Abstract
Two human cDNAs that encode novel vitamin K-dependent proteins have been cloned and sequenced. The predicted amino acid sequences suggest that both are single-pass transmembrane proteins with amino-terminal γ-carboxyglutamic acid-containing domains preceded by the typical propeptide sequences required for posttranslational γ-carboxylation of glutamic acid residues. The polypeptides, with deduced molecular masses of 23 and 17 kDa, are proline-rich within their putative cytoplasmic domains and contain several copies of the sequences PPXY and PXXP, motifs found in a variety of signaling and cytoskeletal proteins. Accordingly, these two proteins have been called proline-rich Gla proteins (PRGP1 and PRGP2). Unlike the γ-carboxyglutamic acid domain-containing proteins of the blood coagulation cascade, the two PRGPs are expressed in a variety of extrahepatic tissues, with PRGP1 and PRGP2 most abundantly expressed in the spinal cord and thyroid, respectively, among those tissues tested. Thus, these observations suggest a novel physiological role for these two new members of the vitamin K-dependent family of proteins.
Keywords: cDNA cloning, cDNA sequence, γ-carboxyglutamic acid, vitamin K
Since the existence of a lipid-soluble antihemorrhagic vitamin was reported by Dam in 1935 (1), the molecular basis for the function of vitamin K in hemostasis has come to be understood in great detail. The identification of γ-carboxyglutamic acid (Gla) residues in bovine prothrombin (2–4) but not in the prothrombin of animals treated with the anticoagulant and vitamin K antagonist dicoumarol (2) clarified the role of vitamin K as a cofactor in the posttranslational γ-carboxylation of selected prothrombin glutamyl residues. Homologous domains with 9–12 Gla residues within the amino-terminal 48 residues have been identified in a number of circulating plasma glycoproteins. The coordination of Ca2+ by several of these Gla residues is required for proper conformation of the Gla domain (5–7) and allows Ca2+-dependent binding of the coagulation factors to anionic phospholipid membrane surfaces at sites of vascular injury (for reviews, see refs. 8–10).
In addition to the four classical vitamin K-dependent coagulation factors, namely, prothrombin and factors VII, IX and X, this protein family includes the anticoagulant factors, proteins C and S, as well as protein Z, a plasma glycoprotein of unknown function. A recent addition to this family is Gas6, a protein expressed in response to serum starvation of cultured cells (11, 12). This vitamin K-dependent protein has been alternatively described as a growth-potentiating factor (13, 14), a cell survival factor (15), or both (16). Unlike other Gla proteins, Gas6 is expressed in a variety of extrahepatic tissues (12) and no role in coagulation has yet been ascribed to it. Rather, Gas6 has been identified as a ligand for the receptor tyrosine kinases Axl (17–19), Rse (alternatively, Sky, Tyro3, Brt, or Tif) (18–20), and Mer (19).
Specific glutamic acid residues within the Gla domains of these proteins are posttranslationally modified by a vitamin K-dependent γ-carboxylase located in the rough endoplasmic reticulum. This reaction requires a γ-carboxylation recognition sequence contained within a propeptide that is flanked by a signal peptide and the amino-terminal domain of the mature protein where the γ-carboxylation occurs.
Given the functional importance of Gla domains in the coagulation factors and gas6, we attempted to identify cDNAs encoding novel Gla domain-containing proteins by searching the dbEST database (21) with a protein query sequence designed from an alignment of all known Gla domain sequences.
Herein, we report the cloning of two cDNAs encoding novel Gla domain-containing proteins. Analysis of the deduced amino acid sequence suggests that these proteins are integral membrane proteins with proline-rich cytoplasmic regions. These proline-rich regions contain the potential WW domain-binding motif, PPXY (22). The WW domain (for reviews, see refs. 23 and 24) is a recent addition to a family of protein modules that include Src homology (SH) 2, SH3, and pleckstrin homology (PH) domains (for reviews, see refs. 25 and 26). Moreover, these proline-rich regions contain several copies of the sequence PXXP, an SH3 domain-binding motif. The established importance of SH3 domain interactions and the emerging significance of WW domain interactions in various cytoskeletal components and signaling molecules suggests potential roles for these two newly identified Gla proteins.
MATERIALS AND METHODS
Expressed Sequence Tag (EST) Database Searches.
The EST database dbEST (21) at the National Center for Biotechnology Information was searched by using the specialized blast algorithm (27) tblastn. The amino acid query sequence LEEXXXXXLERECXEEXCXXEEARE was derived by alignment of all known human Gla domain sequences.
Cloning of Proline-Rich Gla Proteins PRGP1 and PRGP2 cDNAs.
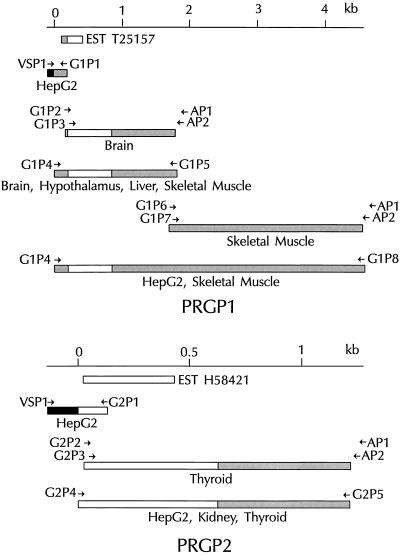
PCR (28) was performed with a Perkin–Elmer/Cetus DNA thermal cycler and the thermostable DNA polymerases Taq (Boehringer Mannheim), KlenTaq (CLONTECH), or Tth (CLONTECH) according to the manufacturer’s instructions. Oligonucleotide primers were designed based on the nucleotide sequences of ESTs. These were used to amplify cDNA fragments from either a human hepatoma cell (HepG2) cDNA plasmid (pPC86) library by conventional PCR or from a variety of Marathon-ready cDNA libraries (CLONTECH) by rapid amplification of cDNA ends (RACE) (29). RACE PCRs were optimized by using a “touchdown” thermal cycling program (30) as well as sequential nested-primer reactions. PCR products were cloned (TA cloning kit, Invitrogen) and manually sequenced using the dideoxynucleotide chain-termination method (Sequenase kit, United States Biochemical) (31). PCR primers were then synthesized based on the extreme 5′ and 3′ ends of newly identified overlapping sequences and used to amplify contiguous cDNAs. In all cases, a minimum of four independent clones were sequenced bidirectionally to ensure the absence of PCR-generated mutations. The strategies used to clone PRGP1 and PRGP2 cDNAs are shown schematically in Fig. 1. The sequences of primers used are listed in Table 1.
Figure 1.
Cloning of PRGP1 (Upper) and PRGP2 (Lower) cDNAs. Overlapping fragments of the cDNAs were generated by the indicated oligonucleotide primers by using PCR. Relative positions of ESTs and their respective accession numbers are indicated. Coding sequences, open bars; 5′ and 3′ noncoding sequences, shaded bars; sequence derived from the plasmid pPC86, solid bars. Origins of each cDNA fragment are indicated below the fragment.
Table 1.
Oligonucleotide primers used in this study
| Primer | Sequence |
|---|---|
| VSP1 | 5′-TGGACGGACCAAACTGCGTATAACGCGTTTGGAATC-3′ |
| G1P1 | 5′-TAGCGTTTTAATATGGAATTGGCTTTTTCTCCCGTGAG-3′ |
| G1P2 | 5′-ACGGGAGAAAAAGCCAATTCC-3′ |
| AP1 | 5′-CCATCCTAATACGACTCACTATAGGGC-3′ |
| G1P3 | 5′-GCCAATTCCATATTAAAACGCTAC-3′ |
| AP2 | 5′-ACTCACTATAGGGCTCGAGCGGC-3′ |
| G1P4 | 5′-TGATGCTCACCCCATTTAGAGAAAG-3′ |
| G1P5 | 5′-CTTAAGGGCTTGAAAATTCTGTGGGAG-3′ |
| G1P6 | 5′-TGGATATATGTGTGGATTAATGACAGGCAG-3′ |
| G1P7 | 5′-CCATTACTCCTTTACTCATAGCTGGTAAAATTATTCCC-3′ |
| G1P8 | 5′-TAGCATACCAAAAACACAGAAACATAAGAAATACCC-3′ |
| G2P1 | 5′-TGGCTACTCAGGAAGCTCTGGGCCTCTGGGGGACCCAG-3′ |
| G2P2 | 5′-CTGCTATATATGGCATTAACCACCTGCCT-3′ |
| G2P3 | 5′-CTGGGTCCCCCAGAGGCCCAGAGCTTCCTGAGTAGCCA-3′ |
| G2P4 | 5′-GTGGAAAATATGAGGGGCCAC-3′ |
| G2P5 | 5′-TTTTTATTTAGGGGAACAGCTCAACTCCAG-3′ |
Primers are listed in the order in which they appear in Fig. 1. G1 primers were used to amplify PRGP1. G2 primers were used for PRGP2. VSP1 is specific for the plasmid pPC86. AP1 and AP2 are adaptor-specific primers used in RACE reactions.
Northern Blot Analysis.
A 1.4-kb HindIII–EcoRI fragment of PRGP1 cDNA and an 850-bp BglI–EcoRI fragment of PRGP2 cDNA were generated by restriction digestion of the plasmid pCR2 (TA cloning kit, Invitrogen) harboring the appropriate cDNA insert and subsequent purification of fragments from agarose gels (Qiaex, Qiagen, Chatsworth, CA). These and β-actin cDNA (CLONTECH) were radio-labeled with [α-32P]dCTP (6000 Ci/mmol; 1 Ci = 37 GBq; Amersham) by random priming (Prime-It II kit, Stratagene) to a specific activity of 109 cpm/μg. Human multiple-tissue Northern blots (CLONTECH) representing 23 different human tissues were prehybridized and hybridized with ExpressHyb solution (CLONTECH) according to the manufacturer’s instructions with a final probe concentration of 106 cpm/ml. The blots were washed for 1 hr at room temperature in 2× standard saline citrate/0.05% SDS and for 1 hr at 65°C in 0.1× standard saline citrate/0.1% SDS and visualized by using a Molecular Dynamics PhosphorImager SF after overnight exposure.
RESULTS AND DISCUSSION
Cloning of PRGP1 cDNA.
A search of the dbEST database with an amino acid query sequence derived from a highly conserved region of all known Gla domains identified EST T25157, which was derived from a human colorectal tumor library (32). Identification of this EST allowed the subsequent cloning of the PRGP1 cDNA (Fig. 1). An oligonucleotide primer based on the EST sequence was used to amplify the 5′ end of the PRGP1 cDNA from a HepG2 cell library by conventional PCR methodology. The 3′ end of the cDNA was amplified by PCR using the technique of RACE from a brain poly(A)+ cDNA library, yielding a contiguous sequence of 1.8 kb with a 3′ polyadenylation consensus sequence followed by a poly(A) tail. Human multiple-tissue Northern blots (see below) using a radiolabeled probe derived from this sequence hybridized with transcripts of approximately 4.6 kb, considerably larger than the expected size of 1.8 kb. This discrepancy in size was clarified by additional 3′ RACE reactions using primers derived from the region of the original cDNA proximal to the polyadenylation site and a skeletal muscle poly(A)+ cDNA library. These PCRs yielded an additional 2.8 kb of 3′ untranslated sequence with the expected polyadenylation consensus sequence followed by a poly(A) tail. A contiguous 4.5-kb cDNA sequence encompassing all previously identified PRGP1 cDNA fragments was amplified from HepG2 and skeletal muscle cDNA libraries and the nucleotide sequence was determined (Fig. 2). Since the 1.8-kb cDNA sequence was identical with the corresponding region of the 4.5-kb cDNA, it was concluded that the different cDNA lengths result from differential usage of polyadenylation sites rather than from differential mRNA splicing. Moreover, since no transcript corresponding to the 1.8-kb band was observed on Northern blots (data not shown), the 4.5-kb transcript was the predominant species.
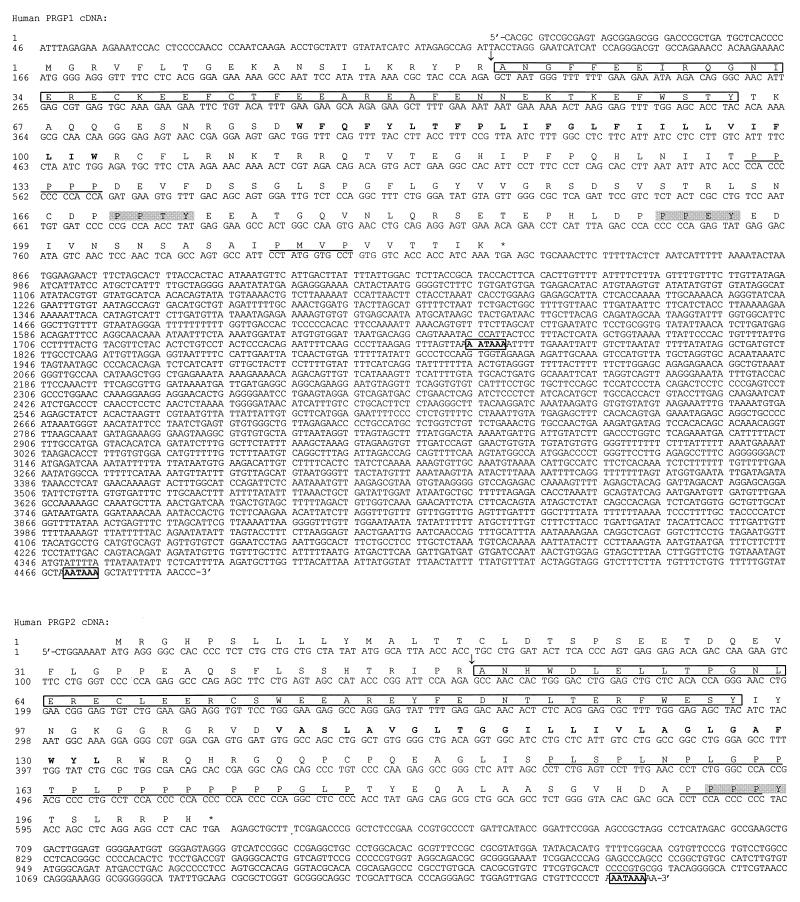
Figure 2.
Nucleotide and predicted amino acid sequence of PRGP1 and PRGP2. Proposed propeptidase cleavage sites are indicated by ↓. Gla domains are outlined. Putative transmembrane regions are shown in boldface type. Potential WW domain interaction motifs (PPXY) are shaded in gray. Potential SH3 domain interaction motifs (PXXP) are underlined. In-frame stop codons are denoted with asterisks. Polyadenylation signals are shown in boldface type and are boxed.
Cloning of PRGP2 cDNA.
The dbEST database search identified a second potential Gla protein sequence, EST H58421, derived from a human fetal liver/spleen library. Oligonucleotide primers based on this sequence were used to clone the PRGP2 cDNA (Fig. 1). The 5′ and 3′ ends of the cDNA were amplified from HepG2 and thyroid cDNA libraries, respectively, and a contiguous 1.2-kb sequence was subsequently amplified from HepG2, kidney, and thyroid cDNA libraries and sequenced (Fig. 2). As in the case of PRGP1, the 3′ RACE product was amplified from a poly(A)+ cDNA library and contained both a polyadenylation sequence and a poly(A) tail.
Sequence Analysis of PRGP1 and PRGP2.
The PRGP1 cDNA contained a 165-bp 5′ untranslated region (UTR), a 657-bp coding sequence, and an unusually long 3.7-kb 3′ UTR. The mature protein of 198 amino acid residues has a deduced molecular mass of 23 kDa after cleavage of the 20-amino acid propeptide. This cleavage site was predicted by alignment of propeptide arginine residues with the −1 and −4 arginine residues of the other Gla domain-containing proteins (Fig. 3). The first methionine codon is encountered at nucleotide 166, immediately proximal to the propeptide-encoding sequence, and is preceded by an in-frame stop codon at nucleotide 49. Therefore, unlike all other Gla domain-containing proteins, PRGP1 lacks a discernible signal peptide. A putative transmembrane region (residues 58 through 83) within the mature protein could, therefore, act as a “signal anchor” to direct the nascent polypeptide to the endoplasmic reticulum lumen. The orientation of the protein within the membrane is largely determined by the relative charges of the residues flanking the transmembrane segment, with the cytoplasmic sequence generally carrying the greater positive charge (33). PRGP1 has relatively more positive charges on the carboxyl-terminal side of the putative transmembrane region and would, therefore, be expected to orient with the carboxyl terminus within the cytoplasm. Such an orientation is consistent with the fact that glutamic acid residues within amino-terminal Gla domains are γ-carboxylated by the endoplasmic reticulum-resident carboxylase. In addition, a disulfide loop within the Gla domain would be expected to form correctly within the oxidizing environment of the endoplasmic reticulum but not within the reducing milieu of the cytoplasm.
Figure 3.
Amino acid sequence alignment of the signal/propeptide and Gla domain regions of known proteins with PRGP1 and PRGP2 deduced amino acid sequences. Highly conserved residues are shaded. Strictly conserved residues are boxed. Highly conserved residues within the propeptide implicated in γ-carboxylation are denoted with an asterisk. Positions at which γ-carboxylation of glutamic acid residues is either known to occur or may occur are indicated by γ. The propeptidase cleavage site/amino terminus of mature protein is indicated with a ↓. The position of the disulfide loop within the Gla domain is also indicated. The query sequence used to search the dbEST database is shown on the bottom line. ProS, protein S; ProC, protein C; ProZ, protein Z; rACNG-RF2, conceptual translation in reading frame 2 of the cDNA encoding the rabbit aortic cyclic nucleotide-gated channel.
The PRGP2 cDNA possesses a short (9 bp) 5′ UTR, a 609-bp coding sequence, and a 548-bp 3′ UTR. The mature protein of 153 amino acid residues has a deduced molecular mass of 17 kDa after cleavage of the 49 residue signal/propeptide. The presence of an amino-terminal signal peptide and a putative transmembrane region defines PRGP2 as a type I single-pass transmembrane protein with the carboxyl terminus oriented to the cytoplasm. As is the case with PRGP1, the PRGP2 propeptide cleavage site can be predicted by alignment of arginine residues within the propeptide with arginine residues within known cleavage sites of the other Gla domain-containing proteins (Fig. 3).
The putative cytoplasmic regions of PRGP1 and PRGP2 are uncharacteristically proline-rich. The proline-containing motifs PPXY and PXXP are found in proteins that interact with WW domains and SH3 domains, respectively. These are modules common to proteins involved in signal transduction and cytoskeletal interactions (22, 25, 26). The amino acid sequence of PRGP1 contains 2 PPXY motifs and 3 PXXP motifs and that of PRGP2 contains 1 PPXY motif and 12 PXXP motifs, many of which overlap (Fig. 2).
Tissue Distribution of PRGP1 and PRGP2.
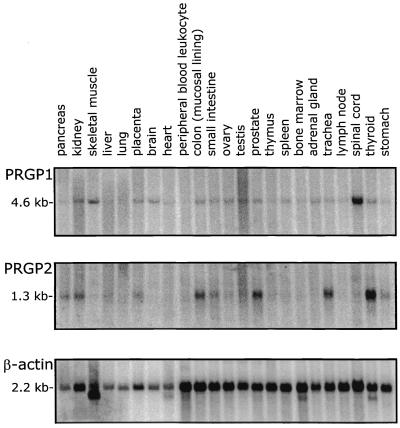
Northern blot analysis of PRGP1 and PRGP2 revealed that both have a broad tissue distribution with PRGP1 showing the highest expression in the spinal cord and PRGP2 showing the highest expression in the thyroid among those tissues tested (Fig. 4). This stands in marked contrast to the Gla domain-containing factors involved in blood coagulation that are expressed exclusively in the liver. This situation is reminiscent of Gas6, which is expressed in a variety of extrahepatic tissues (12) though there is no discernible correlation among the patterns of Gas6, PRGP1, and PRGP2 expression. In addition to the Gla domain-containing proteins, two additional vitamin K-dependent proteins are known to exist in vertebrates, bone Gla protein or osteocalcin and matrix Gla protein; although neither appears to have a homologous Gla domain (for review, see ref. 34), bone Gla protein is expressed exclusively in bone and dentin and matrix Gla protein is expressed in a variety of soft tissues (35). In addition, a broad tissue distribution of the vitamin K-dependent γ-glutamyl carboxylase and uncharacterized endogenous substrates have been identified in a wide variety of bovine tissue by enzymological methods (36). The broad tissue distribution of PRGP1 and PRGP2 presented herein is consistent with the observation that the γ-glutamyl carboxylase and its substrates are ubiquitously expressed.
Figure 4.
Multiple-tissue Northern blots of PRGP1 (Top), PRGP2 (Middle), and a β-actin control (Bottom).
Potential Functions of PRGP1 and PRGP2.
The discovery of PRGP1 and PRGP2 represents the identification of a novel modular context for Gla domains. In these molecules, the Gla domain is followed by a single putative transmembrane stretch and a small proline-rich cytoplasmic region, rather than by epidermal growth factor-like domains, kringle domains, or a disulfide loop, as is the case with all other known Gla domain proteins. It remains to be determined whether either of these two proteins actually interacts within the cytoplasm with WW domain- or SH3 domain-containing proteins and, if so, whether they interact with the cytoskeleton or signal-transduction machinery. In addition, Gla domains have long been known to interact with surfaces rich in anionic phospholipids, particularly phosphatidylserine (9, 10), and such surfaces are attractive candidates for the extracellular target of PRPG1 and PRPG2. However, the possibility of a nonphospholipid ligand cannot be ruled out. Identification of a physiological extracellular target remains an additional challenge in the functional characterization of PRGP1 and PRGP2.
The identification of potential transmembrane sequences in PRGP1 and PRGP2 raises the possibility that these proteins are members of a larger family of transmembrane Gla proteins. A search of the nonredundant DNA sequence database at the National Center for Biotechnology Information has revealed an additional Gla domain-encoding sequence, complete with the expected propeptide-encoding sequence, but lacking that of the signal peptide. The sequence identified was that of the rabbit aortic cyclic nucleotide-gated channel, a multipass transmembrane protein (37). However, the portion of the cDNA that encoded the Gla domain was in a different reading frame than that which encoded the transmembrane regions and the cyclic nucleotide-binding domain. It therefore remains to be demonstrated whether this is truly a novel membrane Gla protein and, if so, how many other members of this class exist.
Acknowledgments
We thank David Yee, Patrick O’Hara, Don Foster, Frank Grant, and Dominic Chung for their advice and helpful discussions. This work was supported, in part, by Research Grant HL-16919 from the National Institutes of Health. J.D.K. was supported by a National Science Foundation Graduate Fellowship.
ABBREVIATIONS
- EST
expressed sequence tag
- RACE
rapid amplification of cDNA ends
- Gla
γ-carboxyglutamic acid
- SH
src homology
- PRGP
proline-rich Gla proteins
- UTR
untranslated region
Footnotes
References
- 1.Dam H. Biochem J. 1935;29:1273–1285. doi: 10.1042/bj0291273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stenflo J. J Biol Chem. 1974;249:5527–5535. [PubMed] [Google Scholar]
- 3.Magnusson S, Sottrup-Jensen L, Petersen T E, Morris H R, Dell A. FEBS Lett. 1974;44:189–193. doi: 10.1016/0014-5793(74)80723-4. [DOI] [PubMed] [Google Scholar]
- 4.Nelsestuen G L, Zytokovicz T H, Howard J B. J Biol Chem. 1974;249:6347–6350. [PubMed] [Google Scholar]
- 5.Soriano-Garcia M, Park C H, Tulinsky A, Ravichandran K G, Skrzypczak-Jankun E. Biochemistry. 1989;28:6805–6810. doi: 10.1021/bi00443a004. [DOI] [PubMed] [Google Scholar]
- 6.Soriano-Garcia M, Padmanabhan K, deVos A M, Tulinsky A. Biochemistry. 1992;31:2554–2566. doi: 10.1021/bi00124a016. [DOI] [PubMed] [Google Scholar]
- 7.Sunnerhagen M, Forsen S, Hoffren A, Drakenberg T, Teleman O, Stenflo J. Nat Struct Biol. 1995;2:504–509. doi: 10.1038/nsb0695-504. [DOI] [PubMed] [Google Scholar]
- 8.Furie B, Furie B C. Cell. 1988;53:505–518. doi: 10.1016/0092-8674(88)90567-3. [DOI] [PubMed] [Google Scholar]
- 9.Mann K G, Nesheim M E, Church W R, Haley P, Krishnaswamy S. Blood. 1990;76:1–16. [PubMed] [Google Scholar]
- 10.Davie E W, Fujikawa K, Kisiel W. Biochemistry. 1991;30:10363–10370. doi: 10.1021/bi00107a001. [DOI] [PubMed] [Google Scholar]
- 11.Schneider C, King R M, Philipson L. Cell. 1988;54:787–793. doi: 10.1016/s0092-8674(88)91065-3. [DOI] [PubMed] [Google Scholar]
- 12.Manfioletti G, Brancolini C, Avanzi G, Schneider C. Mol Cell Biol. 1993;13:4976–4985. doi: 10.1128/mcb.13.8.4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nakano T, Higashino K, Kikuchi N, Kishino J, Nomura K, Fujita H, Ohara O, Arita H. J Biol Chem. 1995;270:5702–5705. doi: 10.1074/jbc.270.11.5702. [DOI] [PubMed] [Google Scholar]
- 14.Li R, Chen J, Hammonds G, Phillips H, Armanini M, Wood P, Bunge R, Godowski P J, Sliwkowski M X, Mather J P. J Neurosci. 1996;16:2012–2019. doi: 10.1523/JNEUROSCI.16-06-02012.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakano T, Kawamoto K, Higashino K, Arita H. FEBS Lett. 1996;387:78–80. doi: 10.1016/0014-5793(96)00395-x. [DOI] [PubMed] [Google Scholar]
- 16.Goruppi S, Ruaro E, Schneider C. Oncogene. 1996;12:471–480. [PubMed] [Google Scholar]
- 17.Varnum B C, Young C, Elliot G, Garcia A, Bartley T D, Fridell Y, Hunt R W, Trail G, Clogston C, Toso R J, Yanagihara D, Bennett L, Sylber M, Merewether L A, Tseng A, Escobar E, Liu E T, Yamane H K. Nature (London) 1995;373:623–626. doi: 10.1038/373623a0. [DOI] [PubMed] [Google Scholar]
- 18.Mark M R, Chen J, Hammonds R G, Sadick M, Godowski P J. J Biol Chem. 1996;271:9785–9789. doi: 10.1074/jbc.271.16.9785. [DOI] [PubMed] [Google Scholar]
- 19.Nagata K, Ohashi K, Nakano T, Arita H, Zong C, Hanafusa H, Mizuno K. J Biol Chem. 1996;271:30022–30027. doi: 10.1074/jbc.271.47.30022. [DOI] [PubMed] [Google Scholar]
- 20.Ohashi K, Nagata K, Toshima J, Nakano T, Arita H, Tsuda H, Suzuki K, Mizuno K. J Biol Chem. 1995;270:22681–22684. doi: 10.1074/jbc.270.39.22681. [DOI] [PubMed] [Google Scholar]
- 21.Boguski M S, Lowe T M J, Tolstoshev C M. Nat Genet. 1993;4:332–333. doi: 10.1038/ng0893-332. [DOI] [PubMed] [Google Scholar]
- 22.Chen H I, Sudol M. Proc Natl Acad Sci USA. 1995;92:7819–7823. doi: 10.1073/pnas.92.17.7819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sudol M, Chen H I, Bougeret C, Einbond A, Bork P. FEBS Lett. 1995;369:67–71. doi: 10.1016/0014-5793(95)00550-s. [DOI] [PubMed] [Google Scholar]
- 24.Einbond A, Sudol M. FEBS Lett. 1996;384:1–8. doi: 10.1016/0014-5793(96)00263-3. [DOI] [PubMed] [Google Scholar]
- 25.Cohen G B, Ren R, Baltimore D. Cell. 1995;80:237–248. doi: 10.1016/0092-8674(95)90406-9. [DOI] [PubMed] [Google Scholar]
- 26.Pawson T. Nature (London) 1995;373:573–580. doi: 10.1038/373573a0. [DOI] [PubMed] [Google Scholar]
- 27.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 28.Mullis K, Faloona F, Scharf S, Saiki R, Horn G, Erlich H. Cold Spring Harbor Symp Quant Biol. 1986;51:263–273. doi: 10.1101/sqb.1986.051.01.032. [DOI] [PubMed] [Google Scholar]
- 29.Frohman M A, Dush M K, Martin G R. Proc Natl Acad Sci USA. 1988;85:8998–9002. doi: 10.1073/pnas.85.23.8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Don R H, Cox P T, Wainwright B J, Baker K, Mattick J S. Nucleic Acids Res. 1991;19:4008. doi: 10.1093/nar/19.14.4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sanger F, Nicklen S, Coulson A R. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Frigerio J, Berthezene P, Garrido P, Ortiz E, Barthellemy S, Vasseur S, Sastre B, Seleznieff I, Dagorn J, Iovanna J. Hum Mol Genet. 1995;4:37–43. doi: 10.1093/hmg/4.1.37. [DOI] [PubMed] [Google Scholar]
- 33.Hartmann E, Rapoport T A, Lodish H F. Proc Natl Acad Sci USA. 1989;86:5786–5790. doi: 10.1073/pnas.86.15.5786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Price P A. Annu Rev Nutr. 1988;8:565–583. doi: 10.1146/annurev.nu.08.070188.003025. [DOI] [PubMed] [Google Scholar]
- 35.Fraser J D, Price P A. J Biol Chem. 1988;263:11033–11036. [PubMed] [Google Scholar]
- 36.Vermeer C. Mol Cell Biochem. 1984;61:17–35. doi: 10.1007/BF00239604. [DOI] [PubMed] [Google Scholar]
- 37.Biel M, Altenhofen W, Hullin R, Ludwig J, Freichel M, Flockerzi V, Dascal N, Kaupp U B, Hofmann F. FEBS Lett. 1993;329:134–138. doi: 10.1016/0014-5793(93)80209-d. [DOI] [PubMed] [Google Scholar]