Abstract
Attempts to calibrate bacterial evolution have relied on the assumption that rates of molecular sequence divergence in bacteria are similar to those of higher eukaryotes, or to those of the few bacterial taxa for which ancestors can be reliably dated from ecological or geological evidence. Despite similarities in the substitution rates estimated for some lineages, comparisons of the relative rates of evolution at different classes of nucleotide sites indicate no basis for their universal application to all bacteria. However, there is evidence that bacteria have a constant genome-wide mutation rate on an evolutionary time scale but that this rate differs dramatically from the rate estimated by experimental methods.
Rates of molecular evolution in higher eukaryotes can be resolved through comparisons of homologous molecules in species for which divergence times have been inferred from fossil and geologic evidence. But without a robust fossil record, how is it possible to establish the age of bacterial species and to calibrate their rates of sequence divergence over an evolutionary time scale? In lieu of direct fossil evidence, divergence times for bacteria have been obtained by three approaches.
Age Inferred by Association with Ecological Events.
By linking the appearance of several bacterial lineages to large-scale events that occurred at known times in the geologic past, Ochman and Wilson (1, 2) constructed a time scale for bacterial evolution. Applying these dates, they calculated the absolute rates of 5S rRNA and 16S rRNA divergence; these were fairly uniform across bacterial species and similar to those observed for the homologous molecules in eukaryotes. Based on an estimated rate of 16S rRNA divergence of nearly 1% per 50 million years, they calculated that Escherichia coli and Salmonella enterica sv. Typhimurium (hereafter referred to as S. enterica)—the most closely related pair of bacterial species for which appreciable genetic information was available at the time—shared a common ancestor between 120 million and 160 million years ago. Comparisons of homologous protein-coding regions from E. coli and S. enterica indicated an average rate of sequence divergence at synonymous sites of 0.90% per million years. (Note that divergence between lineages is twice the substitution rate, and that this yields a substitution rate for E. coli and S. enterica of 0.45% per million years.) Ochman and Wilson (2) could not establish the synonymous substitution rates for bacterial taxa other than E. coli and S. enterica because of insufficient availability of protein-coding sequences for other pairs of closely related bacterial species.
Age Inferred from Host Fossil Record.
The unique association between bacterial endosymbionts and their insect hosts allowed Moran et al. (3) to estimate the dates of divergence for members of the genus Buchnera in the Proteobacteria. Buchnera live within specialized cells of aphids and are essential to the growth and reproduction of their hosts, which inherit the endosymbionts cytoplasmically. The complete concordance between the molecular phylogenies of the bacteria and their hosts indicates that strict vertical transmission has been maintained for over 100 million years and implies synchronous radiation and cospeciation of bacterial and aphid lineages during this time. Calibrating the 16S rRNA evolution of Buchnera with the fossil-based dates of divergence of their aphid hosts, Moran et al. (3) arrived at a rate of 16S rRNA divergence of 1–2% per 50 million years in these bacteria. However, evolutionary rates are accelerated in endosymbionts, presumably because their small population sizes promote the fixation of slightly deleterious mutations through drift (4, 5). As a result, 16S rRNA evolves nearly twice as fast in Buchnera as in free-living relatives based both on calibrated rates and on relative rate tests. Drift also produces an elevated substitution rate at nonsynonymous sites, which are under stronger selective constraints than are synonymous sites. Based on divergence dates derived from fossils, synonymous substitution rates for Buchnera are not more than twice the rates for E. coli/S. enterica, resulting in a ratio of synonymous to nonsynonymous substitutions in Buchnera that is much lower than that observed in E. coli/S. enterica.
Age Inferred from Eukaryotic Molecular Clocks.
Assuming that universally distributed molecules evolve at similar rates in all life forms, it is possible to reconstruct a temporal scale for bacterial evolution by extrapolating from rates calculated for organisms having reliable fossil records (e.g., vertebrate lineages). This method has been used for nearly 30 years to calibrate bacterial evolution (6–8); and in a recent application, Doolittle et al. (9) examined the amino acid sequences of 57 enzymes to obtain divergence times for lineages within and among the major kingdoms. Sequences for eight enzymes were available both for enteric bacteria (E. coli and S. enterica) and for various mammals, and sequence identities were similar for pairwise comparisons between the two enterics and between orders of mammals, averaging 94% and 95%, respectively. If one assumes that the split between E. coli and S. enterica coincided with the interordinal diversification of mammals approximately 100 million years ago, this similarity implies approximately equal substitution rates for amino acids in enterics and mammals.
However, in E. coli/S. enterica, synonymous substitutions were about 20 times more frequent than nonsynonymous substitutions, in contrast to a 5-fold difference in mammalian genes. The relative rarity of nonsynonymous substitutions in enteric bacteria is presumably because of their large population sizes, preventing fixation of slightly deleterious mutations by genetic drift. Whatever the cause, the 4-fold difference in the ratio of synonymous to nonsynonymous substitutions in enteric bacteria and in mammals implies that rate conservation across taxa cannot be true for both protein (i.e., nonsynonymous site) evolution, as presumed by Doolittle et al. (9) and Feng et al. (10), and also for synonymous site evolution, as computed by Ochman and Wilson (1, 2).
The factors that determine substitution rate, including population size, generation time, and mutation rate per generation, are known to differ between bacteria and animals. Thus, any similarities in the substitution rates of such divergent organisms would arise only when these factors happen to balance each other out. Although some genes and proteins evolve at fairly constant rates across animal taxa, there is no theoretical basis for assuming that either synonymous sites or nonsynonymous sites would manifest the same substitution rates in bacteria and eukaryotes, or even that all bacteria lineages would evolve at similar rates. For example, the smaller population sizes of mammals will accelerate substitution rates for slightly deleterious mutations, possibly counterbalancing the effects of their longer generation times, but the rate of evolution at neutral sites will be independent of population size and would be expected to differ across taxa. Indeed, many examples of rates differences among lineages have been reported (11–16).
In certain situations, e.g., where there is complete concordance between the molecular phylogenies of endosymbionts and their hosts, it has been possible to determine the relative rates of evolution for homologous genes of animals and bacteria. For corresponding pairs of aphids and their endosymbionts, which diverged synchronously, synonymous site divergences are approximately the same for symbiont and host mitochondrial genes, whereas divergences of the homologous regions of ribosomal RNA are, on average, 30-fold higher in the bacteria than in host nuclear genes (17–19). Thus, both theoretical and empirical evidence casts serious doubt on the validity of assuming universal rates of sequence evolution among bacterial groups and other organisms. Nonetheless, a more circumspect application of molecular clocks remains the most promising approach for reconstructing the timing of bacterial evolution.
On the positive side, some results do suggest that certain categories of sequence evolution are clock-like. First, under the first two procedures above, which depend on calibrating rates using aspects of bacterial ecology, the rates of sequence evolution are equivalent. Buchnera lineages and E. coli/S. enterica display rather similar rates of divergence for synonymous sites and for 16S rRNA (3–5, 19), suggesting that the absolute rates for some sequences might be approximately constant among bacterial lineages.
In addition, the proposal of Drake et al. (20–22), that DNA-based microbes (including bacteria, viruses and phage) show a constant mutation rate per genome per generation, would imply general constraints governing rates of neutral site evolution. If true, this rule could be used to calibrate absolute rates at synonymous sites by factoring out genome size and generation time of the organism being considered. However, for both synonymous site changes per year and genome changes per generation to be constant among bacterial lineages, the approximately 20-fold variation in bacterial genome size must be balanced exactly by variation in generation time, with bacteria possessing small genomes showing correspondingly slower replication rates. By comparing the relative rates of evolution at different classes of nucleotide sites, the present study reveals that the rates and patterns of substitutions vary widely among bacterial taxa. However, there is evidence that bacteria have a constant genome-wide mutation rate on an evolutionary time scale, but that this rate is not compatible with the genome-wide mutation rate estimated by Drake et al. (20–22) from laboratory studies.
Materials and Methods
To examine relative evolutionary rates over different classes of sites (synonymous, nonsynonymous, and 16S rRNA), we compared homologous sequences from species sufficiently closely related that they are not in saturation for any class of sites. Bacterial species differing by more than 5% in their 16S rRNA sequences are typically at or near saturation at synonymous sites of protein coding regions. After limiting the dataset to include only those pairs of species with 16S rRNAs that were more than more 95% identical, all available coding regions for each pair were extracted from GenBank. In the event that homologous genes were assigned different names in the sister species, homology initially was established through blast similarity searches of all of the coding regions for each species. The resulting pairs of sequences, tentatively assigned as homologs, were aligned by using sequencher 3.0 (Gene Codes, Ann Arbor, MI) and their coding frames were assigned with macclade 3.01 (23). Only those pairs of species having five or more sequenced homologs of more than 100 aa in length were considered for subsequent analysis. The phylogenetic relationships of the taxa included in this study are presented in Fig. 1. (Most of the completely sequenced bacterial genomes currently available in the databases are too distantly related to one another, and to other bacteria for which coding sequences are available, to be useful for this study.) Sequence divergence at synonymous (Ks) and nonsynonymous (Ka) sites were calculated with diverge (GCG), which uses the method of Li (24). Estimates of Ks and Ka for Buchnera spp. (5, 17) and E. coli/S. enterica (25, 26) were obtained from the literature, and values for Mycoplasma spp. (27) were supplied by A. R. Kerr (Marine Biological Laboratories, Woods Hole, MA). For genes where there is no adaptive codon usage bias, substitutions at synonymous sites are neutral, or nearly so, and, hence, reflect the actual rate of spontaneous mutation. The degree of codon usage bias is variable across bacterial taxa; for example, in Buchnera, there is effectively no codon usage bias influencing synonymous site evolution, so this factor does not affect the calculation of mutation rates in this genus (5, 28). At the other extreme, the average divergence at synonymous sites for the E. coli/S. enterica comparisons is depressed because many genes are subject to purifying selection on codon use (29–31). In E. coli, genes with low codon adaptation indices (≤0.3) probably can be considered to have Ks values that approximate the mutation rate, and the average Ks for a set of low biased genes is only 10% higher than the overall average Ks value applied in the present study.
Figure 1.
Phylogenetic tree of bacterial taxa used in this study based on 16S rRNA sequences. Only terminal branch lengths are proportional to sequence divergence. Colors indicate chromosomal G+C content as follows: green, <45% G+C; red, 45–55% G+C; blue, >55% G+C.
For species pairs whose Ks and Ka values are not available from the literature, the following genes (and their corresponding accession numbers) were used in our analysis: Aeromonas hydrophila, exeCDE (X66504), oriC (X89469), pilC (U20255) tapABCD (U20255); Aeromonas salmonicida, exeCDE (X80505), oriC (U65741), pilC (U95640), tapAB (AF059248), tapC (AF059249), tapD (AF059250); Bacillus amyloliquefaciens, alkaline protease (E03694), neutral protease (K02497), penP (Z35653), ribBGH (X95955), thyA (AF004104); Bacillus subtilis, alkaline protease (E00328), neutral protease (E04119), penP (Z35652), ribBGH (X51510), thyA (AF004102); Borrelia afzelii, bmpA (X97241), bmpB (X81519), nlpH (Y08410), fla (X75202), hbb (U48676), ospA (U78301), recA (X87726); Borrelia burgdorferi, bmpB (X81517), ospA (U96253), fla (U96239), hbb (U48683), ospA (U96253); Borrelia garinii, bmpA (X97243), nlpH (Y08413), ospA (X80252), recA (X87727); Mycobacterium leprae, ctaE (Al022602), cysE (Z98741), eno (recovered by blast), folP (AL023093), lipB (Z98741), qcrC (AL022602), sucB (Z98741); Mycobacterium tuberculosis, ctaE (Z70283), cysE (Z83860), eno (Z92539), folP (Z95557), lipB (Z70283), qcrC (Z70283), sucB (Z70283); Neisseria gonorrhoeae, adk (I36471), argF (M34930), aroE (L47159), penA (X59632), por (U17025); Neisseria polysaccharea, adk (U57708), argF (X64870), aroE (U82844), penA (X59626), por (Y09309); Pseudomonas aeruginosa, catA (AF047025), catechol (D83057), cheZ (D373810), himD (L35258), himA (L35259), pahABC (D84146), pyrB (L19649), recA (X05691), rpoS (D26134), rpoA (D90118); Pseudomonas putida, catA (U63511), catechol (D77856), cheZ (AF031898), himD (U5690), himA (U5690), pahABC (M23914), pyrB (M97253), recA (L12684), rpoS (X91654), rpoA (D30045); Rhodobacter capsulatus, bchC (M29966), ccoO (X80134), dorC (U49506), hupL (X13529), rpoH (AF017436), cbbI (U87282); Rhodobacter sphaeroides, bchC (X68795), ccoO (U58092), dorC (AF016236), hupL (L37194), rpoH (U82397), cbbI (J02922); Thermus aquaticus, aspC (AF025665), taqI (M74796), msoD (D84646), fum (D84646), mutS (U33117), recA (I20095), leuB (D10700); Thermus thermophilus, aspC (D38459), taqI (M74795), msoD (AB010884), fum (AB010884), mutS (D63810), recA (D17392), leuB (AB017135). In the case of Heliocobacter pylori, we selected 50 genes at random from the complete genomic sequences of strains J99 (AE001439) and 26695 (AE000511); and for recA comparisons, we also included the recA homologues from the following pairs of species: E. coli (X55552) and S. enterica (recovered by blast), Bifidobacterium animalis (U50266) and B. breve (U50268), B. adolescentitus (U50265) and B. longum (U50270), Paracoccus denitrificans (U59631) and Rhodobacter capsulatus (X82183), Neisseria elongata (AJ223877) and N. flavescens (U57907), Neisseria animalis (U57910) and N. cinera (U57906), Streptococcus pyogenes (U21934) and S. mutans (M61897).
Results and Discussion
Time-Independent Comparisons of Evolutionary Rates in Bacteria.
What is the potential for using molecular sequences to infer a time scale for bacteria evolution? Even though there are very few external reference points (such as those available for Buchnera and their aphid hosts) for calculating the substitution rates in bacteria, we can address the question by determining whether bacteria as a whole are internally consistent with respect to their relative rates of evolution at different classes of sites. For example, are ratios of synonymous and 16S rRNA divergence constant, and do homologous sequences display the same ratios of synonymous to nonsynonymous changes, across pairs of related taxa? If 16S rRNA and synonymous sites can serve as alternative molecular clocks in bacteria, as presumed by various authors, then the relative divergence for these two categories of sites should be fairly constant for different pairs of species. Such comparisons can be made even when divergence dates are unavailable or uncertain and can illuminate the usefulness of calibrating substitution rates as a way of reconstructing the timing of bacterial evolution.
Relative rates of synonymous site and 16S rRNA divergence.
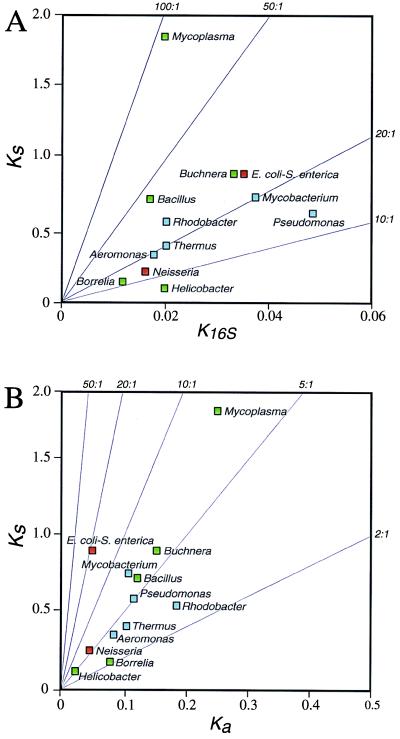
Our studies of enteric bacteria and Buchnera species yielded ratios of synonymous site (Ks) to 16S rRNA (K16S) divergence of approximately 25:1, and other species pairs (Rhodobacter, Mycobacterium) manifest similar ratios of substitutions at these sites (Fig. 2A). In contrast, Bacillus spp. and Mycoplasma spp. display substantially higher relative numbers of synonymous substitutions; and, in fact, in comparisons of all homologous protein coding regions from the Mycoplasma pneumoniae and M. genitalium genomes, synonymous sites are well into saturation despite only 1.8% divergence in their small subunit rRNAs (27). On the other hand, the relative rate of synonymous site and 16S rRNA evolution in the Borrelia, Thermus, Pseudomonas, and Mycobacterium comparisons are about half of that computed for the E. coli/S. enterica comparison, with the lowest Ks/K16S ratio estimated for strains of H. pylori.
Figure 2.
Relative extent of sequence divergence among pairs of bacterial species. Only pairs of species that differ by less than 5% in 16S rRNA sequences and for which sequence information is available for at least five homologous genes are included. Colors correspond to chromosomal G+C contents as in Fig. 1. (A) Divergence at synonymous sites (Ks) vs. divergence at 16S rRNA (K16S). (B) Divergence at synonymous (Ks) vs. nonsynonymous (Ka) sites.
There is no systematic explanation for the differences among organisms in their relative rates of evolution at these two classes of sites. The variation among species in the base composition of 16S rRNAs is very narrow (ranging from 45% GC for the 16S rRNA of Mycoplasma to 55% GC for the 16S rRNA of Thermus) relative to that at synonymous sites (ranging from 10% GC for synonymous sites in Mycoplasma to 90% GC in Thermus). Therefore, the observed Ks to K16S ratios of organisms with extreme base compositions could be depressed because of the fact that divergence at synonymous sites in these genomes are underestimated whereas their 16S rRNAs evolve at some universal rate (see subsequent section for calibration of 16S rRNA). However, genomic base composition is not the major source of the observed variation in evolutionary rates among taxa: species pairs having low GC contents can display very high (Mycoplasma), moderate (Bacillus, Buchnera), or very low (Borrelia, Helicobacter) Ks to K16S ratios. Similarly, there is no obvious relationship between codon biases, bacterial ecology (i.e., free-living, host associated, or intracellular), species phylogenetic position, or gene sample and Ks to K16S ratios. The very low Ks/K16S value observed in Helicobacter pylori could result from usually high levels of gene exchange among strains (32), which could either reduce the relative amount of neutral variation or homogenize 16S rRNA within the species, or from an increase in occurrence of slightly deleterious mutations within populations.
Relative rates of synonymous and nonsynonymous site divergence.
Based on comparisons of homologous genes from E. coli and Salmonella, nonsynonymous substitutions were found to accumulate about four times more slowly in enteric bacteria than in mammals (1, 2). A possible basis for this difference is the relatively large effective population sizes of bacteria, which should slow the fixation of slightly deleterious mutations through genetic drift. The low Ks/Ka ratio of Buchnera genes has been explained as the result of small effective population size in endosymbiotic bacteria (4), with the implication that the E. coli/S. enterica ratios are more typical for bacteria. However, as presented in Fig. 2B, the average Ks/Ka ratio for the E. coli/S. enterica comparison is clearly atypically high. Although Ks/Ka ratios are influenced by effective population sizes, which have not been estimated for most bacterial species, the population sizes of E. coli are probably no larger than that of other free-living species (such as Bacillus) displaying low Ks/Ka values.
Because the functional constraints at nonsynonymous sites vary among genes, the observed variation in Ks/Ka ratios could result from different loci being compared for the different species pairs. In addition, some genes show pronounced codon bias in certain taxa but not in others, and because codon bias reduces Ks values, this factor can dramatically affect Ks/Ka ratios. For example, the universally distributed gene recA is under strong codon usage bias in both E. coli and S. enterica (codon adaptation index = 0.6). The Ks/Ka ratio of the recA homologs for the E. coli/S. enterica comparison is about 4:1, which is lower than the Ks/Ka ratio for recA for any of the other pairs of species considered and sharply contrasts the situation for the pooled sets of genes. Thus, the choice of locus can affect Ks/Ka ratios differentially across taxa, and one cause of such effects is the strength of codon bias, which varies among lineages. Full exploration of the constancy of evolutionary rates at different classes of sites requires nucleotide sequences for several sets of homologous genes common to all taxa, and, currently, such data do not exist.
Estimating the Actual Rate of Molecular Evolution in Bacteria.
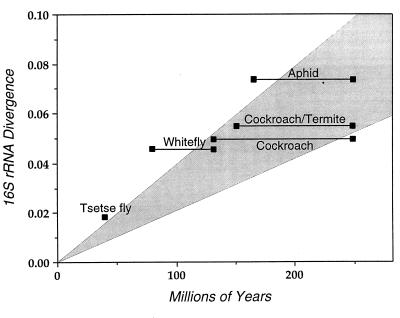
Over the past few years, sequence information has become available for several bacterial endosymbionts whose divergence times can be inferred from the fossil records of their insect hosts (3, 33–36). Although these calculations are limited to bacterial endosymbionts, and to small subunit ribosomal RNAs (aside from Buchnera, only limited information is available on the protein coding sequences for other species pairs), the rates of 16S sequence divergence in these bacteria are fairly similar to those reported for Buchnera and E. coli/S. enterica (Fig. 3). Because Ks/K16S ratios vary widely among taxa (Fig. 2A), the relative constancy in the overall rate of 16S rRNA sequence divergence implies that synonymous substitution rates are highly variable across bacterial species. Therefore, the rates of synonymous site evolution determined for E. coli/S. enterica and Buchnera spp. cannot be extrapolated to other bacteria. The application of these rates could lead to estimates of divergence times that are off by an order of magnitude.
Figure 3.
Rates of 16S rRNA sequence divergence in bacterial endosymbionts of insects. Divergence times are estimated from fossil evidence of insect hosts and were obtained from the literature (3, 33–36). Shaded area denotes a rate of 16S rRNA divergence of 1–2% per 50 million years. Insect and bacterial taxa are follows (host/symbiont): aphids (Schizaphis graminum/Buchnera aphidicola, Schlechtendalia chinesis/Buchnera aphidicola); cockroach-termite (Blattaria germanica/symbiont, Mastotermes darwiniensis/symbiont); cockroach (Blattaria germanica/symbiont, Periplaneta americana/symbiont); whitefly (Trialeurodes vaporariorum/P-endosymbiont, Siphoninus phillyreae/P-endosymbiont); tsetse fly (Glossinia brevipalis/Wigglesworthia glossinidia, Glossinia morsitans/Wigglesworthia glossinidia).
A Universal Genomic Mutation Rate?
Because synonymous sites are under little or no selective constraint, synonymous substitution rates should reflect the rate of mutation. Based on mutation frequencies measured by experimental methods in a few taxa, Drake et al. (20–22) have proposed that the genomic mutation rate is approximately constant across DNA-based microbes. This observation might be in accord with the observation of variation in synonymous substitution rates among bacteria because microbes with smaller genomes (or shorter generation times) would need to have proportionately lower numbers of mutations to yield the same genome-wide rate. The only bacterial species considered by Drake et al. (20–22) was E. coli, for which estimates of mutation rates were determined by scoring the inactivation or reversion of phenotypic markers.
Are the mutation rates based on sequence comparisons in agreement with those determined from laboratory studies? E. coli and S. enterica diverge at a synonymous substitution rate of 0.90% per million years (25, 26), which corresponds to a within lineage rate of 0.0045 mutations per site per million years. Natural populations of E. coli have an estimated 100–300 generations per year (37–39), implying a rate of about 0.0001 to 0.0002 mutations per genome per generation (Table 1). Drake et al.’s estimate (20–22), based on laboratory-derived mutation rates, was 0.003 mutations per genome, more than an order of magnitude higher. Similar calculations for Buchnera, whose generation times can be estimated from the number of replications within the host insect and the number of host generations per year (19, 40–42), result in 0.0001 to 0.0002 mutations per genome per generation, a rate identical to that estimated for E. coli. [Note that E. coli would need to replicate fewer than 10 times per year, and Buchnera fewer than twice per year to attain the genomic mutation rate computed by Drake et al. (20–22).] Hence, the genomic mutation rate may well be constant across bacterial species on an evolutionary time scale, but it is not compatible with the rate based on laboratory studies.
Table 1.
Genomic substitution rates (U) per generation in bacteria
| Species | Genome size, bp | Substitution rate* | Generations per year† | U‡ |
|---|---|---|---|---|
| E. coli | 4.7 × 106 | 4.5 × 10−9 | 100–300 | 0.0001–0.0002 |
| Buchnera | 6.6 × 105 | 8.2 × 10−9 | 30–50 | 0.0001–0.0002 |
Two factors could contribute to the disparity between the laboratory-derived genomic mutation rate and those obtained from sequence comparisons. The first is that nucleotide site divergences must be estimated correctly. This could become a problem when homologous genes are in, or near, saturation, or when base composition is extreme, but even in such cases, the estimated divergences are not more than 50% different from the actual values. The next obstacle is adaptive codon usage bias, which could yield substitution rates at synonymous sites that are lower than the actual rate of mutation, because of purifying selection at these sites. As discussed previously, the effects of codon bias probably has caused us to underestimate the synonymous substitution rate by 10% in E. coli. Cumulatively, the effects of multiple hit corrections and codon usage bias could potentially increase the estimated mutation rate by, at most, 2-fold, which is not sufficient to account for the 30-fold difference between our results and those of Drake et al. (20, 22). It is more likely that measurements of mutation rates based on laboratory culture are elevated and do not reflect the actual, long-term rate of mutation.
The correspondence of genomic mutation rate values for Buchnera and enterics is remarkable in view of their differences in genome size and generation time. The possibility of a constant genomic mutation rate should be further evaluated in other bacterial taxa. The implications of such a pattern, if true, extend well beyond its application to calibrating bacterial evolution.
To further explore the rates and patterns of molecular evolution in bacteria, it will be necessary to examine the sequences of conserved genes among pairs of closely related bacterial species. Data of this sort are clearly lacking, and the availability of complete genomic sequences has only begun to help us resolve these issues. In the next few years, complete sequences of a large number of bacterial genomes will become available, thus creating unprecedented potential for reconstructing the evolutionary history of prokaryotes.
References
- 1.Ochman H, Wilson A C. In: Escherichia coli and Salmonella typhimurium: Molecular and Cellular Aspects, Neidhardt F C, Ingraham J L, Low K B, Magasanik B, Schaechter M, Umbarger H E, editors. Washington, DC: Am. Soc. Microbiol.; 1987. pp. 1649–1654. [Google Scholar]
- 2.Ochman H, Wilson A C. J Mol Evol. 1987;26:74–86. doi: 10.1007/BF02111283. [DOI] [PubMed] [Google Scholar]
- 3.Moran N A, Munson M A, Baumann P, Ishikawa H. Proc R Soc London Ser B. 1993;253:167–171. [Google Scholar]
- 4.Moran N A. Proc Natl Acad Sci USA. 1996;93:2873–2878. doi: 10.1073/pnas.93.7.2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wernegreen J J, Moran N A. Mol Biol Evol. 1999;16:83–97. doi: 10.1093/oxfordjournals.molbev.a026040. [DOI] [PubMed] [Google Scholar]
- 6.McLaughlin P J, Dayhoff M O. Science. 1970;168:1469–1470. doi: 10.1126/science.168.3938.1469. [DOI] [PubMed] [Google Scholar]
- 7.Kimura M, Ohta T. Nat New Biol. 1973;243:199–200. doi: 10.1038/newbio243199a0. [DOI] [PubMed] [Google Scholar]
- 8.Hori H, Osawa M. Proc Natl Acad Sci USA. 1979;76:381–385. doi: 10.1073/pnas.76.1.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doolittle R F, Feng D F, Tsang S. Science. 1996;271:470–477. doi: 10.1126/science.271.5248.470. [DOI] [PubMed] [Google Scholar]
- 10.Feng D F, Cho G, Doolittle R F. Proc Natl Acad Sci USA. 1997;94:13028–13033. doi: 10.1073/pnas.94.24.13028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Britten R J. Science. 1986;231:1393–1398. doi: 10.1126/science.3082006. [DOI] [PubMed] [Google Scholar]
- 12.Wu C-I, Li W-H. Proc Natl Acad Sci USA. 1985;82:1741–1745. doi: 10.1073/pnas.82.6.1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bousquet J, Strauss S H, Doerksen A H, Price R A. Proc Natl Acad Sci USA. 1992;89:7844–7848. doi: 10.1073/pnas.89.16.7844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martin A P, Palumbi S R. Proc Natl Acad Sci USA. 1993;90:4087–4091. doi: 10.1073/pnas.90.9.4087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ohta T. J Mol Evol. 1995;40:56–63. doi: 10.1007/BF00166595. [DOI] [PubMed] [Google Scholar]
- 16.Li W-H. Curr Opin Gen Dev. 1993;3:896–901. doi: 10.1016/0959-437x(93)90011-d. [DOI] [PubMed] [Google Scholar]
- 17.Rouhbakhsh D, Lai C-Y, von Dohlen C D, Baumann L, Baumann P, Moran N A, Voegtlin D J. J Mol Evol. 1996;42:414–421. doi: 10.1007/BF02498635. [DOI] [PubMed] [Google Scholar]
- 18.Moran N A, von Dohlen C D, Baumann P. J Mol Evol. 1995;41:727–731. doi: 10.1007/BF00170675. [DOI] [PubMed] [Google Scholar]
- 19.Clark, M. A., Baumann, P. & Moran, N. A. (1999) Mol. Biol. Evol., in press. [DOI] [PubMed]
- 20.Drake J W. Proc Natl Acad Sci USA. 1991;88:7160–7164. doi: 10.1073/pnas.88.16.7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Drake J W. Proc Natl Acad Sci USA. 1993;90:4171–4175. doi: 10.1073/pnas.90.9.4171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Drake J W, Charlesworth B, Charlesworth D, Crow J F. Genetics. 1998;148:1667–1686. doi: 10.1093/genetics/148.4.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maddison W P, Maddison D R. macclade 3.0: Analysis of Phylogeny and Character Evolution. Sunderland, MA: Sinauer; 1992. [DOI] [PubMed] [Google Scholar]
- 24.Li W-H. J Mol Evol. 1993;36:96–99. doi: 10.1007/BF02407308. [DOI] [PubMed] [Google Scholar]
- 25.Sharp P M. J Mol Evol. 1991;33:23–33. doi: 10.1007/BF02100192. [DOI] [PubMed] [Google Scholar]
- 26.Lawrence J G, Ochman H. J Mol Evol. 1997;44:383–397. doi: 10.1007/pl00006158. [DOI] [PubMed] [Google Scholar]
- 27.Kerr A R, Peden J F, Sharp P M. Mol Microbiol. 1997;6:1177–1179. doi: 10.1046/j.1365-2958.1997.5461902.x. [DOI] [PubMed] [Google Scholar]
- 28.Brynnel E A, Kurland C G, Andersson S G E, Moran N A. Mol Biol Evol. 1998;15:574–582. doi: 10.1093/oxfordjournals.molbev.a025958. [DOI] [PubMed] [Google Scholar]
- 29.Sharp P M, Li W-H. Nucleic Acids Res. 1987;15:1281–1295. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eyre-Walker A, Bulmer M. Genetics. 1995;140:1407–1412. doi: 10.1093/genetics/140.4.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maynard Smith J, Smith N H. Genetics. 1996;142:1033–1036. doi: 10.1093/genetics/142.3.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suerbaum S, Smith J M, Bapumia K, Morelli G, Smith N H, Kunstmann E, Dyrek I, Achtman M. Proc Natl Acad Sci USA. 1998;95:12619–12624. doi: 10.1073/pnas.95.21.12619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Askoy S, Pourhosseni A A, Chow A. Insect Mol Biol. 1995;4:15–22. doi: 10.1111/j.1365-2583.1995.tb00003.x. [DOI] [PubMed] [Google Scholar]
- 34.Bandi C, Sironi M, Damiani G, Margrassi L, Nalepa C A, Laudani U, Sacchi L. Proc R Soc London Ser B. 1995;259:293–299. doi: 10.1098/rspb.1995.0043. [DOI] [PubMed] [Google Scholar]
- 35.Clark M A, Baumann L, Munson M A, Baumann P, Campbell B C, Duffus J E, Osborne J S, Moran N A. Curr Microbiol. 1992;25:119–123. [Google Scholar]
- 36.Munson M A, Baumann P, Clark M A, Baumann L, Moran N A, Voegtlin D J. J Bacteriol. 1991;173:6321–6334. doi: 10.1128/jb.173.20.6321-6324.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Savageau M A. Am Nat. 1983;122:732–744. [Google Scholar]
- 38.Gibbons R J, Kapsimalis B. J Bacteriol. 1967;93:510–512. doi: 10.1128/jb.93.1.510-512.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guttman D S, Dykhuizen D E. Science. 1994;266:1380–1383. doi: 10.1126/science.7973728. [DOI] [PubMed] [Google Scholar]
- 40.Baumann L, Baumann P. Appl Environ Microbiol. 1994;60:3440–3443. doi: 10.1128/aem.60.9.3440-3443.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moran N A. Annu Rev Entomol. 1992;37:321–348. [Google Scholar]
- 42.Hales D F, Tomiuk J, Wohrman K, Sunnucks P. Eur J Entomol. 1997;94:1–55. [Google Scholar]