Abstract
The yeast Saccharomyces cerevisiae contains three proteins (Kap104p, Pse1p, and Kap123p) that share similarity to the 95-kDa β subunit of the nuclear transport factor importin (also termed karyopherin and encoded by KAP95/RSL1 in yeast). Proteins that contain nuclear localization sequences are recognized in the cytoplasm and delivered to the nucleus by the heterodimeric importin complex. A second importin-related protein, transportin, delivers a subset of heterogeneous nuclear ribonucleoproteins (hnRNPs) to the nucleoplasm. We now show that in contrast to loss of importin β (Kap95p/Rsl1p) and transportin (Kap104p), conditional loss of Pse1p in a strain lacking Kap123p results in a specific block of mRNA export from the nucleus. Overexpression of Sxm1p, a protein related to Cse1p in yeast and to the human cellular apoptosis susceptibility protein, relieves the defects of cells lacking Pse1p and Kap123p. Thus, a major role of Pse1p, Kap123p, and Sxm1p may be nuclear export rather than import, suggesting a symmetrical relationship between these processes.
Movement of macromolecules in and out of the nucleus is a highly regulated process essential for proper progression though the cell cycle, response to extracellular signals, and viral maturation. Our knowledge of the mechanism by which proteins are imported into and RNAs exported from the nucleus has grown significantly in recent years. Proteins have been identified that mediate both inward and outward macromolecular movement and both processes appear to be regulated by some of the same factors (reviewed in refs. 1 and 2).
Import of proteins into the nucleus is a highly conserved, multistep process initiated by recognition of a nuclear localization sequence (NLS) by a cognate receptor, followed by docking of the complex at the nuclear pore, and GTP-dependent translocation into the nucleoplasm. The receptor for the canonical NLS is a heterodimeric protein complex variously called importin or karyopherin, composed of an α (NLS-binding) and a β (docking) subunit (3–6). Translocation through the nuclear pore is driven by GTP hydrolysis, catalyzed by the small ras-related GTP-binding protein Ran and its regulators (7–10). Once inside the nucleus, the importin/karyopherin complex is probably dissociated and the transport factors recycled to the cytoplasm (5). This relatively simple model fails to explain the nuclear import of proteins that lack an NLS conforming to the canonical sequences. The recent identification of a role for the importin β homolog transportin, in the nuclear import of heterogeneous nuclear ribonucleoprotein (hnRNP) A1 raised the possibility that different classes of nuclear proteins would be transported by different pathways, defined by the identity of the β subunit involved (11, 12).
In contrast to protein import, how RNAs are exported from the nucleus to the cytoplasm is less well understood. Nascent RNA is subject to a number of posttranscriptional modifications, and it seems likely that RNA moves through the nucleoplasm and the nuclear pore in a complex with a number of specific proteins (13, 14). RNA export does show a number of parallels with nuclear import. It appears to be mediated by nuclear export sequences present in RNA binding proteins such as HIV Rev (15). Poly(A)+ RNA export is dependent on the nucleotide state of Ran (10, 16, 17). Furthermore, binding of a heterodimeric protein complex is critical for export in the case of small nuclear RNAs, and this complex has recently been shown to interact with the α subunit of importin (18, 19). However, little is known about what other factors recognize RNA–protein complexes for export and what drives their outward movement through the pore.
In yeast, the homologs of importin α and β are encoded by the SRP1/KAP60 and RSL1/KAP95 genes, respectively (20, 21). Temperature-sensitive mutants in either of these genes result in defects in NLS-dependent nuclear protein import, but not in RNA export (21, 22). The recently completed yeast genome sequence identified three additional genes encoding proteins with similarity to importin β. One of these, KAP104, encodes the yeast homolog of mammalian transportin (11, 12). PSE1 was previously identified in a screen for effectors of protein secretion but its mechanism of action was not defined (23). The other has not been characterized but is referred to as KAP123 in the yeast database because of its similarity to RSL1/KAP95. Because of their similarity to importin β, we have investigated the potential role of PSE1 and KAP123 in transport of macromolecules across the nuclear envelope in vivo.
MATERIALS AND METHODS
Yeast Strains.
PSE1, KAP123, and SXM1 null strains were generated in the diploid PSY902 (ura3–52/ura3–52, leu2Δ1/leu2Δ1, his3Δ200/his3Δ200, ade2/ADE2, ade3/ADE3, lys2/LYS2, trp1/TRP1) by replacement of the coding region with HIS3 (24). Correct integration was demonstrated by PCR and Southern blot analysis. To confirm that PSE1 is essential for growth, the pse1::HIS3/PSE1 heterozygote was transformed with a URA CEN plasmid (25) containing the PSE1 gene (pPS1066). To confirm that the KAP123 gene is not essential, KAP123 was PCR amplified from genomic DNA, cloned into a URA CEN vector (25) and the resulting plasmid, pPS1067, transformed into the kap123::HIS3/KAP123 heterozygote. After selection of His+ Ura+ transformants and sporulation, none of the four spores were sensitive to 5′-fluoroorotic acid.
Generation of PSE1 Mutations.
Temperature-sensitive mutants were generated by PCR mutagenesis (26, 27). Plasmids were rescued from temperature-sensitive strains and retransformed into the Δpse1 strain to demonstrate plasmid linkage of the temperature-sensitivity. Three pse1 temperature-sensitive alleles were cloned into an integration plasmid (pRS306; ref. 25) and integrated into the haploid strain PSY580 (MATa, ura3–52, leu2Δ1, trp1Δ63, and GAL+; ref. 28) using the pop-in/pop-out replacement method (29). The haploid pse1–1 Δkap123 strain (PSY1042) was generated from a diploid created by a cross between the integrated pse1–1 and Δkap123 strain.
Generation of Anti-Pse1 Antibodies.
A polyclonal anti-Pse1p rabbit antibody was raised against purified recombinant protein containing the N-terminal 480 Pse1p amino acids fused to glutathione S-transferase. To express Pse1p-glutathione S-transferase, PSE1 was PCR amplified and cloned into pGEX4T-1 (Pharmacia).
Localization of Green Fluorescent Protein (GFP) Fusion Proteins.
To generate GFP-tagged proteins, we used a plasmid (pKG67) encoding the NUF2 gene (30) with a C-terminal GFP and the 3′-untranslated region of NUF2. NUF2 was replaced by the PCR-amplified PSE1, KAP123, or the SXM1 genes (pPS1069, pPS1070, and pPS1117). GFP fusion proteins were visualized as described (26).
Overproduction of Pse1p in Yeast.
The PSE1 gene was inserted into pPS293 (26) allowing Pse1p expression under control of the GAL1 promoter. The resulting plasmid, pPS1071, was transformed into PSY580. Cells were grown at 25°C in raffinose-containing medium to a density of 1–2 × 107 cells/ml. Transport of mRNA and proteins was assayed 30 min and 2 hr after Pse1p induction with galactose. Indirect immunofluorescence microscopy (31), using a 1:4,000 dilution of anti-Pse1p antibody, was done 2 hr after galactose induction.
Preparation of Cell Lysates.
Lysates were made from 1 × 108 cells in 1 ml PBSMT (PBS/5 mM MgCl2/0.5% Triton X-100) plus protease inhibitors (0.5 mM phenylmethylsulfonyl fluoride/3 μg/ml each leupeptin, aprotinin, chymostatin, and pepstatin) by glass bead lysis and centrifuged at 13,000 rpm for 10 min at 4°C. Equal amounts of protein were resolved by SDS/PAGE (32) and transferred to nitrocellulose for immunoblot analysis.
Protein and RNA Localization.
Induction of GAL1-driven nuclear reporter proteins was done as described (21). Npl3p was detected with a 1:1,000 dilution of polyclonal antibody (33); Nop1p with a 1:100 dilution of mAb (34); Nab2p and Nab3p with a 1:500 dilution of the mAbs 3F2 and 2F12, respectively (from M. Swanson, College of Medicine, Gainesville, FL); H2B β-galactosidase with a 1:800 dilution of mAb J2F7 (35); and simian virus 40 (SV40)-invertase with a 1:20,000 dilution of polyclonal anti-invertase antibody (36). Indirect immunofluorescence microscopy was done as described (31). mRNA localization was according to ref. 37, with modifications as described (21).
High Copy Suppressor Screen.
A 2-μ URA genomic yeast library (from G. Fink, Whitehead Institute, Cambridge, MA) was introduced into pse1–7 and pse1–21 strains. Approximately 5,000 transformants of each strain were selected on URA dropout media at 25°C for 1 day and shifted for 3 days to 36°C. Plasmids were rescued from temperature-resistant colonies and retransformed into pse1–7 and pse1–21 to demonstrate plasmid linkage. The plasmids were characterized by DNA sequencing.
RESULTS
PSE1 Is Essential for Growth and Has a Redundant Function with KAP123.
Both RSL1/KAP95 and KAP104 genes are necessary for normal growth in yeast. A strain without the RSL1/KAP95 gene is dead (21) and the loss of the KAP104 gene results in slow growth at 25°C and no growth at 36°C (E. Shen and P.A.S., unpublished results, and ref. 11). To study the function of other importin β-like proteins in yeast, we generated a strain missing the PSE1 gene by replacement with HIS3. After sporulation, the pse1::HIS3/PSE1 heterozygote yielded only two viable spores at 25°C. To confirm that PSE1 is essential for growth, the pse1::HIS3/PSE1 heterozygote was transformed with a URA CEN plasmid containing the PSE1 gene. From every four-spore tetrad, two spores were sensitive to 5′-fluoroorotic acid indicating that the pse1::HIS3 spores required a copy of PSE1 to grow.
To test the in vivo function of PSE1, we generated temperature-sensitive alleles by random PCR mutagenesis. Three mutant alleles were introduced back into the genome of haploid yeast cells in place of the wild-type locus. All mutants grow at 25°C, but pse1–1 grows better than pse1–7 and pse1-21 on plates (Fig. 1A). For pse1–21, pse1–7, and pse1–1 cell growth slows within 1 hr after shift to 36°C in solution (data not shown). In contrast to PSE1, a yeast strain missing KAP123 is viable; no growth defects are observed at any temperature (Fig. 1B).
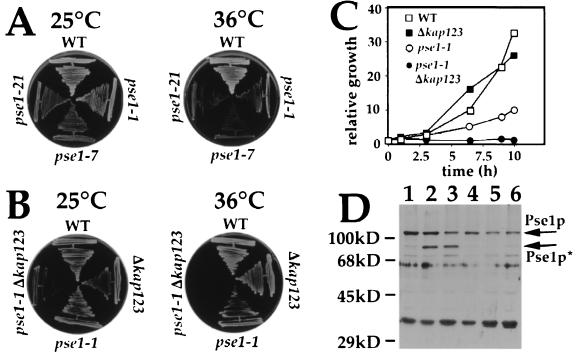
Figure 1.
Effect of mutation in PSE1 and KAP123 on yeast cell growth. (A) PSY580 (WT) and three different temperature-sensitive alleles of pse1 (pse1–1, pse1–7, and, pse1–21) were streaked on rich media and incubated at 25°C or 36°C. (B) PSY580 (WT), a strain without a copy of KAP123 (Δkap123), a strain with a temperature-sensitive allele of PSE1 (pse1–1), and a strain with both mutations (pse1–1 Δkap123) were streaked on rich media and incubated at 25°C or 36°C. (C) PSY580 (WT, □), Δkap123 (▪), pse1–1 (○), and pse1–1 Δkap123 (•) were grown at 25°C in rich liquid medium to a cell density of 1 × 106 cells/ml and shifted to 36°C. Relative cell growth was calculated by counting the cells at indicated time points. (D) Equal amounts of lysate from cells grown at 25°C in rich medium from PSY580 (lane 1), Δkap123 (lane 2), pse1–1 Δkap123 (lane 3), pse1–1 (lane 4), pse1–7 (lane 5), and pse1–21 (lane 6) were resolved on a SDS/PAGE and analyzed by immunoblot using an anti-Pse1p specific antibody.
We tested the possibility that Pse1p and Kap123p might have overlapping functions by creating a new strain containing the pse1–1 temperature-sensitive allele and missing KAP123 (pse1–1 Δkap123). These cells grow less well than pse1–1 at the permissive temperature of 25°C and are temperature sensitive for growth at 36°C (Fig. 1 B and C). As with pse1–1, growth of pse1–1 Δkap123 cells slows within 1 hr after shifting to 36°C (Fig. 1C) and viability of both strains drops to 40–50% of wild-type after 1 hr and to ≈20% after 3 hr at 36°C (data not shown). We were unable to construct strains containing the other two pse1 temperature-sensitive alleles and the deletion of KAP123. In all tetrads from a cross of either pse1–7 or pse1–21 with Δkap123 cells, the pse1–7 Δkap123 and the pse1–21 Δkap123 spores failed to germinate.
In strains with mutant alleles of PSE1 the level of Pse1p is decreased (Fig. 1D). We quantitated the expression of Pse1p by immunoblot using a Pse1p specific polyclonal antibody. Compared with expression of Pse1p in wild-type cells (Fig. 1D, lane 1), the amount of Pse1p is decreased in pse1–1 cells to <50% and in pse1–7 and pse1–21 cells to <25% (Fig. 1D, lanes 4–6). The amount of Pse1p in a Δkap123 strain (Fig. 1D, lane 2) is the same as in a wild-type strain, but in pse1–1 Δkap123 cells Pse1p was reduced to levels as in pse1–7 and pse1–21 cells (Fig. 1D, lanes 3, 5, and 6). However, we detected in Δkap123 and in cells of the double mutant (Fig. 1D, lanes 2 and 3) a second immunoreactive polypeptide with a molecular weight of 90 kDa (Pse1p*). This band might result from degradation of Pse1p suggesting a decrease in the stability of Pse1p in strains missing KAP123. It is also possible that Pse1p* could be a modified form with Kap123p affecting the modification. The reduction of intact Pse1p in the mutants was the same in cells incubated at 25°C as compared with cells shifted for 2 hr to 36°C (data not shown).
Localization of Pse1p and Kap123p.
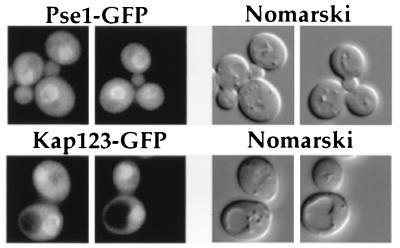
To localize Pse1p and Kap123p, we generated fusion proteins of each to GFP. Both constructs encoded functional proteins because they could restore normal growth to pse1–1 Δkap123 cells when expressed from their respective promoters. Pse1p-GFP is located at the nuclear rim with some intranuclear and cytoplasmic localization (Fig. 2; Upper). The area with the GFP signal was identified as the nucleus by costaining with 4′,6-diamido-2-phenylindole (data not shown). Kap123p-GFP is also nuclear with protein also present in the cytoplasm (Fig. 2, Lower). Similar results were obtained from cells expressing Pse1p under the control of the GAL1 promoter; the intact Pse1p was concentrated at the nuclear envelope and in the nucleus as determined by immunofluorescence microscopy with anti-Pse1p antibody (data not shown).
Figure 2.
Localization of Pse1p and Kap123p. Pse1-GFP was expressed in a Δpse1 strain (Upper). The first and second panels show the GFP signal and the third and fourth panels show the corresponding cells by Nomarski optics. (Lower) The GFP signal of Kap123p tagged with GFP in a Δkap123 strain under the same conditions.
Nuclear Protein Import in pse1 and kap123 Mutants.
The pse1–1, pse1–7, and pse1–21 temperature-sensitive mutants all show normal distribution of several different nuclear proteins after incubation at both permissive and for up to 6 hr at the nonpermissive temperature (data not shown). To assay protein translocation, we used GAL1 driven reporter plasmids encoding the H2B-NLS fused to β-galactosidase (35) and the SV40 NLS fused to normally cytoplasmic invertase (36) as representatives of the canonical NLS containing proteins. We further analyzed the distribution of endogenous Nop1p (38) by indirect immunofluorescence microscopy. As representatives of a second class of nuclear proteins with noncanonical NLSs, we analyzed the distribution of endogenous Nab2p (39), Nab3p (40), Npl3p (33), and a normally nuclear fusion of Hrp1p (41) to GFP. Cells deleted for KAP123 also showed normal distribution of all nuclear proteins tested.
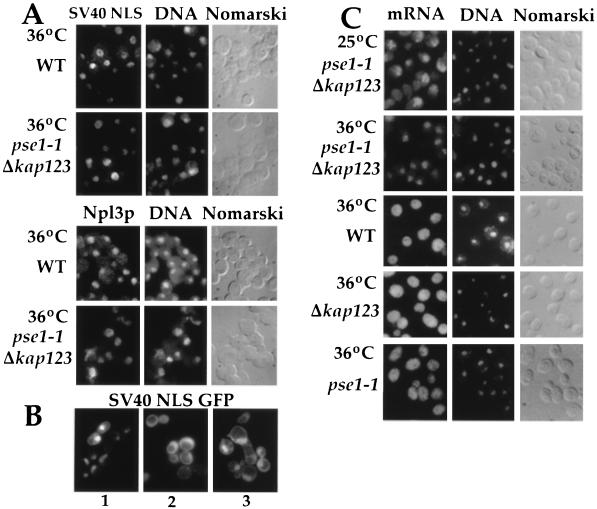
As with the pse1ts alleles, no defect in nuclear protein import was observed in the double pse1–1 Δkap123 mutant. The localization of a normally nuclear, canonical NLS-containing protein, SV40 invertase, was examined at 1 hr (Fig. 3A Upper) and at 3 hr (data not shown) following a shift to the nonpermissive temperature of 36°C. At both time points, SV40 invertase remained completely nuclear in both the double mutant and in wild-type cells (Fig. 3A Upper). Similar results were obtained for the normally nuclear mRNA binding protein, Npl3p (33). After 1 hr at 36°C, Npl3p remained nuclear in both pse1–1 Δkap123 and wild-type cells (Fig. 3A Lower). We also examined the localization of the H2B-NLS fused to β-galactosidase (35), Nop1p (38), Nab2p (39), Nab3p (40), and Hrp1p (41) in pse1–1 Δkap123 cells and found them all to remain nuclear at all temperatures (data not shown). To account for the possibility that these endogenous proteins are not synthesized at sufficiently high levels at the nonpermissive temperature because of the mRNA export block, we also analyzed the re-import of a SV40 NLS-containing GFP fusion protein (NLS-GFP; ref. 42). In pse1–1 Δkap123 cells at 25°C, NLS-GFP localizes in the nucleus (Fig. 3B, panel 1). In cells depleted for ATP, NLS-GFP diffuses out of the nucleus (Fig. 3B, panel 2). However, following a shift to the nonpermissive temperature and re-addition of glucose, the NLS-GFP protein was efficiently re-imported into the nucleus (Fig. 3B, panel 3).
Figure 3.
Protein and mRNA transport in pse1–1 Δkap123 mutants. (A) Cells were grown in selective media and the SV40 NLS invertase was expressed under the control of the ADH1 promotor and visualized by immunofluorescence using an invertase specific antibody (Upper). Npl3p was visualized by immunofluorescence using a Npl3p specific antibody (Lower). (B) pse1–1 Δkap123 cells expressing NLS-GFP were grown at 25°C (panel 1), washed into glucose-free media with 10 mM sodium azide and 2-deoxy-d-glucose for 45 min (panel 2), and then shifted to 36°C for 30 min and then washed into prewarmed glucose-containing media and incubated for 15 min at 36°C (panel 3). (C) Poly(A)+ RNA was detected using an in situ hybridization assay with an oligo dT(50) probe (37).
pse1–1 Δkap123 Mutants Show Defects in mRNA Export.
In contrast to nuclear protein localization, mRNA export from the nucleus is impaired in pse1–1 Δkap123 mutants (Fig. 3C). The distribution of poly(A)+ RNA was examined by in situ hybridization with a poly(dT) probe. In wild-type yeast, Δkap123 mutants, and pse1–1 mutants, mRNA is distributed throughout the cytoplasm indicating normal rapid export from the nucleus. On the other hand, following a 1 hr shift to 36°C, the pse1–1 Δkap123 mutant showed accumulation of poly(A)+ RNA in all nuclei, indicating a block in mRNA export. Even at the permissive temperature, some mRNA accumulates in the nuclei of the double mutant. Similarly, overexpression of PSE1 from the strong GAL1 promoter in wild-type cells also results in cessation of cell growth and accumulation of mRNA in the nucleus in the majority of cells (data not shown). Therefore, both loss and overexpression of Pse1p yield similar phenotypes.
Sxm1p, a Cse1p-Like Protein, Suppresses Mutations in pse1.
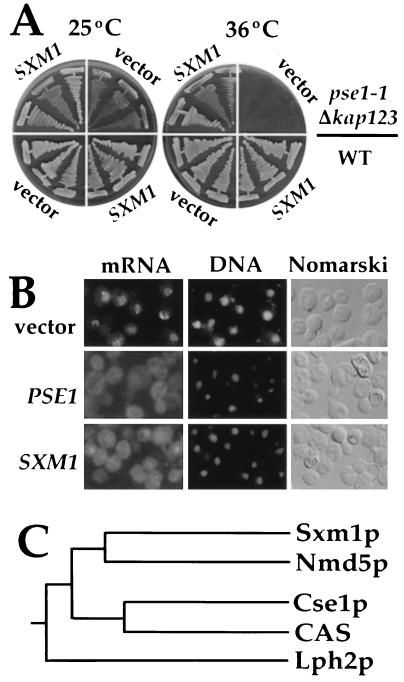
To identify other proteins that might function like or with Pse1p, we introduced a genomic library on multi-copy plasmids into the pse1ts cells and selected for plasmids that allowed cells to grow at 36°C. Three plasmids that suppressed the temperature-sensitive phenotype of both pse1–7 and pse1–21 alleles were characterized by DNA sequencing. One contained the entire PSE1 gene and the other two plasmids were identical. The two identical plasmids contained a 7-kb genomic fragment of yeast chromosome IV. To identify the smallest ORF that suppresses the temperature-sensitive growth, we subcloned a 3.5-kb genomic fragment of the original high copy suppressor clone into a 2 μ-containing plasmid, which we termed SXM1 (suppressor of export machinery). When SXM1 was introduced in multiple copies into pse1–1 Δkap123 double mutants, the ability to grow at 36°C was also restored (Fig. 4A). Moreover, SXM1 in high copy partially restored normal mRNA export to pse1–1 Δkap123 cells (Fig. 4B). SXM1 encodes a yeast protein with 25.3% identity to Nmd5p, 17.3% to Cse1p (43), 15.8% to human cellular apoptosis susceptibility protein (CAS) (44), and 14.5% to Lph2p. The phylogenetic relationship between these four yeast Cse1p-like proteins and a human homolog, the cellular apoptosis susceptibility protein, is shown in Fig. 4C.
Figure 4.
A CSE1 homolog suppresses the mRNA export defect of pse1–1 Δkap123. (A) The wild-type strain PSY580 (Lower) and the pse1–1 Δkap123 strain (Upper) were transformed with a 2-μ plasmid alone (vector) or with a 2-μ plasmid containing the SXM1 gene. Three individual transformants of each strain were streaked on selective media and incubated at 25°C or 36°C. (B) Cells of the pse1–1 Δkap123 strain were grown to 1 × 107 cells/ml in selective media at 25°C and shifted for 1 hr to 36°C. As a control, the pse1–1 Δkap123 strain (Top) was transformed with an empty 2-μ plasmid (vector), a plasmid encoding PSE1 (Middle), or a 2-μ plasmid containing the SXM1 gene (Bottom). The first column shows the intracellular distribution of mRNA. Nuclei, shown in the second column, were visualized with 4′,6-diamidino-2-phenylindole stain. (C) The phylogenetic tree of Sxm1p (designated as YDR395w by the yeast genome project), Nmd5p (YJR132w), Lph2p (YPL282c), Cse1p (chromosome segregation protein 1, YGL238w), and human CAS protein (GenBank database accession no. U33286) was done using the clustal method.
To begin to further characterize the function of Sxm1p, we deleted the SXM1 gene. A yeast strain missing SXM1 is viable and shows no growth defects at any temperature (data not shown).
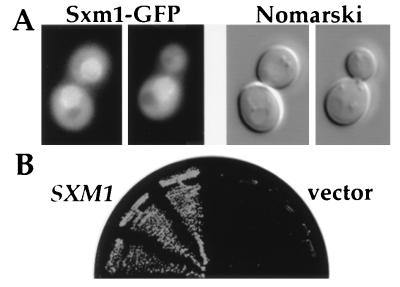
We localized Sxm1p as a GFP fusion in the same manner as for Pse1p and Kap123p. Sxm1p-GFP showed a similar intracellular distribution as Pse1p and Kap123p. The protein is concentrated in the nucleus and at the nuclear envelope. In addition, all cells showed a weaker GFP signal in the cytoplasm (Fig. 5A). To demonstrate that Sxm1p-GFP is a functional protein, we transformed the pse1–1 Δkap123 strain with a low copy plasmid encoding Sxm1p-GFP. The fusion protein was able to support growth of the double mutant at 36°C. This result suggests that SXM1 not only suppresses temperature-sensitive mutations in PSE1 in high copy but can also partially substitute for PSE1. To further test this idea, a Δpse1 strain containing a PSE1 URA3 plasmid was transformed with a SXM1 LEU2 CEN plasmid. The strains containing SXM1 were able to grow on 5′-fluoroorotic acid indicating that Sxm1p can substitute for Pse1p when present in only a few copies per cell (Fig. 5B).
Figure 5.
Sxm1p, a primarily nuclear protein, functionally substitutes for Pse1p. (A) Sxm1p tagged with GFP was expressed in a Δsxm1 strain. Cells were grown at 25°C in selective media to early log phase. The left side shows the GFP signal and the right side the corresponding cells by Nomarski optics. The area with the GFP signal was identified as the nucleus by costaining of DNA with 4′,6-diamidino-2-phenylindole (not shown). (B) A Δpse1 strain covered with a PSE1 URA CEN plasmid was transformed with a low copy LEU 2 plasmid encoding SXM1 or with a LEU 2-containing plasmid alone (vector). Three individual transformants of each were grown at 25°C on 5′-fluoroorotic acid containing plates.
DISCUSSION
By using yeast mutants, we have shown an essential requirement for Pse1p and Kap123p in mRNA export from the nucleus. PSE1 and KAP123 appear to have some functional redundancy because KAP123 is not essential for growth, whereas PSE1 is. However, the lack of KAP123 in the pse1–1 mutant strain results in a strong mRNA export defect. The fact that pse1–1 cells are temperature sensitive could be a result of a very weak RNA export defect in general, or that pse1–1 cells fail to export one specific mRNA. Alternatively, it is possible that Pse1p could be affecting some additional cellular process, with that defect causing the temperature-sensitive growth. However, suppression of both temperature sensitivity and the mRNA export block can be achieved by overexpression of the SXM1 gene.
The results presented here suggest a novel role for proteins in the importin β family. We show that two importin β family members, Pse1p and Kap123p, are required in vivo for nuclear export of mRNA. As such, these proteins may function as adapters between specific recognition/carrier proteins, such as importin α (4, 22) or the cap-binding protein complex (18) and the machinery that moves macromolecules through the pore. We propose a symmetric distribution (and function) of importin β-like proteins on both sides of the nuclear envelope. Importin β (Kap95p) and Kap104p are cytoplasmic proteins, binding the nuclear pore complex (5, 11). GFP-tagged Pse1p and Kap123p are functional and localize to the nucleus, at the nuclear envelope and, to some extent, in the cytoplasm. The cytoplasmic signal may suggest a dynamic distribution of Pse1p and Kap123p; perhaps these proteins move in and out of the nucleus as part of their transport function. An overexpressed N-terminal glutathione S-transferase or GFP-Pse1p fusion protein localizes exclusively to the nucleus (data not shown). However, these N-terminal Pse1p fusion proteins, in contrast to the C-terminal GFP fusion proteins, are not able to support growth of Δpse1. One interesting possibility is that the addition of a moiety to the N-terminus restricts the proteins to the nucleus and thus interferes with their in vivo function.
Importin β was originally identified as an essential cytoplasmic nuclear import factor—required, together with importin α, for recognition of the NLS and delivery to the nuclear pore (45–47). Yeast mutants defective in RSL1/KAP95 only show defects in NLS-dependent protein import (21). Similarly, transportin appears to effect the import of hnRNP A1 in mammalian cells (12). The yeast transportin homolog, Kap104p, also seems to primarily affect import of two RNA binding proteins (11). These observations have led to the proposal that the primary roles of importin β and transportin are to import different substrates into the nucleus.
We suggest that Pse1p and Kap123p may play an essential role in nuclear export. This conclusion is based on the following observations: (i) mutants show a very rapid onset in their mRNA export defect, (ii) no protein import defect is observed, and (iii) Pse1p and Kap123p can be found at the nuclear envelope and in the nucleus. In the absence of the KAP123 gene a mutation in PSE1 causes defects in the ability to export mRNAs. These data are in contrast to what has been observed for Rsl1p/Kap95p and Kap104 (the other two members of the yeast importin β family), which when mutated show no defects in mRNA export (refs. 11 and 21, and E. Shen and P.A.S., unpublished results).
The possibility does remain that we have not found the “right” protein whose import is effected by mutation in PSE1 and KAP123 and that failure to import this protein results in the mRNA export defect. To try to address this, we examined the nuclear import of different classes of nuclear proteins and found them all to remain nuclear even when mRNA export appeared completely compromised. We examined the localization of Npl3p (26) and Hrp1p (41), which we know to be directly involved in mRNA export and processing. These proteins remain nuclear in the double mutant. However, Npl3p and Hrp1p also shuttle between the nucleus and the cytoplasm (ref. 26, E. Shen and P.A.S., unpublished data) and it could be that mutation of PSE1 and KAP123 block their export. This might be expected given the premise that Npl3p must accompany mRNA as it transits through the nuclear pore complex (26).
Interestingly, we identified SXM1 by its ability to compensate for the loss of PSE1. SXM1 is related to the uncharacterized NMD5, LPH2, and to CSE1. Furthermore, a number of other proteins related to Cse1p/importin β have recently been identified (48). CSE1 was originally implicated in chromosome maintenance (43) and more recently in regulating the cell cycle-dependent degradation of cyclins by the anaphase promoting complex/cyclosome (49). Depletion of a human homolog, CAS, has been shown to prevent apoptosis in certain cancer cell lines (44, 50). It is tempting to speculate that these effects may be connected to nuclear transport events. Other nuclear transport factors, such as importin α in yeast and Drosophila have been shown to alter the ability of cells to transit through mitosis (22, 51). Taken together, nuclear import and export may be a way for the cell to control its growth cycle, for instance in regulating the function of the anaphase promoting complex/cyclosome. Thus, nuclear transport factors such as the ones characterized here are novel candidate targets for growth control.
Acknowledgments
We thank P. Ferrigno, J. A. Kahana, A. Corbett, D. Wong, and M. Lee for advice and comments on the manuscript, and E. Shen for communication of unpublished data. This work was supported by the National Institutes of Health (P.A.S.) and the Deutsche Forschungsgemeinschaft (M.S.).
ABBREVIATIONS
- NLS
nuclear localization sequence
- GFP
green fluorescent protein
- SV40
simian virus 40
- hnRNP
heterogeneous nuclear ribonucleoprotein
- CAS
cellular apoptosis susceptibility protein
References
- 1.Görlich D, Mattaj I W. Science. 1996;271:1513–1518. doi: 10.1126/science.271.5255.1513. [DOI] [PubMed] [Google Scholar]
- 2.Koepp D M, Silver P A. Cell. 1996;87:1–4. doi: 10.1016/s0092-8674(00)81315-x. [DOI] [PubMed] [Google Scholar]
- 3.Adam S A, Gerace L. Cell. 1991;66:837–847. doi: 10.1016/0092-8674(91)90431-w. [DOI] [PubMed] [Google Scholar]
- 4.Görlich D, Prehn S, Laskey R A, Hartmann E. Cell. 1994;79:767–778. doi: 10.1016/0092-8674(94)90067-1. [DOI] [PubMed] [Google Scholar]
- 5.Görlich D, Vogel F, Mills A D, Hartmann E, Laskey R A. Nature (London) 1995;377:246–248. doi: 10.1038/377246a0. [DOI] [PubMed] [Google Scholar]
- 6.Moroianu J, Blobel G, Radu A. Proc Natl Acad Sci USA. 1995;92:2008–2011. doi: 10.1073/pnas.92.6.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moore M S, Blobel G. Nature (London) 1993;365:661–663. doi: 10.1038/365661a0. [DOI] [PubMed] [Google Scholar]
- 8.Melchior F, Guan T, Yokoyama N, Nishimoto T, Gerace L. J Cell Biol. 1995;131:571–581. doi: 10.1083/jcb.131.3.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Corbett A H, Koepp D M, Lee M S, Schlenstedt G, Hopper A K, Silver P A. J Cell Biol. 1995;130:1017–1026. doi: 10.1083/jcb.130.5.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schlenstedt G, Saavedra C, Loeb J D J, Cole C N, Silver P A. Proc Natl Acad Sci USA. 1995;92:225–229. doi: 10.1073/pnas.92.1.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aitchinson J D, Blobel G, Rout M P. Science. 1996;274:624–627. doi: 10.1126/science.274.5287.624. [DOI] [PubMed] [Google Scholar]
- 12.Pollard V W, Michael W M, Nakielny S, Siomi M C, Wang F, Dreyfuss G. Cell. 1996;86:985–994. doi: 10.1016/s0092-8674(00)80173-7. [DOI] [PubMed] [Google Scholar]
- 13.Visa N, Alzhanova-Ericsson A T, Sun X, Kiseleva E, Bjorkroth B, Wurtz T, Daneholt B. Cell. 1996;84:253–264. doi: 10.1016/s0092-8674(00)80980-0. [DOI] [PubMed] [Google Scholar]
- 14.Visa N, Izaurralde E, Ferreira J, Daneholt B, Mattaj I W. J Cell Biol. 1996;133:5–14. doi: 10.1083/jcb.133.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gerace L. Cell. 1995;82:341–344. doi: 10.1016/0092-8674(95)90420-4. [DOI] [PubMed] [Google Scholar]
- 16.Kadowaki T, Goldfarb D, Spitz L M, Tartakoff A M, Ohno M. EMBO J. 1993;12:2929–2937. doi: 10.1002/j.1460-2075.1993.tb05955.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saavedra C, Tung K-S, Amberg D C, Hopper A K, Cole C N. Genes Dev. 1996;10:1608–1620. doi: 10.1101/gad.10.13.1608. [DOI] [PubMed] [Google Scholar]
- 18.Görlich D, Kraft R, Kostka S, Vogel F, Hartmann E, Laskey R A, Mattaj I W. Cell. 1996;87:21–32. doi: 10.1016/s0092-8674(00)81319-7. [DOI] [PubMed] [Google Scholar]
- 19.Izaurralde E, Lewis J, Gamberi C, Jarmalowski A, McGuigan C, Mattaj I W. Nature (London) 1995;376:709–712. doi: 10.1038/376709a0. [DOI] [PubMed] [Google Scholar]
- 20.Yano R, Oakes M, Tabb M M, Nomura M. Mol Cell Biol. 1992;12:5640–5651. doi: 10.1128/mcb.12.12.5640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koepp D M, Wong D H, Corbett A H, Silver P A. J Cell Biol. 1996;133:1163–1176. doi: 10.1083/jcb.133.6.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Loeb J D L, Schlenstedt G, Pellman D, Kornitzer D, Silver P A, Fink G. Proc Natl Acad Sci USA. 1995;92:7647–7651. doi: 10.1073/pnas.92.17.7647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chow T Y-K, Ash J J, Dignard D, Thomas D Y. J Cell Sci. 1992;101:709–719. doi: 10.1242/jcs.101.3.709. [DOI] [PubMed] [Google Scholar]
- 24.Jones J S, Prakash L. Yeast. 1990;6:363–666. doi: 10.1002/yea.320060502. [DOI] [PubMed] [Google Scholar]
- 25.Sikorski R S, Hieter P. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee M S, Henry M, Silver P A. Genes Dev. 1996;10:1233–1246. doi: 10.1101/gad.10.10.1233. [DOI] [PubMed] [Google Scholar]
- 27.Orr-Weaver T L, Szostak J W. Proc Natl Acad Sci USA. 1983;80:4417–4421. doi: 10.1073/pnas.80.14.4417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Winston F, Dollard C, Ricupero-Hovasse S L. Yeast. 1995;11:53–55. doi: 10.1002/yea.320110107. [DOI] [PubMed] [Google Scholar]
- 29.Rothstein R. In: Guide to Yeast Genetics and Molecular Biology. Guthrie C, Fink G R, editors. San Diego: Academic; 1991. pp. 281–301. [Google Scholar]
- 30.Kahana J A, Schnapp B J, Silver P A. Proc Natl Acad Sci USA. 1995;92:9707–9711. doi: 10.1073/pnas.92.21.9707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Corbett A H, Silver P A. J Biol Chem. 1996;271:18477–18484. doi: 10.1074/jbc.271.31.18477. [DOI] [PubMed] [Google Scholar]
- 32.Laemmli U K. Nature (London) 1970;277:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 33.Bossie M A, DeHoratius C, Barcelo G, Silver P A. Mol Biol Cell. 1992;3:875–893. doi: 10.1091/mbc.3.8.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Flach J, Bossie M, Vogel J, Corbett A H, Jinks T, Willins D A, Silver P A. Mol Cell Biol. 1994;14:8399–8407. doi: 10.1128/mcb.14.12.8399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moreland R B, Langevin G L, Singer R H, Garcea R L, Hereford L M. Mol Cell Biol. 1987;7:4048–4057. doi: 10.1128/mcb.7.11.4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nelson M, Silver P A. Mol Cell Biol. 1989;9:384–389. doi: 10.1128/mcb.9.2.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Amberg D C, Goldstein A L, Cole C N. Genes Dev. 1992;6:1173–1189. doi: 10.1101/gad.6.7.1173. [DOI] [PubMed] [Google Scholar]
- 38.Schimmang T, Tollervey D, Kern H, Frank R, Hurt E C. EMBO J. 1989;8:4015–4024. doi: 10.1002/j.1460-2075.1989.tb08584.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Anderson J T, Wilson S M, Datar K V, Swanson M S. Mol Cell Biol. 1993;13:2730–2741. doi: 10.1128/mcb.13.5.2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wilson S M, Datar K V, Paddy M R, Swedlow J R, Swanson M S. J Cell Biol. 1994;127:1173–1184. doi: 10.1083/jcb.127.5.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Henry M, Borland C Z, Bossie M, Silver P A. Genetics. 1996;142:103–115. doi: 10.1093/genetics/142.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shugula N, Roberts P, Gu Z, Spitz L, Tabb M M, Nomura M, Goldfarb D S. J Cell Biol. 1996;135:329–339. doi: 10.1083/jcb.135.2.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xiao Z, McGrew J T, Schroeder A J, Fitzgerald-Hayes M. Mol Cell Biol. 1993;13:4691–4702. doi: 10.1128/mcb.13.8.4691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brinkmann U, Brinkmann E, Gallo M, Pastan I. Proc Natl Acad Sci USA. 1995;92:10427–10431. doi: 10.1073/pnas.92.22.10427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chi N C, Adam J H E, Adam S A. J Cell Biol. 1995;130:265–274. doi: 10.1083/jcb.130.2.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Imamoto N, Shimamoto T, Takao T, Tachibana T, Kose S, Matsubae M, Sekimoto T, Shimonshi Y, Yoneda Y. EMBO J. 1995;14:3617–3626. doi: 10.1002/j.1460-2075.1995.tb00031.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Radu A, Blobel G, Moore M S. Proc Natl Acad Sci USA. 1995;92:1769–1773. doi: 10.1073/pnas.92.5.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fornerod M, van Dreusen J, van Baal S, Reynolds A, Davis D, Murti K G, Fransen J, Grosveld G. EMBO J. 1997;16:807–816. doi: 10.1093/emboj/16.4.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zachariae W, Shin T H, Galova M, Obermaier B, Nasmyth K. Science. 1996;274:1201–1204. doi: 10.1126/science.274.5290.1201. [DOI] [PubMed] [Google Scholar]
- 50.Brinkmann U, Brinkmann E, Gallo M, Scherf U, Pastan I. Biochemistry. 1996;35:6891–6899. doi: 10.1021/bi952829+. [DOI] [PubMed] [Google Scholar]
- 51.Küssel P, Frasch M. Mol Gen Genet. 1995;248:351–363. doi: 10.1007/BF02191602. [DOI] [PubMed] [Google Scholar]