-Awar O S, Fares H, De Virgilio C, Pringle J R. Curr Opin Cell Biol. 1996;8:106–119. doi: 10.1016/s0955-0674(96)80054-8. [DOI] [PubMed] [Google Scholar]
- 4.Neufeld T P, Rubin G M. Cell. 1994;77:371–379. doi: 10.1016/0092-8674(94)90152-x. [DOI] [PubMed] [Google Scholar]
- 5.Field C M, Alberts B M. J Cell Biol. 1995;131:165–178. doi: 10.1083/jcb.131.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Field C M, Oegema K F, Doberstein S, Alberts B M, Mitchison T J. Mol Biol Cell. 1996;7:229a. (abstr.). [Google Scholar]
- 7.Castrillon D H, Wasserman S A. Development (Cambridge, UK) 1994;120:3367–3377. doi: 10.1242/dev.120.12.3367. [DOI] [PubMed] [Google Scholar]
- 8.Faix J, Steinmetz M, Boves H, Kammerer R A, Lottspeich F, Mintert U, Murphy J, Stock A, Aebi U, Gerisch G. Cell. 1996;86:631–642. doi: 10.1016/s0092-8674(00)80136-1. [DOI] [PubMed] [Google Scholar]
- 9.de Hostos E L, Rehfuess C, Bradtke B, Waddell D R, Albrecht R, Murphy J, Gerisch G. J Cell Biol. 1993;120:163–173. doi: 10.1083/jcb.120.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sato N, Yonemura S, Obinata T, Tsukita S, Tsukita S. J Cell Biol. 1991;113:321–330. doi: 10.1083/jcb.113.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bretscher M S. Science. 1984;224:681–686. doi: 10.1126/science.6719108. [DOI] [PubMed] [Google Scholar]
- 12.Bretscher M S. Cell. 1996;85:465–467. doi: 10.1016/s0092-8674(00)81246-5. [DOI] [PubMed] [Google Scholar]
- 13.Brodsky F M. Science. 1988;242:1396–1402. doi: 10.1126/science.2904698. [DOI] [PubMed] [Google Scholar]
- 14.Robinson M S, Watts C, Zerial M. Cell. 1996;84:13–21. doi: 10.1016/s0092-8674(00)80988-5. [DOI] [PubMed] [Google Scholar]
- 15.Ruscetti T, Cardelli J A, Niswonger M L, O’Halloran T J. J Cell Biol. 1994;126:343–352. doi: 10.1083/jcb.126.2.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seeger M, Payne G S. EMBO. 1992;11:2811–2818. doi: 10.1002/j.1460-2075.1992.tb05348.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Niswonger M L, O’Halloran T J. Development (Cambridge, UK) 1997;124:443–451. doi: 10.1242/dev.124.2.443. [DOI] [PubMed] [Google Scholar]
- 18.Moores S L, Sabry J H, Spudich J A. Proc Natl Acad Sci USA. 1996;93:443–446. doi: 10.1073/pnas.93.1.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O’Halloran T, Anderson R G W. J Cell Biol. 1992;118:1371–1377. doi: 10.1083/jcb.118.6.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Manstein D J, Titus M A, De Lozanne A, Spudich J A. EMBO J. 1989;8:923–932. doi: 10.1002/j.1460-2075.1989.tb03453.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Knecht D A, Loomis W F. Science. 1987;236:1081–1086. doi: 10.1126/science.3576221. [DOI] [PubMed] [Google Scholar]
- 22.De Lozanne A, Spudich J A. Science. 1987;236:1086–1091. doi: 10.1126/science.3576222. [DOI] [PubMed] [Google Scholar]
- 23.Pasternak C, Spudich J A, Elson E L. Nature (London) 1989;341:549–551. doi: 10.1038/341549a0. [DOI] [PubMed] [Google Scholar]
- 24.Fukui Y, De Lozanne A, Spudich J A. J Cell Biol. 1990;110:367–378. doi: 10.1083/jcb.110.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Neujahr R, Heizer C, Gerisch G. J Cell Sci. 1997;110:123–137. doi: 10.1242/jcs.110.2.123. [DOI] [PubMed] [Google Scholar]
- 26.Michaely P, Bennett V. Mol Biol Cell. 1995;6:412a. (abstr.). [Google Scholar]