Abstract
Fragile sites are specific loci that form gaps, constrictions, and breaks on chromosomes exposed to partial replication stress and are rearranged in tumors. Fragile sites are classified as rare or common, depending on their induction and frequency within the population. The molecular basis of rare fragile sites is associated with expanded repeats capable of adopting unusual non-B DNA structures that can perturb DNA replication. The molecular basis of common fragile sites was unknown. Fragile sites from R-bands are enriched in flexible sequences relative to nonfragile regions from the same chromosomal bands. Here we cloned FRA7E, a common fragile site mapped to a G-band, and revealed a significant difference between its flexibility and that of nonfragile regions mapped to G-bands, similar to the pattern found in R-bands. Thus, in the entire genome, flexible sequences might play a role in the mechanism of fragility. The flexible sequences are composed of interrupted runs of AT-dinucleotides, which have the potential to form secondary structures and hence can affect replication. These sequences show similarity to the AT-rich minisatellite repeats that underlie the fragility of the rare fragile sites FRA16B and FRA10B. We further demonstrate that the normal alleles of FRA16B and FRA10B span the same genomic regions as the common fragile sites FRA16C and FRA10E. Our results suggest that a shared molecular basis, conferred by sequences with a potential to form secondary structures that can perturb replication, may underlie the fragility of rare fragile sites harboring AT-rich minisatellite repeats and aphidicolin-induced common fragile sites.
Fragile sites are specific loci that appear as constrictions, gaps, or breaks on chromosomes from cells exposed to partial inhibition of DNA replication (16). They are classified as rare or common, depending on their frequency within the population and their specific mode of induction. Rare fragile sites (n = 28, as listed in the Genome Database [GDB]) appear in <5% of the human population and segregate in specific families. Most rare fragile sites are induced by folate deficiency, and others are induced by DNA minor groove binders (52). Common fragile sites (n = 89, as listed in the GDB), on the other hand, are considered to be an intrinsic part of the chromosomal structure and are thought to be present in all individuals. Most common fragile sites (n = 76) are induced by aphidicolin (16), an inhibitor of DNA polymerases α and δ. After induction with replication inhibitors these sites are involved in sister chromatid exchange, deletions and translocations, gene amplification, and plasmid integration (reviewed in reference 15). Common fragile sites also correlate with chromosomal breakpoints in tumors (22, 61) and were shown to play a role in the in vivo occurrence of deletions and translocations (reviewed in reference 44), gene amplification (24), and integration of foreign DNA (37, 54, 59). Despite their inherent instability, several common fragile sites are conserved between mice and humans (17, 29, 49), indicating the important biological role of these sites.
Seven rare fragile sites have been characterized at the molecular level: five folate sensitive fragile sites were cloned and found to consist of expanded tandem CGG microsatellite repeats (>200 copies) (28; reviewed in reference 53). These repeats are capable of adopting unusual, non-B DNA structures, such as hairpins (12), slipped strand (S)-DNA (41, 42), or quadruplex DNA (11). These various DNA secondary structures can perturb the elongation of DNA replication in vitro and in vivo (47, 55). Two non-folate-sensitive rare fragile sites were cloned as FRA16B and FRA10B, induced by distamycin A and/or bromodeoxyuridine (BrdU) (25, 60). They are comprised of polymorphic AT-rich minisatellite repeats, and their expression is associated with expansion of one or more of the repeats, up to several kilobases. The expanded FRA16B and FRA10B repeats are highly similar and contain inverted repeats able to form hairpin structures (reviewed in reference 19).
Six common fragile sites have been cloned and characterized: FRA3B (3, 40, 43, 58, 59, 62), FRA7G (24, 27), FRA7H (37), FRA16D (34, 45), FRAXB (1), and FRA6F (38). The cytogenetic expression (gaps and constrictions) of these sites is visible along large genomic regions spanning hundreds to thousands of kilobases. Studies of replication time revealed a perturbed elongation of DNA replication along common fragile regions (23, 24, 31, 56), indicating that the fragile sequences have intrinsic features that might delay replication. However, no expanded repeats, nor any other specific sequences that could perturb replication, were identified.
Mammalian chromosomes are organized into regions, R- and G-bands, which differ in their structure and function. Generally, G-bands are AT-rich and gene-poor and undergo DNA replication late in the S phase of the cell cycle. R-bands are GC-rich and gene-rich and undergo DNA replication early in the S phase (13). G- and R-bands differ also in the organization and localization of replication foci (9, 46), as well as in the organization and level of condensation of their chromatin (13). In the absence of any obvious DNA sequences that could account for the fragility at common fragile sites, we previously studied DNA helix flexibility, a structural characteristic of the DNA that might affect DNA replication and chromatin condensation. We found that G-bands are enriched in clusters of flexible sequences compared to R-bands (36). Furthermore, the cloned fragile sites, all mapped to R-bands, were found to be enriched in clusters of sequences with high DNA flexibility relative to nonfragile sequences from R-bands, resembling the flexibility of G-bands (35-38, 45). Thus, in order to better understand the contribution of DNA flexibility to the fragility at common fragile sites, it was important to analyze the flexibility of common fragile sites mapped to G-bands.
Here we describe the cloning of FRA7E, a common fragile site mapped to the G-band 7q21.11. We found a significant higher DNA flexibility in fragile regions mapped to G-bands relative to nonfragile regions mapped to the same bands. These results support the hypothesis that the flexible sequences contribute to the mechanism of fragility. Moreover, we show that flexible sequences are composed of interrupted AT-dinucleotide repeats, highly similar to the AT-rich repeats expanded in the rare fragile sites FRA16B and FRA10B. We further show that nonexpanded alleles of these rare fragile sites span the same genomic regions as the aphidicolin-induced common fragile sites, FRA16C and FRA10E, respectively. These results suggest that a shared mechanism, conferred by sequences with a potential to form secondary structures, can perturb replication and lead to fragility at both rare fragile sites harboring AT-rich minisatellite repeats and aphidicolin-induced common fragile sites.
MATERIALS AND METHODS
Cells and growth conditions.
The simian virus 40-transformed human fibroblast cell line GM00847 (Coriell Cell Repository, Camden, N.J.) was grown in Eagle minimal essential medium supplemented with 10% fetal calf serum.
Physical map of DNA clones at the FRA7E region.
A DNA sequence-based map of chromosome 7q21.11 was constructed from the finished sequences of the GenBank database. The assembled map with no physical (clone) gaps represents a consistent presentation of the order of DNA markers in comparison to our other studies, which used additional technologies of radiation and somatic cell hybrid mapping, as well as fluorescence in situ hybridization (FISH). (Additional information is available online [http://www.genet.sickkids.on.ca/chromosome7/].)
Preparation of chromosomes and induction of fragile sites.
Cells were grown on coverslips, and common fragile sites were induced by growing the cells in M-199 medium in the presence of 0.4 μM aphidicolin and 0.5% ethanol, with or without 2.2 mM caffeine, for 24 h prior to the fixation of chromosomes by standard procedures. Induction of rare fragile sites was performed by adding 32.5 μM BrdU for 24 h, as previously described (52).
FISH.
DNA clones (YAC, PAC, and BAC) were labeled with digoxigenin (DIG)-11-dUTP (Boehringer Manheim) by nick translation. DIG-labeled probes were detected with fluorescein isothiocyanate (FITC)-conjugated sheep anti-DIG specific antibodies (Boehringer Mannheim). FISH on metaphase chromosomes was performed as previously described (32).
Cytogenetic analysis of hybridization signals and fragile sites.
Green and red fluorescence were visualized by using a Nikon B-2A filter cube. For weak signals a modified Chromatech HQ-FITC (Chroma Technology, Brattleboro, Vt.) filter set was used (excitation band, 460 to 500 nm; emission band, 520 to 600 nm). Images were captured with an intensified charge-coupled device imager (Paultek Imaging, Grass Valley, Calif.) and digitized with a frame grabber (Imascan/MONO-D; Imagraph, Chelmsford, Mass.). The Image-Pro PLUS program (Media Cybernetics, Silver Spring, Md.) was used to measure the fragile site-telomere distance relative to the total length of the chromosome and compared to the GDB mapping of the fragile sites, as previously described (37).
PCR of FRA10B alleles.
PCR across the FRA10B locus was performed on DNA from GM00847 cells with primers F1, F2, and R (25). The PCR was performed as previously described (25).
DNA sequences.
Large genomic sequences were retrieved by the UCSC Genome Browser (http://genome.ucsc.edu/cgi-bin/hgGateway; obtained from the November 2002 Freeze of the Human Genome Database); sequences of specific clones and of short segments within clones were retrieved from GenBank. Fragile-site sequences for flexibility analysis (see below) include the entire regions reported to span the cloned common fragile sites. Controls for flexibility analysis were chosen from large, fully assembled (phase 3 high-throughput genomic sequence data only) genomic sequences, mapped at the 850-band resolution to chromosomal bands harboring no fragile sites. All of the sequences that were used for the flexibility and PileUp analyses (see below) are listed in Tables 1 and 2, respectively.
TABLE 1.
Sequences analyzed by TwistFlex
| Chromosomal region | Chromosomal positiona | First DNA markerb | Last DNA markerb | Length (Mb) | Source or reference |
|---|---|---|---|---|---|
| G-bands | |||||
| FRA7E | chr7:79-83.5Mb | D7S1934 | SHGC-104456 | 4.5 | This study |
| FRA71c | chr7:142.9-144.4Mb | SHGC-153624 | SWSS2627 | 1.5 | 7 |
| Controls (nonfragile) | chr10:9.5-11.15Mb (10p14) | SHGC-147592 | WI-4120 | 1.65 | |
| chr10:11.25-12.1Mb (10p14) | RH27102 | SHGC-57578 | 0.85 | ||
| chr12:16.5-18.5Mb (12p12.3) | B224C2/T7 | UT2022 | 2 | ||
| chrX:41.8-42.9Mb (Xp11.3) | HUMUT1223 | DXS1708 | 1.1 | ||
| chrX:115.4-116.9Mb (Xq25) | SWXD1380 | AFM164TG1 | 1.5 | ||
| chrX:117.1-119Mb (Xq25) | DXS7597 | HUMSWX2784 | 1.9 | ||
| R-bands | |||||
| FRA7G | chr7:110.7-115Mb | SHGC-143971 | RH44861 | 4.3 | 24 |
| FRA3B | chr3:59.3-60Mb | SHGC-86352 | RH41625 | 0.7 | 3 |
| chr3:60.4-63.3Mb | SHGC-102025 | RH26408 | 2.9 | ||
| FRA6F | chr6:111600834-112591783bp | SHGC-144121 | SHGC-82095 | 0.99 | 38 |
| FRAXB | chrX:5855178-6311942bp | DXS7731 | DXS9036 | 0.46 | 1 |
| FRA16Dd | chr16:79329316-79691715bp | SHGC-150973 | WI-2755 | 0.36 | 45 |
| FRA7He | chr7:128848390-129012733bp | D7S614 | STSG33533FS | 0.16 | 37 |
| Controls (nonfragile) | chr20:0.1-5Mb (20p13) | D20S210 | RH70061 | 4.9 | |
| chr14:50-51.5Mb (14q22.3) | SHGC-149912 | RH53548 | 1.5 | ||
| chr14:90.2-92Mb (14q32.31) | RH44545 | SHGC-107994 | 1.8 | ||
| Total | 33.1 |
TABLE 2.
Sequences analyzed by PileUp
| Sequence | Region | Clone | Base positions | Length (bp) |
|---|---|---|---|---|
| Expandeda FRA10B 17b-3′ | FRA10B | AF053530 | 1-153 | 153 |
| Expanded FRA10B 17-5′ | FRA10B | AF053529 | 36-446 | 411 |
| Expanded FRA16B | FRA16B | U85253 | 347-558 | 212 |
| Flexc FRA7E | FRA7E | AC004903 | 63367-927 | 561 |
| Flex FRA3B | FRA3B | AC098482 | 38111-468 | 358 |
| Flex FRA7G | FRA7G | AC079621 | 72472-74661 | 2,190 |
| Flex, nonfragile 1 | Nonfragile (10p14) | AL136318 | 37617-38537 | 921 |
| Flex, nonfragile 2 | Nonfragile (20p13) | AL161656 | 90364-91192 | 829 |
| Non-flexd FRA7E | FRA7E | AC004880 | 24701-910 | 210 |
| Non-flex, nonfragile | Nonfragile (12p12.3) | AC007528 | 109441-660 | 220 |
Expanded, minisatellite repeats expanded in the rare fragile-site alleles.
Allele no. 17 of the FRA10B locus.
Flex, an AT-dinucleotide-rich flexibility island.
Non-flex, a nonflexible sequence with >78% A/T.
Computational analysis of DNA flexibility.
To evaluate DNA flexibility, we used a measure of the potential local variations in the DNA structure, expressed as fluctuations in the twist angle (48). This measure provides average fluctuations in the twist angle for each of the possible dinucleotides and thus enables the evaluation of the flexibility of a DNA sequence by summation of these values. To carry out the analysis, we developed a computer program, TwistFlex (http://www.jail.cs.huji.ac.il/∼netab/index.html), based on our previous program, FlexStab (37), which can calculate flexibility measures of DNA sequences of any size. The analysis was performed in overlapping windows of 100 bp. Dinucleotide values were summed along the window and averaged by the window length. Windows with values of >13.7° were considered as flexibility peaks. Since the window length is 100 bp, flexibility peaks that were <100 bp apart were considered one flexibility peak. TwistFlex can also analyze clusters of flexibility peaks, which we defined as at least three flexibility peaks in which the distance between any two adjacent peaks is ≤5 kb.
Computational analysis of DNA sequence and structure.
Multiple sequence alignment was performed by using the PileUp program. Alignments of pairs of DNA sequences were performed by using the Gap program. Secondary structure analyses of single-stranded DNA sequences were carried out by using the MFold program with DNA free-energy parameters. All of these programs are from the GCG package.
Secondary structure assessment.
We assessed whether the stable secondary structures, predicted for the single-stranded DNA of the flexibility peaks, stem from their high A/T composition per se or due to their specific sequence organization. For this, we compared the free-energy values of each potential structure to those of same-length random sequences with identical base compositions. From each flexible sequence we generated by computer shuffling 100 random sequences with equal base compositions. Each random sequence was submitted to MFold, and its secondary structure was predicted along with its free-energy value. The significance of the original structure was assessed by the fraction of times that the shuffled sequences' structures had lower free-energy values than the original structure.
Analysis of flexibility clusters.
Fragile-site sequences and nonfragile sequences were divided into regions 500 kb in length. For each such region the number of clusters of flexibility peaks was counted. The significance of the difference in the number of clusters between fragile and nonfragile regions was assessed by using the Mann-Whitney test.
Analysis of base composition.
The composition of A/T bases and of AT-dinucleotides was computed for the sequences of flexibility peaks and for their flanking sequences. The difference in base composition (dinucleotide composition) between these two types of sequences was evaluated by a median test.
RESULTS
FRA7E spans a large genomic region at the G-band 7q21.11.
The G-band mapped common fragile site cloned and characterized in the present study is FRA7E. Our preliminary results showed that FRA7E is proximal to the PGY genes, which are mapped to the G-band 7q21.1 (data not shown). Thus, we constructed a physical map proximal to PGY at 7q21.1 (Fig. 1). We determined the location of clones from this region relative to the FRA7E gaps and constrictions by FISH on metaphase chromosomes from GM00847 cells induced by aphidicolin to express fragility. Clones were considered as spanning FRA7E if, on different chromosomes from the same preparation, their hybridization signals appeared proximal to, distal to, or on both sides of FRA7E gaps and constrictions. Since there are several common fragile sites along 7q, we used computational image analysis to identify FRA7E (see Materials and Methods). Our analysis revealed that genomic clones from 7q21.11 span the FRA7E region (Fig. 1 and 2 and Table 3). Interestingly, clones AC004866 and AC004960 hybridized mostly distal to the gaps and constrictions of FRA7E, although they are flanked by clones that equally hybridized distally and proximally to the fragile region. This pattern might reflect the existence of two adjacent fragility “hot spots” that flank these clones, which are too close to be distinguished cytogenetically. Hence, induction of FRA7E can lead to an unusual chromatin organization exhibited as gaps and constrictions along a large genomic region of at least 4.5 Mb of DNA. The distal boundary of the FRA7E region is yet to be determined.
FIG. 1.
Physical map across the FRA7E region at 7q21.11. The order and extent of overlap of BAC, PAC, and YAC clones (shown as lines or solid bars) was based on their DNA marker content. Clones used for FISH analysis are named and marked as solid bars. The full information on this contig can be found in the Genome Database and at http://www.genet.sickkids.on.ca/chromosome7/.
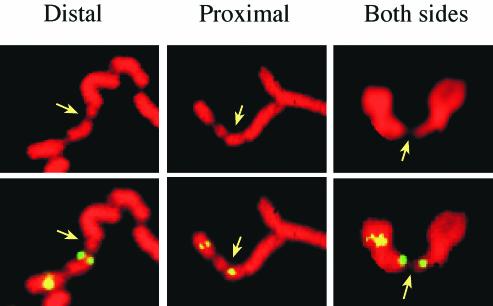
FIG. 2.
Examples of FISH signals relative to FRA7E. FISH analysis was performed on metaphase chromosomes from GM00847 cells following aphidicolin treatment. FITC-labeled RPCI11-105J12 (left and middle panels) shows signals distal and proximal to FRA7E on different chromosomes. FITC-labeled AC004892 (right panel) shows signals on both sides of FRA7E. Probes from the FRA7E region were cohybridized with a probe distal to FRA7E to identify chromosome 7. The images show propidium staining (upper panel) and FISH with FITC-labeled probes (lower panel). The arrows point to FRA7E.
TABLE 3.
FISH analysis of chromosomes expressing FRA7E
| Clone | No. of chromosomes with signals (%)
|
||
|---|---|---|---|
| Proximal | Both sides | Distal | |
| AC003988 | 24 (100) | 0 (0) | |
| AC004880 | 18 (95) | 1 (5) | |
| AC004972 | 9 (53) | 8 (47) | |
| AC004866 | 2 (6) | 1 (3) | 32 (91) |
| AC004960 | 2 (11) | 16 (89) | |
| AC004892 | 17 (49) | 1 (2) | 17 (49) |
| AC005159 | 11 (61) | 3 (17) | 4 (22) |
| AC004894/AC004006 | 25 (71) | 1 (3) | 9 (26) |
| NH_0099P20/RPCI11-105J12 | 20 (34) | 2 (3) | 38 (63) |
| AC006322/AC004848 | 8 (18) | 2 (5) | 34 (77) |
| AC004907 | 9 (35) | 17 (65) | |
| HSC7E596/AC006151 | 4 (13) | 26 (87) | |
Common fragile sites mapped to G-bands exhibit significantly higher DNA flexibility compared to nonfragile regions.
The analysis of the FRA7E sequence did not reveal any clear feature that could account for its fragility. Thus, we performed DNA flexibility analysis, previously suggested to play a role in the mechanism of fragility. The analysis was performed by using TwistFlex, a new computer program based on our previous program, FlexStab (37), which identifies DNA sequences with potential high flexibility (flexibility peaks), as well as clusters of such sequences (see Materials and Methods), at genomic regions of any size. The analysis was performed on the entire regions spanning FRA7E and FRA7I, a recently cloned common fragile site assigned to the G-band 7q35 (7), and on nonfragile sequences mapped to G-bands (Table 1 and Materials and Methods). The analysis revealed significantly more clusters of flexibility peaks in FRA7E and FRA7I than in the nonfragile regions (P < 0.01; Table 4). These results indicate that common fragile sites mapped to G-bands are highly enriched in clusters of sequences with high DNA flexibility relative to nonfragile regions from these bands. A similar significant difference was previously identified between fragile and nonfragile sequences mapped to R-bands (36). Since the former analysis was performed on relatively short genomic sequences (cosmids, BACs, and PACs), we extended the analysis of R-bands to sequences of large, fully assembled genomic contigs (Table 1). A significant difference in the number of flexibility clusters was identified between the fragile and nonfragile regions mapped to R-bands (P < 0.05; Table 4). Thus, in both G- and R-bands the fragile regions are enriched in clusters of sequences with high DNA flexibility relative to nonfragile regions.
TABLE 4.
Flexibility analysis of genomic regions from G- and R-bands
| Chromosomal region | Sequence | No. of flexibility islands/Mb | No. of flexibility clusters/Mb (P)a |
|---|---|---|---|
| G-bands | Fragile sites | 57 | 3.3 (<0.01) |
| Nonfragile regions | 33 | 0.8 | |
| R-bands | Fragile sites | 34 | 1.3 (<0.05) |
| Nonfragile regions | 18 | 0.2 |
Flexibility clusters are defined as ≥3 flexibility islands in which the distance between any two adjacent islands is ≤5 kb. P values were calculated for the difference between fragile and nonfragile sequences.
The sequences of flexibility peaks are similar to AT-rich repeats in rare fragile sites.
In order to further understand the possible contribution of sequences with high DNA flexibility to the molecular basis of fragility at common fragile sites, we analyzed the sequence composition and organization of the flexibility peaks found in >33 Mb of the studied genomic DNA (fragile and nonfragile regions; see Table 1). The analysis revealed that the flexible sequences, ranging in size from 100 bp to several hundred base pairs, are composed of a very high A/T content (78% ± 1.4%), significantly different from that of their nonflexible flanking sequences (61% ± 3.6%) (P < 0.001). In addition, the flexible sequences are particularly rich in AT-dinucleotides (21% ± 0.5%), a finding significantly different from the frequency found in nonflexible sequences (8% ± 1%) (P < 0.001). These results were expected from the thermodynamic calculations used by the TwistFlex program (48). However, the flexible sequences are enriched also in TA-dinucleotides (20% ± 0.5), compared to nonflexible sequences (7% ± 1%) (P < 0.001). Moreover, the AT/TA-dinucleotides appear in the flexibility peaks as interrupted runs (see an example in Fig. 3A). We therefore termed the flexibility peaks AT-dinucleotide-rich flexibility islands.
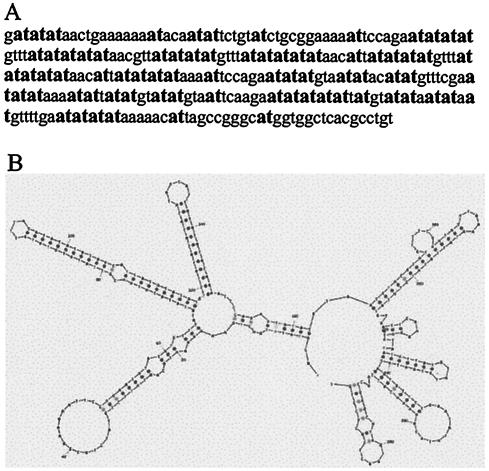
FIG. 3.
Sequence and secondary structure of an AT-dinucleotide-rich flexibility island. (A) Sequence of a 294-bp AT-dinucleotide-rich flexibility island (clone AC079799, 146129 to 146422 bp) from FRA7E. AT-dinucleotides are in boldface. (B) Predicted secondary structure by MFold of the AT-dinucleotide-rich flexibility island shown in panel A.
We found the sequence composition (high A/T content) and organization (high level of AT-dinucleotides) of these islands reminiscent of the AT-rich repeats in the BrdU and/or distamycin A-induced rare fragile sites, FRA16B and FRA10B (25, 60). Each of these rare fragile sites contains a long (>1-kb) AT-rich sequence comprised of several repeats, of which one or more might expand and lead to fragility. In order to examine the sequence similarity among the AT-rich sequences in common and rare fragile sites, we performed a multiple sequence alignment of the following sequences: the repeats expanded in the FRA16B and FRA10B rare fragile site alleles (“expanded,” Table 2), AT-dinucleotide-rich flexibility islands (flex) from several common fragile sites (FRA7G, FRA3B, and FRA7E) and from nonfragile regions, and nonflexible sequences with a similar A/T content (non-flex). As shown in Fig. 4, when the sequences were grouped by their similarities, no higher intrasequence similarity in the groups of rare and common fragile sites was revealed. Moreover, the analysis showed that the repeat sequences from FRA16B and FRA10B, previously shown to be highly similar (19), might have even a higher similarity to AT-dinucleotide-rich flexibility islands (Fig. 4, expanded FRA16B to flex FRA7G and expanded FRA10B to flex nonfragile 2). Further analysis of the level of similarity between pairs of sequences revealed that the FRA16B repeats are 70 and 78% similar to the 3′ and 5′ sequences of the FRA10B allele, respectively, and 77 and 76% similar to the flexibility islands from FRA7G and from nonfragile region 2, respectively. In comparison, the similarity between the FRA16B and FRA10B repeats and the nonflexible sequences with the same A/T content is only ∼50%. Thus, AT-rich sequences from rare and common fragile sites have a similar sequence composition and organization.
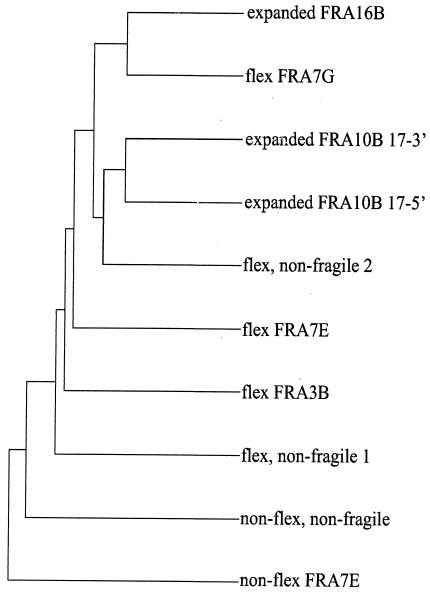
FIG. 4.
Multiple sequence alignment of AT-dinucleotide flexibility islands and sequences from FRA16B and FRA10B. The dendrogram shows the clustering relationships used to determine the order of pairwise alignments that together create the final multiple sequence alignment. The distance along the vertical axis is proportional to the difference between sequences. Shown are sequences from the FRA16B and FRA10B expanded repeats (note that the fully expanded FRA10B sequence is not available; thus, the 5′ and 3′ sequences of the expanded region were analyzed), five AT-dinucleotide-rich flexibility islands from three fragile regions and two nonfragile regions, and two nonflexible sequences with an A/T content (>78%) similar to that of the flexibility islands. For the genomic localization and lengths of these sequences, see Table 2.
AT-rich rare fragile sites and common fragile sites span the same genomic regions.
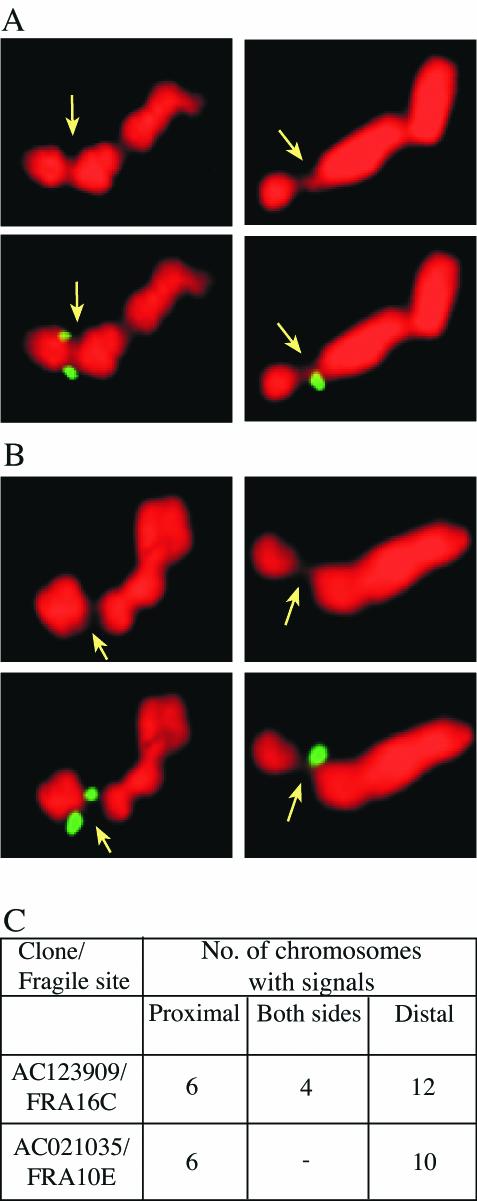
The similarity between AT-rich sequences from rare and common fragile sites raises the question whether the normal alleles of these rare fragile sites could also be the sites of aphidicolin-induced common fragile sites. Thus, we examined the cytogenetic mapping of the seven BrdU- and/or distamycin A-inducible rare fragile sites reported in the GDB and found that six of them are mapped to the same chromosomal band as aphidicolin-induced common fragile sites (Table 5). In order to investigate the possibility that the rare and common fragile sites actually span the same genomic regions at the molecular level, we analyzed the localization of the currently cloned rare fragile sites, FRA16B and FRA10B, relative to the common fragile sites FRA16C and FRA10E, respectively. First, we sought to exclude the remote possibility that the analyzed GM00847 cells carry expanded alleles at the FRA16B or FRA10B loci. Hence, we treated the cells with BrdU to induce fragility. No gaps or constrictions were found on 100 chromosomes 16 or 10. Furthermore, we performed PCR and sequence analysis of the FRA10B locus and found that it comprises a small normal allele identical to the small normal allele 2 reported by Hewett et al. (25; data not shown). Together, these results indicate that GM00847 chromosomes carry normal alleles at the FRA10B and FRA16B loci. Subsequently, we performed FISH analysis with the clones AC123909 and AC021035, each containing a normal allele of FRA16B and FRA10B, respectively, on metaphase chromosomes treated with aphidicolin to induce fragility. The analysis showed that these clones span the gaps and constrictions of the common fragile sites FRA16C and FRA10E, respectively (Fig. 5). Thus, the BrdU and/or distamycin A-inducible rare fragile sites FRA16B and FRA10B and the aphidicolin-induced common fragile sites FRA16C and FRA10E span the same genomic regions, respectively.
TABLE 5.
Colocalization of BrdU and/or distamycin A-inducible rare fragile sites with aphidicolin-induced common fragile sites
| Rare fragile site | Common fragile site | Chromosomal mapping |
|---|---|---|
| FRA8E | FRA8C | 8q24.1 |
| FRA10B | FRA10E | 10q25.2 |
| FRA11I | FRA11C | 11p15.1 |
| FRA12C | FRA12E | 12q24 |
| FRA16B | FRA16C | 16q22.1 |
| FRA16E | 16p12.1 | |
| FRA17A | Unnameda | 17p12 |
Reported by Simonic and Gericke (50).
FIG. 5.
Colocalization of rare and common fragile sites at 16q22.1 and 10q25.2. Examples of hybridization signals proximal and distal to aphidicolin-induced gaps and constrictions at FRA16C (A) and FRA10E (B) in GM00847 cells are shown. FISH analyses were performed with FITC-labeled BACs as indicated in panel C. Signals distal (left panels) and proximal (right panels) to the fragile sites (indicated with an arrow) are shown. The numbers of chromosomes with signals proximal, on both sides and distal to the fragile sites, are given in panel C.
Based on the previous result that aphidicolin-induced common fragile sites are large genomic regions enriched in clusters of flexibility islands, we performed TwistFlex analysis on 1.1 Mb of contiguous sequence from the FRA16C/FRA16B region (between markers SHGC-141023 and RH102921) and 2.4 Mb of contiguous sequence from the FRA10E/FRA10B region (between markers RH51139 and RH17163). No flexibility clusters were found in these regions. However, in the FRA16C/FRA16B region an exceptional >3.4-kb AT-rich flexibility island was identified (at 134256 to 137689 bp in AC123909), which contains the FRA16B minisatellite repeats and their flanking AT-rich repeats (60). In the FRA10E/FRA10B region, two long, >1.1-kb AT-dinucleotide rich flexibility islands (180 kb apart) were found (at 144218 to 145400 bp in AC021035 and at 24605 to 25779 bp in AL607043). The flexibility island in the AC21035 clone harbors the FRA10B repeats (25). These very long AT-dinucleotide flexibility islands might contribute to the fragility at the FRA16B/FRA16C and FRA10B/FRA10E genomic regions.
AT-dinucleotide flexibility islands can readily fold into DNA secondary structures.
The repeats of AT-rich rare fragile sites were shown to have the potential to form hairpin structures, which have been suggested to perturb the elongation of DNA replication (18). Based on the similarity between these repeats and the AT-dinucleotide-rich flexibility islands of common fragile sites, we analyzed the potential of the latter to form such structures. For that purpose, we used the MFold program to predict the optimal folding of single-stranded AT-dinucleotide-rich flexibility islands into DNA secondary structures (hairpins). We found that AT-dinucleotide-rich flexibility islands of >200 bp may readily fold into secondary structures (Fig. 3B), which are significantly more stable than same-length random sequences with the same base composition (P < 0.01; see Materials and Methods). Hence, the sequence of the AT-dinucleotide-rich flexibility islands, rather than their base composition per se, contributes to the stability of the secondary structures.
DISCUSSION
Here we show that common fragile sites and rare fragile sites harboring AT-rich minisatellite repeats may share the same molecular basis. Our results suggest that the sequences in rare and common fragile sites share features that contribute to their sensitivity to replication perturbation, leading to fragility. Since their discovery, fragile sites have been classified as rare or common, depending on their frequency within the population and their mode of induction. Based on the results presented here, we suggest that fragile sites can be classified according to the sequences that contribute to their fragility. Several fragile sites harbor CGG repeats, whereas others harbor AT-dinucleotide-rich sequences. In rare fragile sites, these sequences are organized as long uninterrupted repeats, whereas in common fragile sites they are organized as interrupted shorter repeats (Fig. 3A). We base our suggestion on molecular analysis, which showed that normal alleles of the FRA16B and FRA10B rare fragile sites colocalize with the aphidicolin-induced common fragile sites FRA16C and FRA10E, respectively. Moreover, we found that four of five yet-uncloned rare fragile sites induced by BrdU and/or distamycin A are cytogenetically colocalized with aphidicolin-induced common fragile sites (Table 5), suggesting that these rare and common fragile sites also span the same genomic regions.
Identification of the sequences that confer fragility at rare fragile sites was relatively straightforward, since these sites appear in a small fraction of the population and thus could be identified by positional cloning. Elucidating the molecular basis of aphidicolin-induced common fragile sites was more difficult since common fragile sites are part of the normal chromosomal structure. Previous studies have shown that common fragile sites are enriched in clusters of sequences with high DNA flexibility (35-38, 45). In the current study we show that these flexible sequences are composed of AT-dinucleotide-rich sequences of various lengths. The sequence of these AT-dinucleotide-rich flexibility islands show high similarity to the FRA16B and FRA10B expanded repeats. It is no surprise, therefore, that the AT-rich sequences of rare fragile sites were found by TwistFlex to be highly flexible. However, the sequence organization of rare and common alleles differs. Expanded alleles of rare fragile sites harbor tens of kilobases of uninterrupted repeats, while the common fragile regions are enriched in clusters of short AT-dinucleotide-rich islands or contain single (several-kilobase-long) such islands, as in the case of the common fragile sites FRA16C and FRA10E. These results suggest that common fragile sites that harbor uninterrupted AT-dinucleotide repeats might in rare cases evolve (by expansion of these repeats) into rare fragile site.
What can be the role of the AT-dinucleotide-rich flexibility islands in the mechanism of fragility? The folate sensitive rare fragile sites consist of expanded CGG repeats in tandem. These alleles replicate very late, at G2, which is later than the normal alleles at these sites (20, 21, 51). The CGG repeats can adopt non-B DNA structures (quadruplex DNA and hairpins) that inhibit replication fork movement both in vitro and in vivo and thus cause the delayed replication (47, 55). Similar late replication was found for the expanded AT-rich repeats in FRA16B and FRA10B (18), further supporting their involvement in the mechanism of fragility. Furthermore, the expanded AT-rich repeats at FRA16B and FRA10B were suggested to form hairpin structures that contribute to their expansion and fragility (25, 60). In common fragile sites, aphidicolin causes a delay in replication accomplishment along the fragile regions; hence, a significant portion of the fragile regions is unreplicated in G2 (23, 24, 31, 56). Furthermore, the replication along common fragile regions is perturbed even under normal growth conditions (23), indicating that these sites harbor sequences with intrinsic features that might lead to delay in replication. Recently, Casper et al. (4) provided evidence that aphidicolin-induced common fragile sites on metaphase chromosomes represent unreplicated DNA resulting from stalled replication forks that escape the ATR-dependent replication checkpoint (4). The results of the present study indicate that the AT-dinucleotide-rich flexibility islands might lead to replication perturbation in common fragile sites. Two features of these sequences might contribute to their sensitivity to replication perturbation. The first is their high DNA flexibility. AT-dinucleotide runs were experimentally shown to be more flexible than random DNA (6) and thus can act as sinks for the superhelical density generated ahead of the replication fork in its progress. Accumulating superhelical density can hinder efficient topoisomerase activity and decrease the processivity of the polymerase complex (2, 14). This perturbation is expected to be enhanced in the presence of low levels of aphidicolin, which inhibits the activity of polymerases α and δ. The second feature of the AT-dinucleotide-rich flexibility islands that might contribute to fragility is their potential to form secondary structures upon unwinding of the double helix (Fig. 3B). Interestingly, several studies have shown that, upon replication arrest by aphidicolin, the separation of the DNA strands ahead of the replication fork can continue up to several kilobases (8, 33, 57). This might facilitate the formation of DNA secondary structures in the AT-dinucleotide-rich sequences. Such secondary structures are expected to perturb the progression of the replication fork (5, 26, 30). The appearance of gaps and constrictions at common fragile sites after replication stress might reflect incomplete or delayed resolution of stalled replication forks. In the present study we pinpoint the sequences that might be involved in replication perturbation. However, the reason why these sequences might escape the ATR-dependent replication checkpoint as shown by Casper et al. (4) has not yet been investigated.
As mentioned above, the generation of secondary structures was suggested for the AT-rich minsatellite repeats of FRA16B and FRA10B (25, 60). Importantly, distamycin A, as well as other minor groove binders that can induce the expression of several rare fragile sites, was shown to inhibit the activity of the Werner and Bloom helicases known to unwind unusual DNA structures (reviewed in reference 10). This indicates that unusual DNA structures at fragile regions play an important role in the replication perturbation, leading to fragility.
The flexibility analysis of large genomic regions performed in the present study clearly showed that fragile sites are significantly enriched in clusters of AT-dinucleotide-rich flexibility islands. Their potential to perturb replication might depend on the length of these islands, their number within the clusters, and the number of clusters along the regions. The effect of very long AT-dinucleotide flexibility islands, such as those found in FRA16C and FRA10E, might be comparable to that of flexibility clusters, sufficient to cause replication perturbation upon aphidicolin induction, leading to fragility. Interestingly, spontaneous expression of FRA16B and FRA10B can be found in cells of individuals with expanded AT-rich repeats, indicating that these repeats are sufficient for replication perturbation, which can be further enhanced by the inducers of fragility.
It is important to note that there are other regions in the human genome, such as those harboring expanded non-CGG trinucleotide repeats, that are highly flexible (2) and that have the potential to form DNA secondary structures (39), which were not found, thus far, to express fragility. This might indicate that such DNA structures are necessary but not sufficient for fragile-site expression. However, these regions might have the potential to express fragility under conditions which are yet unidentified.
Previous DNA flexibility analysis (36) and the analysis performed here on sequences mapped to R-bands revealed that fragile regions are enriched in flexibility clusters compared to nonfragile regions. The inconsistency in replication progression between fragile and flanking nonfragile regions might contribute to fragility. In the current study a similar difference in the flexibility pattern was found for fragile and nonfragile regions mapped to G-bands.
In summary, the results presented in the present study pinpoint the sequences that may contribute to the fragility of common fragile sites and indicate a general basis of fragility for rare and common fragile sites induced by different replication inhibitors.
Acknowledgments
This study was supported by the ISF grant to B.K.
REFERENCES
- 1.Arlt, M. F., D. E. Miller, D. G. Beer, and T. W. Glover. 2002. Molecular characterization of FRAXB and comparative common fragile site instability in cancer cells. Genes Chromosomes Cancer 33:82-92. [DOI] [PubMed] [Google Scholar]
- 2.Bacolla, A., R. Gellibolian, M. Shimizu, S. Amirhaeri, S. Kang, K. Ohshima, J. E. Larson, S. C. Harvey, B. D. Stollar, and R. D. Wells. 1997. Flexible DNA: genetically unstable CTG.CAG and CGG.CCG from human hereditary neuromuscular disease genes. J. Biol. Chem. 272:16783-16792. [DOI] [PubMed] [Google Scholar]
- 3.Becker, N. A., E. C. Thorland, S. R. Denison, L. A. Phillips, and D. I. Smith. 2002. Evidence that instability within the FRA3B region extends four megabases. Oncogene 21:8713-8722. [DOI] [PubMed] [Google Scholar]
- 4.Casper, A. M., P. Nghiem, M. F. Arlt, and T. W. Glover. 2002. ATR regulates fragile site stability. Cell 111:779-789. [DOI] [PubMed] [Google Scholar]
- 5.Challberg, M. D., and T. J. Kelly, Jr. 1979. Adenovirus DNA replication in vitro: origin and direction of daughter strand synthesis. J. Mol. Biol. 135:999-1012. [DOI] [PubMed] [Google Scholar]
- 6.Chen, H. H., D. C. Rau, and E. Charney. 1985. The flexibility of alternating dA-dT sequences. J. Biomol. Struct. Dyn. 2:709-719. [DOI] [PubMed] [Google Scholar]
- 7.Ciullo, M., M. A. Debily, L. Rozier, M. Autiero, A. Billault, V. Mayau, S. El Marhomy, J. Guardiola, A. Bernheim, P. Coullin, D. Piatier-Tonneau, and M. Debatisse. 2002. Initiation of the breakage-fusion-bridge mechanism through common fragile site activation in human breast cancer cells: the model of PIP gene duplication from a break at FRA7I. Hum. Mol. Genet. 11:2887-2894. [DOI] [PubMed] [Google Scholar]
- 8.Droge, P., J. M. Sogo, and H. Stahl. 1985. Inhibition of DNA synthesis by aphidicolin induces supercoiling in simian virus 40 replicative intermediates. EMBO J. 4:3241-3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferreira, J., G. Paolella, C. Ramos, and A. I. Lamond. 1997. Spatial organization of large-scale chromatin domains in the nucleus: a magnified view of single chromosome territories. J. Cell Biol. 139:1597-1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Franchitto, A., and P. Pichierri. 2002. Protecting genomic integrity during DNA replication: correlation between Werner's and Bloom's syndrome gene products and the MRE11 complex. Hum. Mol. Genet. 11:2447-2453. [DOI] [PubMed] [Google Scholar]
- 11.Fry, M., and L. A. Loeb. 1994. The fragile X syndrome d(CGG)n nucleotide repeats form a stable tetrahelical structure. Proc. Natl. Acad. Sci. USA 91:4950-4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gacy, A. M., G. Goellner, N. Juranic, S. Macura, and C. T. McMurray. 1995. Trinucleotide repeats that expand in human disease form hairpin structures in vitro. Cell 81:533-540. [DOI] [PubMed] [Google Scholar]
- 13.Gardiner, K. 1995. Human genome organization. Curr. Opin. Genet. Dev. 5:315-322. [DOI] [PubMed] [Google Scholar]
- 14.Gellibolian, R., A. Bacolla, and R. D. Wells. 1997. Triplet repeat instability and DNA topology: an expansion model based on statistical mechanics. J. Biol. Chem. 272:16793-16797. [DOI] [PubMed] [Google Scholar]
- 15.Glover, T. W. 1998. Instability at chromosomal fragile sites. Rec. Results Cancer Res. 154:185-199. [DOI] [PubMed] [Google Scholar]
- 16.Glover, T. W., C. Berger, J. Coyle, and B. Echo. 1984. DNA polymerase alpha inhibition by aphidicolin induces gaps and breaks at common fragile sites in human chromosomes. Hum. Genet. 67:136-142. [DOI] [PubMed] [Google Scholar]
- 17.Glover, T. W., A. W. Hoge, D. E. Miller, J. E. Ascara-Wilke, A. N. Adam, S. L. Dagenais, C. M. Wilke, H. A. Dierick, and D. G. Beer. 1998. The murine Fhit gene is highly similar to its human orthologue and maps to a common fragile site region. Cancer Res. 58:3409-3414. [PubMed] [Google Scholar]
- 18.Handt, O., E. Baker, S. Dayan, S. M. Gartler, E. Woollatt, R. I. Richards, and R. S. Hansen. 2000. Analysis of replication timing at the FRA10B and FRA16B fragile site loci. Chromosome Res. 8:677-688. [DOI] [PubMed] [Google Scholar]
- 19.Handt, O., G. R. Sutherland, and R. I. Richards. 2000. Fragile sites and minisatellite repeat instability. Mol. Genet. Metab. 70:99-105. [DOI] [PubMed] [Google Scholar]
- 20.Hansen, R. S., T. K. Canfield, A. D. Fjeld, S. Mumm, C. D. Laird, and S. M. Gartler. 1997. A variable domain of delayed replication in FRAXA fragile X chromosomes: X inactivation-like spread of late replication. Proc. Natl. Acad. Sci. USA 94:4587-4592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hansen, R. S., T. K. Canfield, M. M. Lamb, S. M. Gartler, and C. D. Laird. 1993. Association of fragile X syndrome with delayed replication of the FMR1 gene. Cell 73:1403-1409. [DOI] [PubMed] [Google Scholar]
- 22.Hecht, F., and T. W. Glover. 1984. Cancer chromosome breakpoints and common fragile sites induced by aphidicolin. Cancer Genet. Cytogenet. 13:185-188. [DOI] [PubMed] [Google Scholar]
- 23.Hellman, A., A. Rahat, S. W. Scherer, A. Darvasi, L. C. Tsui, and B. Kerem. 2000. Replication delay along FRA7H, a common fragile site on human chromosome 7, leads to chromosomal instability. Mol. Cell. Biol. 20:4420-4427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hellman, A., E. Zlotorynski, S. W. Scherer, J. Cheung, J. B. Vincent, D. I. Smith, L. Trakhtenbrot, and B. Kerem. 2002. A role for common fragile site induction in amplification of human oncogenes. Cancer Cell 1:89-97. [DOI] [PubMed] [Google Scholar]
- 25.Hewett, D. R., O. Handt, L. Hobson, M. Mangelsdorf, H. J. Eyre, E. Baker, G. R. Sutherland, S. Schuffenhauer, J.-I. Mao, and R. I. Richards. 1998. FRA10B structure reveals common elements in repeat expansion and chromosomal fragile site genesis. Mol. Cell 1:773-781. [DOI] [PubMed] [Google Scholar]
- 26.Huang, C. C., and J. E. Hearst. 1981. Fine mapping of secondary structures of fd phage DNA in the region of the replication origin. Nucleic Acids Res. 9:5587-5599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang, H., J. Qian, J. Proffit, K. Wilber, R. Jenkins, and D. I. Smith. 1998. FRA7G extends over a broad region: coincidence of human endogenous retroviral sequences (HERV-H) and small polydispersed circular DNAs (spcDNA) and fragile sites. Oncogene 16:2311-2319. [DOI] [PubMed] [Google Scholar]
- 28.Jones, C., L. Penny, T. Mattina, S. Yu, E. Baker, L. Voullaire, W. Langdon, G. Sutherland, R. Richards, and A. Tunnacliffe. 1995. Association of a chromosome deletion syndrome with a fragile site within the proto-oncogene CBL2. Nature 376:145-149. [DOI] [PubMed] [Google Scholar]
- 29.Krummel, K. A., S. R. Denison, E. Calhoun, L. A. Phillips, and D. I. Smith. 2002. The common fragile site FRA16D and its associated gene WWOX are highly conserved in the mouse at Fra8E1. Genes Chromosomes Cancer. 34:154-167. [DOI] [PubMed] [Google Scholar]
- 30.LaDuca, R. J., P. J. Fay, C. Chuang, C. S. McHenry, and R. A. Bambara. 1983. Site-specific pausing of deoxyribonucleic acid synthesis catalyzed by four forms of Escherichia coli DNA polymerase III. Biochemistry 22:5177-5188. [DOI] [PubMed] [Google Scholar]
- 31.Le Beau, M. M., F. V. Rassool, M. E. Neilly, R. Espinosa III, T. W. Glover, D. I. Smith, and T. W. McKeithan. 1998. Replication of a common fragile site, FRA3B, occurs late in S phase and is delayed further upon induction: implications for the mechanism of fragile site induction. Hum. Mol. Genet. 7:755-761. [DOI] [PubMed] [Google Scholar]
- 32.Lichter, P., T. Cremer, J. Borden, L. Manuelidis, and D. C. Ward. 1988. Delineation of individual human chromosomes in metaphase and interphase cells by in situ suppression hybridization using recombinant DNA libraries. Hum. Genet. 80:224-234. [DOI] [PubMed] [Google Scholar]
- 33.Lonn, U., and S. Lonn. 1988. Extensive regions of single-stranded DNA in aphidicolin-treated melanoma cells. Biochemistry 27:566-570. [DOI] [PubMed] [Google Scholar]
- 34.Mangelsdorf, M., K. Ried, E. Woollatt, S. Dayan, H. Eyre, Finnis, M., L. Hobson, J. Nancarrow, D. Venter, E. Baker, and R. I. Richards. 2000. Chromosomal fragile site FRA16D and DNA instability in cancer. Cancer Res. 60:1683-1689. [PubMed] [Google Scholar]
- 35.Mimori, K., T. Druck, H. Inoue, A. H., L. Berk, M. Mori, K. Huebner, and C. M. Croce. 1999. Cancer-specific chromosome alterations in the constitutive fragile region FRA3B. Proc. Natl. Acad. Sci. USA 96:7456-7461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mishmar, D., Y. Mandel-Gutfreund, H. Margalit, and B. Kerem. 1999. Common fragile sites: G-band characteristics within an R-band. Am. J. Hum. Genet. 64:908-910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mishmar, D., A. Rahat, S. W. Scherer, G. Nyakatura, B. Hinzmann, Y. Kohwi, Y. Mandel-Gutfroind, J. R. Lee, B. Drescher, D. E. Sas, H. Margalit, M. Platzer, A. Weiss, L.-C. Tsui, A. Rosenthal, and B. Kerem. 1998. Molecular characterization of a common fragile site (FRA7H) on human chromosome 7 by the cloning of an SV40 integration site. Proc. Natl. Acad. Sci. USA 95:8141-8146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morelli, C., E. Karayianni, C. Magnanini, A. J. Mungall, E. Thorland, M. Negrini, D. I. Smith, and G. Barbanti-Brodano. 2002. Cloning and characterization of the common fragile site FRA6F harboring a replicative senescence gene and frequently deleted in human tumors. Oncogene 21:7266-7276. [DOI] [PubMed] [Google Scholar]
- 39.Ohshima, K., and R. D. Wells. 1997. Hairpin formation during DNA synthesis primerrealignment in vitro in triplet repeat sequences from human hereditary disease genes. J. Biol. Chem. 272:16798-16806. [DOI] [PubMed] [Google Scholar]
- 40.Paradee, W., C. M. Wilke, L. Wang, R. Shridhar, C. M. Mullins, A. Hoge, T. W. Glover, and D. I. Smith. 1996. A 350-kb cosmid contig in 3p14.2 that crosses the t(3;8) hereditary renal cell carcinoma ranslocation breakpoint and 17 aphidicolin-induced FRA3B breakpoints. Genomics 35:87-93. [DOI] [PubMed] [Google Scholar]
- 41.Pearson, C. E., and R. R. Sinden. 1996. Alternative structures in duplex DNA formed within the trinucleotide repeats of the myotonic dystrophy and fragile X loci. Biochemistry 35:5041-5053. [DOI] [PubMed] [Google Scholar]
- 42.Pearson, C. E., Y. H. Wang, J. D. Griffith, and R. R. Sinden. 1998. Structural analysis of slipped-strand DNA (S-DNA) formed in (CTG)n •(CAG)n repeats from the myotonic dystrophy locus. Nucleic Acids Res. 26:816-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rassool, F. V., M. M. Le Beau, M.-L. Shen, M. E. Neilly, R. Espinosa III, S. T. Ong, F. Boldog, H. Drabkin, R. McCarroll, and T. W. McKeithan. 1996. Direct cloning of DNA sequences from the common fragile Site region at chromosome band 3p14.2. Genomics 35:109-117. [DOI] [PubMed] [Google Scholar]
- 44.Richards, R. I. 2001. Fragile and unstable chromosomes in cancer: causes and consequences. Trends Genet. 17:339-345. [DOI] [PubMed] [Google Scholar]
- 45.Ried, K., M. Finnis, L. Hobson, M. Mangelsdorf, S. Dayan, J. K. Nancarrow, E. Woollatt, G. Kremmidiotis, A. Gardner, D. Venter, E. Baker, and R. I. Richards. 2000. Common chromosomal fragile site FRA16D sequence: identification of the FOR gene spanning FRA16D and homozygous deletions and translocation breakpoints in cancer cells. Hum. Mol. Genet. 9:1651-1663. [DOI] [PubMed] [Google Scholar]
- 46.Sadoni, N., S. Langer, C. Fauth, G. Bernardi, T. Cremer, B. M. Turner, and D. Zink. 1999. Nuclear organization of mammalian genomes: polar chromosome territories build up functionally distinct higher order compartments. J. Cell Biol. 146:1211-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Samadashwily, G. M., R. Raca, and S. M. Mirkin. 1997. Trinucleotide repeats affect DNA replication in vivo. Nat. Genet. 17:298-304. [DOI] [PubMed] [Google Scholar]
- 48.Sarai, A., J. Mazur, R. Nussinov, and R. L. Jernigan. 1989. Sequence dependence of DNA conformational flexibility. Biochemistry 28:7842-7849. [DOI] [PubMed] [Google Scholar]
- 49.Shiraishi, T., T. Druck, K. Mimori, J. Flomenberg, L. Berk, H. Alder, W. Miller, K. Huebner, and C. M. Croce. 2001. Sequence conservation at human and mouse orthologous common fragile regions, FRA3B/FHIT and Fra14A2/Fhit. Proc. Natl. Acad. Sci. USA 8:5722-5727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Simonic, I., and G. S. Gericke. 1996. The enigma of common fragile sites. Hum. Genet. 97:524-531. [DOI] [PubMed] [Google Scholar]
- 51.Subramanian, P. S., D. L. Nelson, and A. C. Chinault. 1996. Large domains of apparent delayed replication timing associated with triplet repeat expansion at FRAXA and FRAXE. Am. J. Hum. Genet. 59:407-416. [PMC free article] [PubMed] [Google Scholar]
- 52.Sutherland, G. R., P. B. Jacky, and E. G. Baker. 1984. Heritable fragile sites on human chromosomes. XI. Factors affecting expression of fragile sites at 10q25, 16q22, and 17p12. Am. J. Hum. Genet. 36:110-122. [PMC free article] [PubMed] [Google Scholar]
- 53.Sutherland, G. R., and R. I. Richards. 1995. The molecular basis of fragile sites in human chromosomes. Curr. Opin. Genet. Dev. 5:323-327. [DOI] [PubMed] [Google Scholar]
- 54.Thorland, E. C., S. L. Myers, D. H. Persing, G. Sarkar, R. M. McGovern, B. S. Gostout, and D. I. Smith. 2000. Human papillomavirus type 16 integrations in cervical tumors frequently occur in common fragile sites. Cancer Res. 1:5916-5921. [PubMed] [Google Scholar]
- 55.Usdin, K., and K. J. Woodford. 1995. CGG repeats associated with DNA instability and chromosome fragility form structures that block DNA synthesis in vitro. Nucleic Acids Res. 23:4202-4209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang, L., J. Darling, J. S. Zhang, H. Huang, W. Liu, and D. I. Smith. 1999. Allele-specific late replication and fragility of the most active common fragile site, FRA3B. Hum. Mol. Genet. 8:431-437. [DOI] [PubMed] [Google Scholar]
- 57.Wiekowski, M., P. Droge, and H. Stahl. 1987. Monoclonal antibodies as probes for a function of large T antigen during the elongation process of simian virus 40 DNA replication. J. Virol. 61:411-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wilke, C. M., S. W. Guo, B. K. Hall, F. Boldog, R. M. Gemmill, S. C. Chandrasekharappa, C. L. Barcroft, H. A. Drabkin, and T. W. Glover. 1994. Multicolor FISH mapping of YAC clones in 3p14 and identification of YAC spanning both FRA3B and the t(3;8) associated with hereditary renal cell carcinoma. Genomics 22:319-326. [DOI] [PubMed] [Google Scholar]
- 59.Wilke, C. M., B. K. Hall, A. Hoge, W. Pardee, D. l. Smith, and T. W. Glover. 1996. FRA3B extends over a broad region and contains a spontaneous HPV16 integration site: direct evidence for the coincidence of viral integrastion sites and fragile sites. Hum. Mol. Genet. 5:187-195. [DOI] [PubMed] [Google Scholar]
- 60.Yu, S., M. Mangelsdorf, D. Hewett, L. Hobson, E. Baker, H. J. Eyre, N. Lapsys, D. Le Paslier, N. A. Doggett, G. R. Sutherland, and R. I. Richards. 1997. Human chromosomal fragile site FRA16B is an amplified AT-rich minisatellite repeat. Cell 88:367-374. [DOI] [PubMed] [Google Scholar]
- 61.Yunis, J. J., and A. Soreng. 1984. Constitutive fragile sites and cancer. Science 226:1199-1204. [DOI] [PubMed] [Google Scholar]
- 62.Zimonjic, D. B., T. Druck, M. Ohta, K. Kastury, C. M. Croce, N. C. Popescu, and K. Huebner. 1997. Positions of chromosome 3p14.2 fragile sites (FRA3B) within the FHIT gene. Cancer Res. 57:1166-1170. [PubMed] [Google Scholar]