Abstract
The yeast translation factor eIF4G associates with both the cap-binding protein eIF4E and the poly(A)-binding protein Pab1p. Here we report that the two yeast eIF4G homologs, Tif4631p and Tif4632p, share a conserved Pab1p-binding site. This site is required for Pab1p and poly(A) tails to stimulate the in vitro translation of uncapped polyadenylylated mRNA, and the region encompassing it is required for the cap and the poly(A) tail to synergistically stimulate translation. This region on Tif4631p becomes essential for cell growth when the eIF4E binding site on Tif4631p is mutated. Pab1p mutations also show synthetic lethal interactions with eIF4E mutations. These data suggest that eIF4G mediates poly(A) tail stimulated translation in vitro, and that Pab1p and the domain encompassing the Pab1p-binding site on eIF4G can compensate for partial loss of eIF4E function in vivo.
Both the cap and the poly(A) tail on eukaryotic mRNA serve as translational enhancers. Each of these elements work by associating with a specific RNA-binding protein, and together function synergistically to stimulate translation (reviewed in ref. 1). The translation initiation factor eIF4E binds to the cap structure, whereas the poly(A) tail binding protein (Pab1p) is bound to the poly(A) tail. It has been previously shown that eIF4E is part of a complex of proteins in eukaryotes called eIF4F (2). This complex contains a large subunit, eIF4G, and in higher eukaryotes the RNA-dependent ATPase eIF4A. The association of the eIF4F complex with the cap structure leads to the recruitment of the ribosome to the RNA, presumably via an association of the eIF4G subunit with the ribosome-associated eIF3 complex (2).
The association of Pab1p with the poly(A) tail also leads to translational stimulation of mRNA in vivo and in vitro (3, 4). Like eIF4E and the cap structure, Pab1p and the poly(A) tail stimulate the recruitment of the 40S ribosomal subunit to mRNA (4). Pab1p and poly(A) tail-dependent in vitro translation can be measured by two distinct assays. First, the amount of translation of uncapped, polyadenylylated mRNA versus uncapped, poly(A)-deficient mRNA can be quantified. Second, the degree of synergistic interaction between the cap/eIF4E complex and the poly(A) tail/Pab1p complex can be measured by comparing the amount of translation of capped, polyadenylylated mRNA to that of the sum of translation of capped and of polyadenylylated mRNA (4, 5).
We recently reported that Pab1p associates with yeast eIF4G (6). This finding raised several interesting questions. For instance, is the association of Pab1p with eIF4G responsible for the stimulation of translation by the poly(A) tail in translation extracts? If so, is this association responsible for both assayable measures of translational stimulation by the poly(A) tail? Furthermore, what are the effects of destroying the ability of Pab1p to bind to eIF4G in vivo? Is cell growth compromised, and are there other mutations that exacerbate this growth alteration? The experiments described here detail the identification, mutagenesis, and in vitro and in vivo consequences of mutating the Pab1p-binding region of the two yeast eIF4G homologs Tif4631p and Tif4632p. The results from this work indicate that the association of Pab1p with eIF4G mediates the ability of the poly(A) tail to stimulate translation in vitro, and that this association is not an essential one in vivo unless the function of the cap-binding protein eIF4E is compromised.
MATERIALS AND METHODS
Recombinant Protein Production and Analysis.
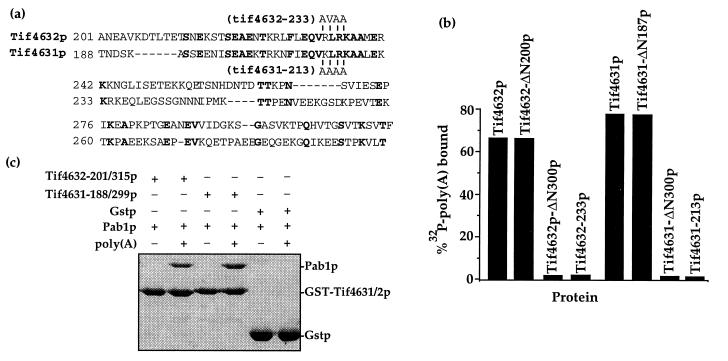
For Fig. 1b, the PCR products of a TIF4631 or TIF4632 amplification using different mutagenic oligonucleotides were digested and subcloned into a similarly digested pAS466 or pAS476, pGEX2T (Pharmacia) vectors containing either the Tif4631 or Tif4632 ORF flanked by BamHI and EcoRI sites (6), respectively. These plasmids were placed into bacterial strain BL21 to yield the following glutathione S-transferase fusion protein expressing strains: BAS2027, Tif4631p; BAS3125, Tif4631–213p; BAS3110, Tif4631-ΔN187p; BAS3111, Tif4631-ΔN300p; BAS2030, Tif4632p; BAS3032, Tif4632–233p; BAS3002, Tif4632-ΔN200p; BAS3003, Tif4632-ΔN300p. Equal amounts (approximately 1 μg) of full-length and mutated Tif4631p and Tif4632p were immobilized on glutathione resin and incubated with approximately 50,000 cpm of [32P]poly(A) with or without 10 μg of Pab1p. After extensive washing, resin associated [32P]poly(A) was detected by direct scintillation counting. In all cases, no [32P]poly(A) was retained on the resin in the absence of Pab1p (see ref. 6 for details). For Fig. 1c, the PCR product of a TIF4631 amplification using OAS227 (CGCGGATCCACTAATGA CTCTAAGGCCAGT) and OAS228 (CCGGAATTCTTAGGTTAACACCTTTG GAGTGGATTC), which encoded the Pab1p-binding domain of this protein, was digested with BamHI/EcoRI, subcloned into the similarly digested Gstp-expression vector pGEX2T, and placed into bacterial strain BL21 to yield BAS3035. Induced fusion protein from this strain, BAS3024 (containing the Tif4632p fragment; ref. 6), and BAS2028 (containing only the vector) was immobilized on glutathione resin and then incubated with 10 μg of recombinant Pab1p (7) with or without 10 μg of poly(A) (Sigma). After extensive washing, resin-associated proteins were eluted in SDS and analyzed by 10% SDS/PAGE. Note that all amino acid numbering is based on the modified TIF4631 and TIF4632 genes (6), which contain a six-nucleotide BamHI site insertion (GGATCC) after the initiator codon and therefore two extra amino acids.
Figure 1.
Identification and mutagenesis of a Pab1p binding site on eIF4G. (a) Alignment of the Pab1p-binding domain of Tif4632p (6) and the homologous region of Tif4631p. Identical residues are highlighted, the positions and type of substitutions in Tif4632–233p and Tif4631–213p are indicated above or below the sequences, and the positions of the residues in the full-length proteins are shown. (b) Mutations within the Tif4631p and Tif4632p Pab1p binding domains destroy Pab1p binding. Each eIF4G protein containing the indicated mutation was fused to glutathione S-transferase, immobilized on glutathione resin, and assayed for its ability to bind to a [32P]poly(A)/Pab1p complex, as previously described (6). [32P]poly(A) retention reflects the amount of Pab1p binding to the eIF4G proteins. (c) The Pab1p-binding domain of Tif4631p resides between amino acids 188 and 299. The Tif4631p and Tif4632p fragments diagrammed in a were fused to glutathione S-transferase, immobilized on glutathione resin, and assayed for their ability to bind Pab1p in the presence or absence of poly(A), as previously described (6). Bound proteins were resolved in a 10% SDS/PAGE gel and visualized by Coomasie blue staining. The position of each recombinant protein is indicated.
Yeast Genetic Methods.
For Table 1, EcoRI/BamHI fragments containing the mutated TIF4631 or TIF4632 genes were subcloned into pAS481, a yeast TRP1CEN plasmid bearing the entire TIF4632 gene and artificial BamHI and EcoRI sites at the start and stop codons (6). TIF4631 and TIF4632 mutations were created by PCR-directed mutageneses. After transformation of these plasmids into the indicated strains and growth on yeast minimal (YM)-Trp medium (8), cells were restruck onto 5-fluoroorotic acid plates (8) and incubated for up to 2 weeks at 30°C. No visible growth after 2 weeks was scored as a nonfunctional gene.
Table 1.
Viability of tif4631 and tif4632 mutants
| Gene | Strain | Generation time, hr
|
Pab1p binding | |
|---|---|---|---|---|
| 30°C | 37°C | |||
| TIF4631 | YAS2069 | 2.0 | 2.0 | Yes |
| tif4631-ΔN187 | YAS2070 | 2.0 | 2.4 | Yes |
| tif4631-ΔN300 | YAS2071 | 2.4 | 4.8 | No |
| tif4631-ΔN400 | YAS2076 | 2.4 | 4.5 | No |
| tif4631-ΔN500 | Dead | Dead | No | |
| tif4631-213 | YAS2075 | 2.4 | 2.9 | No |
| tif4631-459 | YAS2074 | 4.1 | Dead | ND |
| tif4631-ΔN187,459 | YAS2104 | 2.1 | Dead | ND |
| tif4631-ΔN300,459 | Dead | Dead | ND | |
| TIF4632 | YAS1981 | 2.0 | 2.0 | Yes |
| tif4632-ΔN200 | YAS1983 | 2.0 | 2.2 | Yes |
| tif4632-ΔN300 | YAS1984 | 2.3 | 2.6 | No |
| tif4632-ΔN400 | YAS1985 | 2.4 | 3.4 | No |
| tif4632-ΔN500 | Dead | Dead | No | |
| tif4632-233 | YAS2001 | 2.4 | 2.4 | No |
Yeast strain YAS1948 (a ade2 his3 leu2 trp1 ura3 pep4::HIS3 tif4631::LEU2 tif4632::ura3 pTIF4632URA3CEN) or YAS1947 (α ade2 his3 leu2 trp1 ura3 tif4631::LEU2 tif4632::ura3 pTIF4632URA3CEN) were transformed with TRP1CEN plasmids (6) bearing the indicated tif4631 or tif4632 genes, respectively. Lack of growth on 5-fluoroorotic acid plates after 2 weeks was indicative of no viability. Growth rates were measured in yeast extract/peptone/dextrose (YPD) medium at the indicated temperatures. In vitro Pab1p-binding data are summarized from previous data (6) and from Fig. 1. ND, not determined.
In Vitro Translation.
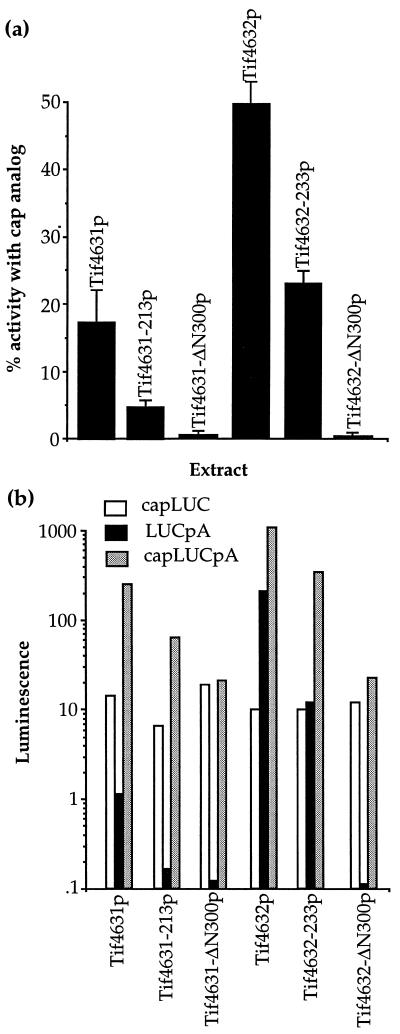
Translation extracts (4) (OD260 ≈ 100) were treated with 60 units/100 μl extract micrococcal nuclease for Fig. 2a, or left untreated but made 2 mM EGTA for Figs. 2b and 3b. For [35S]methionine incorporation studies in Fig. 2a, 22.2 μl of a mixture containing 15 μl of extract and 7.2 μl of the other translation components (4) were incubated for 5 min, and then supplemented with 8 μl of a mixture containing 10 uci [35S]methionine (translation grade, NEN), with or without 60 ng of yeast poly(A)+ mRNA, with or without 15.4 nmols of 7mGpppG (NEB, Beverly, MA). The amount of [35S]methionine incorporation in 8.5-μl aliquot, as judged using a trichloroacetic acid precipitation assay (9), was determined at 10, 20, and 30 min post [35S]methionine addition. Incorporation was linear out to 40 min, it linearly increased with increasing amounts of mRNA, and it was stimulated at least 4-fold by the RNA. Synthesis values at each point were calculated by subtracting the amount of incorporation found in the absence of mRNA with or without analog from that found in its presence with or without analog. Rates of incorporation were determined from the slopes of the line derived from the three measurements and normalized for the slight variation in the protein concentration in each extract. The ratio of these slopes are depicted in Fig. 2a. Typically, these rates were in the 1,000–3,000 cpm/min per 8.5-μl aliquot range. All incubations were at 26°C. For Fig. 2b, approximately 90 ng of the indicated luciferase mRNAs (5) in a 7.5-μl mixture containing all the required translation components was added to 7.5 μl of extract and analyzed for luciferase production as previously described (4). All mutants were analyzed minimally three times from two or more independent extract preparations.
Figure 2.
Mutations within the Pab1p-binding domain of Tif4631p and Tif4632p inhibit poly(A)-dependent translation. (a) Equal amounts of nuclease-treated translation extracts were programmed with [35S]methionine and yeast mRNA with or without the cap analog 7mGpppG. The percent of translational activity remaining in each extracts in the presence of the analog versus its absence is shown. This value represents poly(A)-dependent translation (4). Translational rates of incorporation in the absence of the analog, relative to the wild-type extracts were: Tif4631-213p (88%), Tif4631-ΔN300p (115%), Tif4632-233 (70%)p, and Tif4632-ΔN300p (72%). (b) Equal amounts of translation extracts prepared from yeast strains harboring the indicated TIF4631 or TIF4632 genes were analyzed for their ability to translate luciferase (LUC) mRNA containing either a cap (capLUC), a poly(A) tail (LUCpA), or both (capLUCpA) as previously described (4, 5). Luciferase enzyme production was monitored by a luminescence assay. The data shown are representative of at least three independent assays. LUC mRNA lacking a cap and a poly(A) tail was not detectably translated in these extracts (data not shown).
Figure 3.
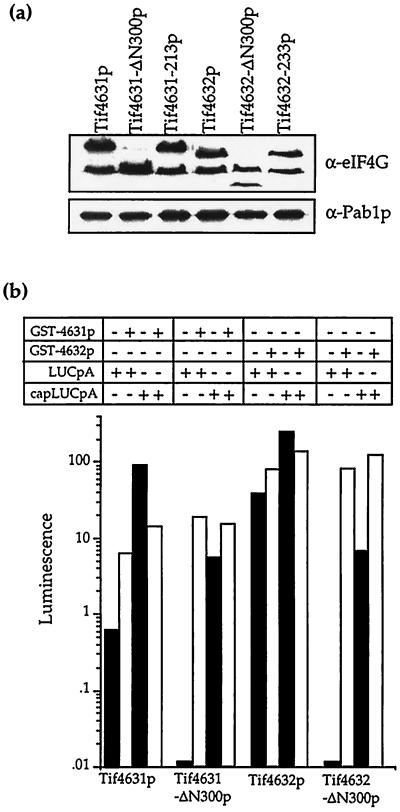
Reconstitution of poly(A)-dependent translation in extracts containing mutant eIF4G. (a) Western analysis for Pab1p and the various forms of Tif4631p and Tif4632p in each of the extracts. Rat polyclonal antibodies against the C-terminus of Tif4631p and mouse mAbs to Pab1p were used to probe equal total amounts of protein from each extract that had been resolved by SDS/PAGE. The upper band in each of the samples probed for eIF4G represents the intact protein, while the lower band presumably represents a proteolytic fragment. (b) Recombinant eIF4G rescues the translational deficiency of the mutant extracts. The expression levels of LUCpA or capLUCpA mRNA in the indicated extracts containing or lacking recombinant Tif4631p or Tif4632p are shown.
For Fig. 3b, approximately 17 ng of full-length, recombinant glutathione S-transferase–Tif4631p (from BAS2027) or 33 ng of glutathione S-transferase–Tif4632p (from BAS2030) purified by glutathione-Sepharose chromatography (6) and dialyzed against translation buffer A with 10% glycerol (5) were added in 1 μl to 7.5 μl of translation extract before the addition of the mRNA and other components in 6.5 μl. For the Western analysis in Fig. 3a, equal amounts of the indicated extracts were resolved by 10% SDS/PAGE, immobilized on nitrocellulose, probed with either rat α-Tif4631-ΔN489p (kindly provided by M. Altmann, University of Bern, Bern, Switzerland) or the Pab1p mAb 1G1 (10), and visualized by chemiluminescence using the ECL detection system (Amersham).
RESULTS
We have previously mapped the Pab1p-binding site on yeast Tif4632p to a 114-aa region (6). Alignment of this region of Tif4632p with the corresponding region of Tif4631p revealed it to be conserved between the two homologs (Fig. 1a). Deleting this conserved region from recombinant Tif4631p fused to the glutathione S-transferase protein yielded a Tif4631p fragment (Tif4631-ΔN300) that was unable to bind to Pab1p in vitro (Fig. 1b). In contrast, Tif4631-ΔN187 still bound to Pab1p. Similar data were previously reported for deletions within Tif4632p (6). A series of point mutations within a conserved part of this region (Fig. 1a) in either Tif4631p (Tif4631-213p) or Tif4632p (Tif4632-233p) also resulted in a full-length protein that also was unable to bind to Pab1p in vitro (Fig. 1b). Finally, the 111-aa Tif4631p fragment containing the Pab1p binding site, like the 114-aa fragment from Tif4632p (6), was shown to be capable of binding to the Pab1p/poly(A) complex in vitro (Fig. 1c). These data show that the yeast eIF4G homologs Tif4631p and Tif4632p contain a conserved site that is both necessary and sufficient for in vitro Pab1p binding.
In vitro translation assays were performed using extracts from strains (see below) harboring the various forms of the eIF4G proteins shown in Fig. 1b. In the first set of translation assays, [35S]methionine incorporation was used to measure the translational stimulation of the extracts by exogenous yeast poly(A)+ mRNA. The degree of inhibition of methionine incorporation in these extracts by the cap analog 7mGpppG was used to evaluate the extract’s ability to be stimulated by the poly(A) tail. This assay is based on previous observations that cap-dependent translation is inhibited in these extracts by this analog (4, 5). Extracts containing Tif4632p incorporated methionine at 50% of their normal rates in the presence of the cap analog, whereas extracts containing Tif4631p incorporated at 15% of their normal rates (Fig. 2a). In contrast to these wild-type extracts, those containing Tif4631-ΔN300 or Tif4632-ΔN300 proteins were completely inhibited for translation in the presence of the cap analog. Tif4631-213p and Tif4632-233p extracts showed milder degrees of inhibition in the presence of the cap analog, suggesting that these mutations result in only partial loss of function in vitro.
As an alternative means to measure the effects of the eIF4G mutations on poly(A) tail dependent translation, the ability of the mutant extracts to translate luciferase mRNA containing a cap (capLUC), a poly(A) tail (LUCpA), or both (capLUCpA) was investigated (4). As shown in Fig. 2b, extracts with the point-mutated Tif4631p (Tif4631-213p) exhibited an approximately 7-fold decrease in their ability to translate LUCpA mRNA versus capLUC mRNA. Extracts containing the point-mutated Tif4632p (Tif4632-233p) exhibited a 17-fold decrease in their ability to translate LUCpA mRNA. Extracts containing either Tif4631-ΔN300p or Tif4632-ΔN300p were unable to translate LUCpA mRNA. In contrast, the translation of the capLUC mRNAs in each of the extracts were nearly equal. As also observed in Fig. 2a, extracts containing Tif4631 were much less competent than Tif4632 extracts to be stimulated by the poly(A) tail. This may reflect differences in the affinities of these proteins for Pab1p in the absence of other cap-associated initiation factors, or it may reflect a lowered ability of Tif4631p to drive initiation on mRNA lacking a cap structure. Together with the above 35S-incorporation studies, these data show that point mutations that alter the conserved Pab1p-binding site in either of the yeast eIF4G proteins results in substantial decreases in the translation of uncapped, polyadenylylated mRNA. These data support the conclusion that the Pab1p-binding sites on Tif4631p and Tif4632p serve a functional role in vitro.
The ability of the cap and the poly(A) tail to synergistically stimulate translation (capLUCpA data, Fig. 2b) (see ref. 4) was only partially inhibited in extracts containing either Tif4631-213p or Tif4632-233p, and nearly completely destroyed in extracts containing Tif4631-ΔN300p or Tif4632-ΔN300p. For the extracts containing the point-mutated proteins, the partial loss of capLUCpA expression was much smaller than expected from the results for the LUCpA mRNA. These data suggest that the synergism between the cap and the poly(A) tail, which is mediated by eIF4E and Pab1p (4), also requires the N terminus of eIF4G. They also suggest that the Pab1p-binding site on eIF4G that was defined by our mutageneses is not solely responsible for mediating the synergism between the cap and the poly(A) tail.
Results from several other experiments examining the mutant and wild-type extracts confirm the validity of the conclusion that the translational deficiencies described above were due to a specific loss of eIF4G’s ability to be stimulated by Pab1p and poly(A). First, both the ability of the extracts to incorporate methionine (Fig. 2a legend) and to translate capLUC mRNA (Fig. 2b) were nearly identical. This finding argues that the extracts were not lacking in activity for any other translation factor. Second, the amount of Pab1p in each of the extracts were nearly equal (Fig. 3a), indicating that the eIF4G mutations did not indirectly lead to a loss of Pab1p. Third, the amount of each of the mutant eIF4G proteins in the extracts were essentially identical to the wild type’s level, except for Tif4632-ΔN300p, which was present at approximately 50% of the wild type’s level. These data suggest that the observed results were not due to an indirect effect of loss of eIF4G proteins from the extract. Finally, the deficiencies of the mutant extracts could be restored by the addition of recombinant eIF4G (Fig. 3b). This shows that the extracts failed to be stimulated by poly(A) due to the loss of an eIF4G function. We note that the addition of recombinant Tif4631p to wild-type extracts is inhibitory for capLUCpA translation, possibly because it is still capable of binding some initiation factors but is only partially capable of performing one of its translation functions.
The in vivo effects of the eIF4G mutations were also analyzed. The capacity of various mutant eIF4G proteins to support yeast cell viability was examined (Table 1). In this assay, a yeast strain that contains a disruption of both its TIF4631 and TIF4632 genes and carries TIF4632 on a URA3CEN plasmid was transformed with each of the mutant TIF4631/2 genes on TRP1CEN plasmids. The ability of the resulting transformant to grow on 5-fluoroorotic acid indicates that the mutant gene is capable of supporting yeast cell viability (8). As shown in Table 1, the point mutations in the Pab1p-binding site of Tif4631p (tif4631-213) and Tif4632p (tif4632-233) that destroyed Pab1p binding in vitro partially impaired but did not destroy the essential activity of these proteins. Deletion of up to 400 amino acids from the N-termini of either Tif4631p or Tif4632p (tif4631-ΔN400, tif4632-ΔN400) also did not impair this essential activity. However, removing the domain encompassing the Pab1p-binding site from Tif4631p (i.e., Tif4631-ΔN300p) did lead to impaired growth rates at 37°C, and synthetically lethal interactions with mutations in the Tif4631p eIF4E-binding site (see below). We note that the failure of some mutant eIF4G genes to support viability could result from reduced expression of the protein instead of reduced function. Why mutations in Tif4632p did not lead to identical phenotypes remains to be explored (see Discussion). These data show that the region of yeast eIF4G encompassing the Pab1p-binding site is not essential for yeast cell viability.
Further genetic analysis was performed to understand in more detail why loss of the region encompassing the Pab1p-binding site on eIF4G had such dramatic consequences on the in vitro translation assays but still allowed for cell viability. Because Pab1p and eIF4E share the common function of recruiting 40S subunits to mRNA in vitro (4), it seemed possible that the Pab1p-binding region of eIF4G was dispensable in vivo because eIF4E could still function with eIF4G to recruit the ribosome. This hypothesis predicted that phenotypes associated with loss of the Pab1p-binding region on eIF4G would become apparent when eIF4E function became impaired, and that Pab1p mutations would shown synthetic lethality with eIF4E mutations. To test the first of these predictions, the eIF4E-binding site on Tif4631, which has been previously identified (11), was altered by site-specific mutagenesis. Mutating a highly conserved tyrosine to alanine (Y454A) within this site on Tif4631p led to a nonfunctional protein (data not shown). This suggests that eIF4E’s association with Tif4631p may be an essential interaction. Mutating two leucine residues to alanine (L459AL460A) within this site on Tif4631p (Tif4631-459p) yielded a protein that conferred temperature-sensitive growth to the yeast (Table 1). The L459AL460A mutation in Tif4631p then was combined with either the ΔN187 or ΔN300 mutation, or the point mutations within the Pab1p-binding site (Tif4631-213). Only the combination of the L459AL460A mutation with the ΔN300 led to cell inviability (Table 1). The level of expression of the Tif4631-ΔN300,459 protein in a strain bearing wild-type Tif4632p was found by Western blot analysis to be no less than 2-fold below that of Tif4631p in a similar strain (data not shown). These data are consistent with the hypothesis that the Pab1p-binding region of eIF4G becomes essential under conditions where eIF4E function is decreased.
A second set of experiments was performed to test the prediction that Pab1p mutations would show synthetic lethality with eIF4E mutations. We found that a specific loss of function allele of the PAB1 gene (pab1-16), which was generated as part of a systematic effort to destroy the RNA-binding capacity of each of Pab1p’s four RNA-binding domains (12), showed synthetic lethal interactions with the eIF4E mutation cdc33-1 (Table 2). Interestingly, this pab1-16 gene was also unable to stimulate poly(A)-tail dependent in vitro translation (S. Kessler and A.B.S., unpublished work). Whether the pab1-16 mutation also shows synthetic lethal interactions with the tif4631-459 mutation remains unanswered due to the absence of usable markers in strains bearing all of the appropriate chromosomal deletions to perform this test. These genetic data show that Pab1p and eIF4E do have some functional redundancy, and together with the above data suggest that the N terminus of eIF4G compensates for loss of eIF4E function in vivo via its interaction with Pab1p.
Table 2.
Synthetic lethal interactions between pab1 and cdc33-1 mutations
| Gene, RRM mutation | Strain | Doubling time, hr | Viable with cdc33-1 |
|---|---|---|---|
| PAB1-1 | YAS2043 | 2.6 | Yes |
| pab1-5 (RRM1) | YAS2144 | 3.6 | Yes |
| pab1-6 (RRM2) | YAS2145 | 3.1 | Yes |
| pab1-16 (RRM1,2) | YAS2151 | 5.5 | No |
| pab1-22 (RRM1,4) | YAS2149 | 4.1 | Yes |
| pab1-23 (RRM2,3) | YAS2150 | 5.7 | Yes |
Yeast strain YAS1881 (α ade2 his3 leu2 trp1 ura3 cdc33-1 pab1::HIS3 pPAB1URA3CEN) was transformed with the listed mutant pab1 genes on TRP1CEN plasmids (12). Which of Pab1p’s four RNA-recognition motifs (RRM) contains a valine substitution in its highly conserved RNP1 sequence (described fully in ref. 12) is noted for each mutant. The ability of the transformed yeast strains to grow on 5-fluoroorotic acid medium at 26°C is shown. No growth after 2 weeks was indicative of no viability. The doubling time of a yeast strain (YAS) containing wild-type CDC33 and the mutant pab1 gene is also shown.
DISCUSSION
The above experiments provide strong evidence that yeast eIF4G mediates poly(A) tail dependent translation in vitro. We found that point mutations in the Pab1p-binding sites of Tif4631p or Tif4632p diminished the ability of yeast extracts to translate uncapped, polyadenylylated mRNA. Deletion of the region containing this Pab1p-binding site further led to the loss of synergistic stimulation of translation by the cap and the poly(A) tail. The deleterious in vivo effects of removing this region became apparent when the eIF4E binding site on eIF4G was mutated. Similarly, mutations in PAB1 and the eIF4E gene CDC33 showed synthetic lethal interactions. In summary, these biochemical data indicate that eIF4G is needed for the in vitro stimulation of translation by poly(A) tails, and the genetic data suggest that Pab1p’s association with eIF4G can compensate for eIF4E loss of function in vivo.
Saccharomyces cerevisiae contains two forms of eIF4G that are redundant for their essential functions (13). In this and past work, we have identified several differences between them. For instance, Tif4631p copurifies with eIF4E by conventional column chromatography, whereas Tif4632p does not (6). Furthermore, extracts from strains harboring Tif4632p are more capable of translating uncapped, polyadenylylated mRNA than extracts from strains containing Tif4631p. In addition, N-terminal deletions of Tif4631p lead to significant impairment of growth rates at high temperature, whereas similar deletions of Tif4632p do not. Finally, a deletion of TIF4631 leads to a synthetic lethal interaction with cdc33-1 whereas a deletion of TIF4632 does not (A.B.S., unpublished observation). Based on these observations, we hypothesize that Tif4631p and Tif4632p are not functionally identical in yeast. This lack of identity may in part explain why the most severe phenotypes we have observed in our eIF4G mutagenesis studies are found in strains containing mutated TIF4631. Perhaps Tif4631p is more sensitive to alterations in its association with Pab1p than Tif4632p because it is functionally limiting in amounts in the translation initiation pathway, whereas Tif4632p may be in excess and therefore not as sensitive an indicator when its activity is diminished.
Is the Pab1p-binding site on eIF4G that was destroyed by point mutagenesis solely responsible for mediating poly(A)-tail dependent translation in vivo? In our in vitro assays, this site was shown to be required for poly(A) tails to stimulate the translation of uncapped mRNA but not for the ability of poly(A) to synergistically activate translation in conjunction with the cap structure. Because very little uncapped mRNA exists within eukaryotic cells, we assume that our measures of translational synergy between the cap and the poly(A) tail most accurately reflect how poly(A) tails function in vivo. Based on this, we conclude that the Pab1p binding site on eIF4G defined by our mutagenesis (Fig. 1a) cannot mediate all of the stimulation of translation by Pab1p and the poly(A) tail in vivo.
Based on this conclusion, we assume that Pab1p is making other contacts within the translation initiation complex in vivo. These contacts could be with other regions within eIF4G that become exposed in the presence of the cap. Alternatively, they could be with other proteins that are stabilized within the initiation complex by the cap structure and the N terminus of eIF4G. In this model, the Pab1p-binding site on eIF4G would be only one of several contact points for Pab1p on the initiation complex. We hypothesize that one or more of these other contacts are primarily responsible for mediating Pab1p’s synergistic stimulation of translation. Furthermore, we suggest that Pab1p’s interaction with the Pab1p-binding site on eIF4G is responsible for Pab1p’s translational stimulation of uncapped, polyadenylylated mRNA because eIF4G cannot be recruited to the mRNA via the other, cap-dependent interactions. The identification of the putative binding targets for Pab1p and poly(A) in the translation initiation complex assembled on capped and polyadenylylated mRNA awaits further investigation.
The data presented here highlight the central role eIF4G may play in coordinating the interactions between the two ends of the mRNA molecule. Previous experiments suggesting that mRNA decapping and degradation in yeast is stimulated by the loss of the poly(A) tail (14) can be interpreted in light of our findings. For instance, it seems likely that the relative affinity of eIF4E for the cap structure would be decreased when the interaction of Pab1p with eIF4G and perhaps other initiation factors is disrupted due to the loss of the poly(A) tail. This exposure of the cap structure could lead to enhanced rates of decapping. The ability of mRNA 3′ untranslated regions (UTR) to repress or activate translation (reviewed in ref. 15) also can be interpreted with our data. Perhaps these elements work by enhancing or repressing the protein interactions between the two ends of the RNA. Due to the proximity of the 3′ UTR with the 5′ end of the mRNA, it is also possible that 3′ UTR elements could modulate the interactions between eIF4E and other translation factors. Future studies will be needed to address these and other implications of our findings.
Acknowledgments
This work was supported by grants to A.B.S. from the American Cancer Society, the National Institutes of Health, and the Hellman Family Fund and to S.E.W. from the Jane Coffin Childs Memorial Fund for Medical Research.
References
- 1.Sachs A B, Sarnow P, Hentze M W. Cell. 1997;89:831–838. doi: 10.1016/s0092-8674(00)80268-8. [DOI] [PubMed] [Google Scholar]
- 2.Hershey J W B, Mathews M B, Sonenberg N, editors. Translational Control. Vol. 30. Plainview, NY: Cold Spring Harbor Lab. Press; 1996. [Google Scholar]
- 3.Sachs A B, Davis R W. Cell. 1989;58:857–867. doi: 10.1016/0092-8674(89)90938-0. [DOI] [PubMed] [Google Scholar]
- 4.Tarun S, Sachs A B. Genes Dev. 1995;9:2997–3007. doi: 10.1101/gad.9.23.2997. [DOI] [PubMed] [Google Scholar]
- 5.Iizuka N, Najita L, Franzusoff A, Sarnow P. Mol Cell Biol. 1994;14:7322–7330. doi: 10.1128/mcb.14.11.7322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tarun S Z, Sachs A B. EMBO. 1996;15:7168–7177. [PMC free article] [PubMed] [Google Scholar]
- 7.Sachs A B, Davis R W, Kornberg R D. Mol Cell Biol. 1987;7:3268–3276. doi: 10.1128/mcb.7.9.3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guthrie C, Fink G. In: Guide to Yeast Genetics and Molecular Biology. Abelson J, Simon M I, editors. Vol. 194. San Diego: Academic; 1991. [Google Scholar]
- 9.Sachs A B, Deardorff J A. Cell. 1992;70:961–973. doi: 10.1016/0092-8674(92)90246-9. [DOI] [PubMed] [Google Scholar]
- 10.Anderson J, Paddy M, Swanson M. Mol Cell Biol. 1993;136:102–112. doi: 10.1128/mcb.13.10.6102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mader S, Lee H, Pause A, Sonenberg N. Mol Cell Biol. 1995;15:4990–4997. doi: 10.1128/mcb.15.9.4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deardorff J A, Sachs A B. J Mol Biol. 1997;269:67–81. doi: 10.1006/jmbi.1997.1013. [DOI] [PubMed] [Google Scholar]
- 13.Goyer C, Altmann M, Lee H S, Blanc A, Deshmukh M, Woolford J L, Trachsel H, Sonenberg N. Mol Cell Biol. 1993;13:4860–4874. doi: 10.1128/mcb.13.8.4860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beelman C A, Parker R. Cell. 1995;81:179–183. doi: 10.1016/0092-8674(95)90326-7. [DOI] [PubMed] [Google Scholar]
- 15.Curtis D, Lehmann R, Zamore P. Cell. 1995;81:171–178. doi: 10.1016/0092-8674(95)90325-9. [DOI] [PubMed] [Google Scholar]