Abstract
The 14-3-3σ (sigma) protein, a negative regulator of the cell cycle, is a human mammary epithelium-specific marker that is downregulated in transformed mammary carcinoma cells. It has also been identified as a p53-inducible gene product involved in cell cycle checkpoint control after DNA damage. Although 14-3-3σ is linked to p53-regulated cell cycle checkpoint control, detailed mechanisms of how cell cycle regulation occurs remain unclear. Decreased expression of 14-3-3σ was recently reported in several types of carcinomas, further suggesting that the negative regulatory role of 14-3-3σ in the cell cycle is compromised during tumorigenesis. However, this possible tumor-suppressive role of 14-3-3σ has not yet been characterized. Here, we studied the link between 14-3-3σ activities and p53 regulation. We found that 14-3-3σ interacted with p53 in response to the DNA-damaging agent adriamycin. Importantly, 14-3-3σ expression led to stabilized expression of p53. In studying the molecular mechanism of this increased stabilization of p53, we found that 14-3-3σ antagonized the biological functions of Mdm2 by blocking Mdm2-mediated p53 ubiquitination and nuclear export. In addition, we found that 14-3-3σ facilitated the oligomerization of p53 and enhanced p53's transcriptional activity. As a target gene of p53, 14-3-3σ appears to have a positive feedback effect on p53 activity. Significantly, we also showed that overexpression of 14-3-3σ inhibited oncogene-activated tumorigenicity in a tetracycline-regulated 14-3-3σ system. These results defined an important p53 regulatory loop and suggested that 14-3-3σ expression can be considered for therapeutic intervention in cancers.
14-3-3 family proteins have many diverse functions in a broad range of organisms, including critical roles in signal transduction pathways and cell cycle regulation (1, 10, 41). The 14-3-3σ gene, one of seven isoform members of the 14-3-3 family (1), was originally characterized as a human mammary epithelium-specific marker 1 (HME1) (33), which is downregulated in mammary carcinoma cells. We previously cloned 14-3-3σ by expression cloning and characterized its gene product as a negative regulator of cyclin-dependent kinases (CDKs) (21). Independently, the gene was also identified as a p53-inducible gene involved in cell cycle checkpoint control after DNA damage (13).
Because of the negative roles of p53 and 14-3-3σ in cell cycle progression, it is possible that p53 and 14-3-3σ are functionally linked. However, the detailed mechanism that underlies the link between p53 and 14-3-3σ remains elusive. Some target genes of p53, such as Mdm2, can regulate p53 activity (24). Because 14-3-3σ is a target gene of p53 and the 14-3-3 family members may act as possible transcriptional coactivators (10, 31, 40) or molecular chaperone (38), there is a possibility that 14-3-3σ, like Mdm2, regulates the activity or function of p53.
Downregulation of 14-3-3σ can make primary human epithelial cells grow indefinitely in a single step without the need for exogenous oncogenes or oncoviruses, suggesting that the 14-3-3σ decrease leads to tumor formation (6). In addition, 14-3-3σ expression levels are also significantly reduced in v-Ha-ras-transformed mammary epithelial cells, simian virus 40-transformed human keratinocytes (7), head and neck squamous cell carcinoma cell lines (39), primary bladder tumors (29), and colonic polyp specimens (26). Importantly, the 14-3-3σ gene is not expressed in most breast cancers (9), gastric cancer (37), or hepatocellular carcinoma (15). These findings suggest that loss of 14-3-3σ function is crucial in the development of cancer.
p53's tumor-suppressive function is lost in almost half of all human cancers (28), yet the defects in the other half remain elusive. It is possible that p53-independent regulatory mechanisms are lost in these cases, but it may also be that different points of p53-dependent pathways are inactivated. Since 14-3-3σ is regulated by p53 to guard genomic stability (8, 13) and is required to prevent mitotic catastrophe in response to DNA damage (3), dysregulation of 14-3-3σ may account for some of the cancers that do not have p53 mutations. Therefore, p53 may act as the center of a complex network of signaling pathways, and the other components of these pathways, such as 14-3-3σ, pose alternative targets for inactivation.
We previously showed that 14-3-3σ interacts with CDKs and can inhibit CDK activities to block cell cycle progression (21). Overexpression of 14-3-3σ also inhibits cell proliferation and prevents anchorage-independent growth of breast cancer cell lines (21). These findings demonstrate that the tumor-suppressive role of 14-3-3σ has been compromised during tumorigenesis.
Here, we assessed the roles of 14-3-3σ to determine whether 14-3-3σ regulates p53 transcriptionally and functionally. We found that 14-3-3σ specifically increased p53 stability and enhanced p53 transcriptional activity. We discovered that 14-3-3σ antagonized Mdm2-mediated p53 degradation and p53 nuclear export. Also, we examined the effectiveness of using 14-3-3σ as an anticancer therapeutic agent. We showed that 14-3-3σ expression reduced the tumorigenicity of oncogene-expressing cells in nude mice. These results indicate that 14-3-3σ is an important component of the p53 network to positively regulate p53 and that manipulating its tumor-suppressive activity may be useful for rational cancer therapy.
MATERIALS AND METHODS
Cell culture and reagents.
R1B/L17 (mink lung epithelial cell line derivative) (22), NIH 3T3 cells, H1299 cells, A549 cells (from the American Type Culture Collection), Rat1-akt cells (48), NIH 3T3/HER2 cells (47), and p53 null mouse embryo fibroblast (MEF) cells were cultured in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum. A DEAE-dextran method was used to transiently transfect R1B/L17 cells (more than 70% transfection efficiency) as described previously (22). H1299 cells were transfected with the Fugene 6 transfection reagent (Boehringer Mannheim) following the manufacturer's guidelines. The NIH 3T3/HER2/tTA cell line was generated by transfecting pUHD15-1(neo) (12) into NIH 3T3/HER2 cells with the Fugene 6 transfection reagent. Two days after transfection, cells were selected in 600 μg of G418 per ml for 2 to 3 weeks.
Positive clones were selected and assayed by transient transfection of pUHC13-3 carrying tetO, the human cytomegalovirus immediate-early promoter, and the luciferase gene (12). Flag-tagged 14-3-3σ cloned in the pUHD10-3 hygromycin resistance vector (34) was introduced into NIH 3T3/HER2/tTA cells with the Fugene 6 reagent, and 300 μg of hygromycin B per ml plus 2 μg of tetracycline per ml was used to select positive clones over 2 to 3 weeks. NIH 3T3/HER2/tTA-14-3-3σ cells were cultured in medium without 2 μg of tetracycline per ml for 16 to 20 h for induced expression. The glutathione S-transferase (GST)-p53 construct was a kind gift from Cheng-Ming Chiang. Adriamycin at 0.2 μg per ml (Sigma) was prepared in dimethyl sulfoxide for use. Cells were irradiated with a 137Cs source emitting a fixed dose rate of 10 Gy (irradiator model 0103; U.S. Nuclear Corp.).
Western blot analysis.
Total cell lysates were solubilized in lysis buffer (20 mM Tris-HCl [pH 8.0], 100 mM NaCl, 1 mM EDTA, 0.5% NP-40, 0.5% Triton X-100, 1 mM phenylmethylsulfonyl fluoride, 1 mM NaF, 1 mM sodium orthovanadate, 1 μg each of aprotinin, leupeptin, and pepstatin per ml) and processed as previous described (22). Cells can be immunoprecipitated with anti-Flag antibody (M2; Sigma), anti-p53 antibody (DO1; Oncogene Science), or incubated with His-Bind resin (Novagen). Western analysis was performed on 12% polyacrylamide gels with a 5% polyacrylamide stacking gel. After electrophoretic transfer (Amersham Pharmacia) of protein from sodium dodecyl sulfate (SDS)-polyacrylamide gels to polyvinylidene difluoride membranes (Millipore), the membranes were blocked with buffer containing 20 mM Tris-HCl (pH 7.5), 150 mM NaCl, 0.05% Tween 20, 5% Blotto (Bio-Rad) for 1 h at room temperature and incubated for 1 h at room temperature with primary polyclonal anti-14-3-3σ antibody (Santa Crutz), antiactin antibody (Sigma), anti-p53 antibody (DO1; Oncogene Science), anti-green fluorescent protein (GFP) (Invitrogen) antibody, and monoclonal anti-Flag antibody (M2; Sigma). Subsequently membranes were washed and incubated for half an hour at room temperature with peroxidase-conjugated secondary antibodies. Following several washes, membranes were incubated with the chemiluminescence (ECL) system (Roche Molecular Biochemicals) according to the manufacturer's instructions.
In vitro binding assay.
For the in vitro binding assay, a T7 RNA polymerase-driven pET vector containing the coding region of the 14-3-3σ domain cDNA was transcribed in vitro and translated with a TNT kit (Promega). These products were labeled with [35S]methionine and were then incubated with immobilized GST-p53. The retained proteins were detected by autoradiography.
Metabolic labeling, immunoprecipitations, and half-life determination.
Cells were transfected, and the cellular proteins were pulse-labeled with [35S]methionine (100 Ci per ml) for 30 min in methionine-free medium and chased with unlabeled methionine for the indicated times; Mdm2 or p53 was immunoprecipitated from each lysate with Mdm2 (SMP14, Santa Crutz) or p53 (DO-1, Santa Cruz) antibody, separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and analyzed by phosphorimaging (Molecular Dynamics) for quantitation. The intensity of radiolabeled Mdm2 or p53 was quantitated by the Imagequant program of the Phosphorimager. The half-life of the protein was determined graphically according to procedures described previously (45).
Northern blot analysis.
Total RNAs were isolated with Qiagen RNesay kits. Each sample containing 20 μg of total RNAs was used for Northern blot analysis. RNAs were transferred to Genescreen Plus membranes (NEN) with a Turboblotter system (Schleicher & Schuell). The p21 and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) cDNA probes were labeled by a random-primed DNA labeling kit (Roche Molecular Biochemicals). The GAPDH probe was used to indicate the integrity and equal amounts of loading for each RNA sample.
Ubiquitination assay.
H1299 cells were cotransfected with pCMV-p53 (1 μg), Mdm2 (3 μg), histidine-tagged ubiquitin (1 μg), and increasing amount of Flag-14-3-3σ. After 24 h posttransfection, cells were treated with 200 μM ALLN (N-acetyl-Leu-Leu-norleucine-al; Sigma) for 12 h. The cell lysates were harvested with lysis buffer (50 mM Tris [pH 7.5], 150 mm NaCl, 0.5% NP-40, 0.5% Triton X-100, 5 mM N-ethylmaleimide). The histidine-tagged ubiquitin-containing protein complexes were brought down with His-Bind resin (Novagen). The protein complexes were then resolved by a DS-12% polyacrylamide gel and probed with goat anti-p53 (FL393, Santa Cruz Biotechnology) to observe the ubiquitinated p53.
Immunofluorescence.
Endogenous p53 or Mdm2 subcellular localization was detected in R1b/L17 or Rat1-akt cells. Cells were prepared and seeded onto chamber slides with 2 × 105 cells per well 1 day prior to staining. Cells were then fixed with methanol/acetone (1:1v/v) at room temperature for 2 min and stained for 1 h with rabbit anti-p53 (FL293, Santa Cruz) followed by 1 h of incubation with indocarbocyanine-conjugated anti-rabbit immunoglobulin antibody (Zymed).
For studying the subcellular localization of exogenous 14-3-3σ, cells were infected with an adenovirus construct, Ad-14-3-3σ (hemagglutinin [HA] tagged) or Ad-β-gal at a multiplicity of infection of 5. Twenty-four hours after infection, 2 × 105 cells were seeded onto tissue culture chamber slide (Nunc). Two days later, cells were fixed as mentioned above. Monoclonal anti-HA polyclonal antibody (Santa Cruz) was used in detecting 14-3-3σ expression. In this case, the fluorescein isothiocyanate-conjugated anti-rabbit immunoglobulin secondary antibody (Jackson Research Laboratories) was used. Mouse anti-Mdm2 (SMP14, Santa Cruz) antibody was used, followed by Texas red-conjugated anti-goat immunoglobulin antibody (Jackson Research Laboratories) to stain Mdm2. For all staining, the cells were incubated with 0.1 μg of 4,6-diamidino-2-phenylindole (DAPI) per ml (Sigma) to stain the nuclei. Immunofluorescence was detected with an Axioplan 2 fluorescence microscope (Zeiss).
Luciferase reporter gene assay.
The BDS2-3X-luc reporter containing a p53-responsive element or the BDS2-mutant-luc reporter with a mutated p53-responsive element (13) was transfected with the pCMV-p53- or pCMV-14-3-3σ-expressing vectors into R1B/L17 cells or p53 null MEF cells. Luciferase activity was assayed with the dual luciferase assay system (Promega) according to the manufacturer's instructions.
5-Bromo-2-deoxyuridine incorporation assay.
NIH 3T3/HER2/tTA and NIH 3T3/HER2/tTA-14-3-3σ cells were cultured in medium with or without 2 μg of tetracycline per ml for 16 to 20 h prior to performing the bromodeoxyuridine incorporation assay. Cells were trypsinized and plated onto four-well chamber slides 24 h after transfection. Bromodeoxyuridine incorporation was performed 48 h after transfection with a bromodeoxyuridine labeling and detection kit (Boehringer Mannheim) following the manufacturer's guidelines. For all staining, cells were incubated with 0.1 mg of 4′,6′-diamidino-2-phenylindole (DAPI) per ml (Sigma) to stain the nuclei. Immunofluorescence was detected with an Axioplan 2 fluorescent microscope (Zeiss).
Tumor growth in nude mice.
Female 4- to 5-week-old nude mice (Charles River Laboratories, Wilmington, Mass.) were maintained in the animal facility at the University of Texas M. D. Anderson Cancer Center. Mice were divided into two experimental groups, four for each. NIH 3T3/HER2/tTA-14-3-3σ cells (1 × 106 cells in 0.2 ml of phosphate-buffered saline per injection with 2 μg of doxycycline per ml for one group and without doxycycline for other) were injected subcutaneously into the flanks of mice, and each mouse was inoculated with cells at two sites. After cell inoculation, animals were fed drinking water containing 5% sucrose in the presence or absence of 200 μg of doxycycline per ml (Sigma) (19). Drinking water was changed every other day for a 3-week period of tumor formation. Tumor volumes were measured and recorded three times a week from day 5 of cell inoculation. At the end of 3 weeks, the mice were sacrificed, and the tumors were removed for detection of 14-3-3σ gene expression.
RESULTS
14-3-3σ interacted with p53.
The 14-3-3 protein family has been shown to act as adapter proteins, binding to many signal proteins to exert their biological function (38). It is possible that 14-3-3σ has such an adapter role in regulating the p53 network. We first examined whether 14-3-3σ interacted with p53 physically. Here, we showed that 14-3-3σ interacted with p53 in vivo as characterized by coimmunoprecipitation experiments. p53 can be stabilized by the DNA-damaging agent adriamycin in A549 cells, which have wild-type p53 (Fig. 1A). Importantly, 14-3-3σ was detected in the anti-p53 immunoprecipitation complex after treatment with adriamycin in A549 cells, suggesting that 14-3-3σ associated with p53 following DNA damage.
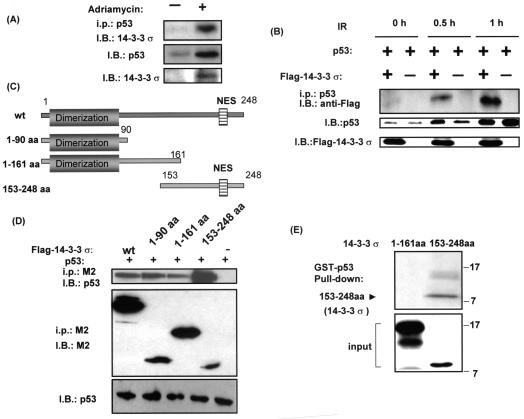
FIG. 1.
14-3-3σ interacted with p53. (A) Interaction between 14-3-3σ and p53 after adriamycin treatment. A549 cells were treated with 0.2 μg of adriamycin per ml (+) or not (−) for 24 h. Equal amounts of cell lysates were immunoblotted (I.B.) with anti-p53 antibody or anti-14-3-3σ to show the expression of p53 or 14-3-3σ after adriamycin treatment. Equal amounts of cell lysates were also immunoprecipitated (i.p.) with anti-p53 antibody, then resolved in SDS-polyacrylamide gel electrophoresis, and immunoblotted with anti-14-3-3σ antibody to observe the association between p53 and endogenous 14-3-3σ. (B) Interaction between 14-3-3σ and p53 after ionizing radiation (IR). R1B/L17 cells were transfected with the indicated Flag-tagged 14-3-3σ constructs and p53 expression vectors for 48 h and irradiated with 10 Gy. After the indicated hours of ionizing radiation, equal amounts of cell lysates were immunoprecipitated with anti-p53 antibody, and the immunoprecipitates were immunoblotted with anti-Flag antibody (M2) to observe the association between p53 and Flag-tagged 14-3-3σ. Cell lysates were also immunoblotted (I.B.) with anti-p53 antibody or anti-14-3-3σ to show the expression of p53 or 14-3-3σ. (C) Schematic representation of 14-3-3σ deletion constructs. (D) Interaction between the domains of 14-3-3σ and p53. R1B/L17 cells were transfected with the indicated Flag-tagged 14-3-3σ constructs and p53 expression vectors. Cell lysates were immunoprecipitated (i.p.) with anti-Flag antibody (M2), and the immunoprecipitates were immunoblotted (I.B.) with anti-p53 (DO-1; Santa Cruz) to observe the interaction. The same blot was stripped and immunoblotted with anti-Flag antibody (M2, Sigma) to demonstrate the amount of 14-3-3σ deletion constructs expressed and immunoprecipitated. Equal amounts of protein from the cell lysates were immunoblotted with anti-p53 (DO-1) to indicate the expression of p53. (E) p53 specifically interacts with 14-3-3σ in vitro. GST-p53 immobilized on glutathione beads was incubated with in vitro-transcribed and translated 14-3-3σ domains (amino acids 1 to 161 or 153 to 248), which were 35S labeled. Bound domains were detected by autoradiography. The bottom panel shows 10% of the in vitro-translated 35S-labeled 14-3-3σ domain inputs.
We also examined whether another genotoxic condition such as ionizing radiation increased the interaction between these two proteins in a transfection assay. Cells were transfected with equal amounts of p53-expressing plasmid in the presence or absence of 14-3-3σ-expressing plasmid. Importantly, increasing amounts of 14-3-3σ were detected in the anti-p53 immunoprecipitation complex (Fig. 1B) from irradiated cells harvested after 0.5 or 1 h of treatment compared with cells harvested immediately after irradiation (0 h), demonstrating that ionizing radiation increased the interaction between these two proteins.
The observation that 14-3-3σ interacted with p53 prompted us to map the possible region of 14-3-3σ that is involved in binding to p53. Cells were cotransfected with various deletion mutants of 14-3-3σ (Fig. 1C) and p53 expression vectors, and coimmunoprecipitation experiments were performed to determine the interaction. We showed that the C-terminal region of 14-3-3σ (14-3-3σ amino acids 153 to 248) bound to p53 very efficiently compared with other domains (Fig. 1D), as determined by the ratio of bound p53 to 14-3-3σ constructs (quantitated by densitometer; data not shown), demonstrating that different domains of 14-3-3σ bound to p53 differentially inside the cell.
We then tested the specificity of the interaction between 14-3-3σ and p53 in a defined system with the GST pull-down assay in vitro. GST-tagged p53 was able to bind to in vitro-translated, 35S-labeled 14-3-3σ (amino acids 153 to 248) but not 14-3-3σ (amino acids 1 to 161) (Fig. 1E), suggesting that the C-terminal sequence comprising residues 153 to 248 of 14-3-3σ might contain the p53 interaction domain. In conclusion, these results indicated that 14-3-3σ interacted with p53 physically.
14-3-3σ increased p53's stability.
Since some p53-interacting proteins, such as Mdm2 and BRCA1 (30), can affect either the stability or the transcriptional activity of p53, we hypothesized that 14-3-3σ would have similar activities toward p53. To determine whether 14-3-3σ affects p53's stability, we used H1299 cells (p53 null) to examine the role of 14-3-3σ activity in regulating the protein level of p53 by transfection. Cells were cotransfected with equal amounts of p53 and increasing amounts of 14-3-3σ. Significantly, the expression level of p53 was increased when a high level of 14-3-3σ was present (Fig. 2A), suggesting that 14-3-3σ had a positive impact on p53 stability.
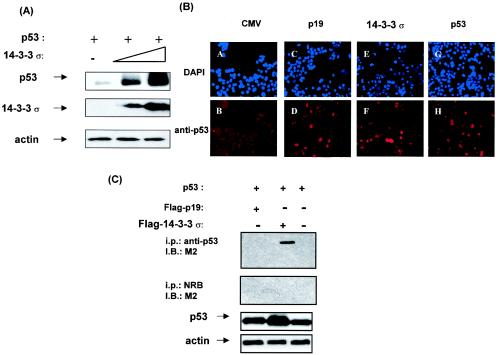
FIG. 2.
14-3-3σ increased p53 stabilization. (A) Ectopic expression of 14-3-3σ increased the stabilization of p53. H1299 cells were transfected with equal amounts of p53 expression vector and increasing amounts of the Flag-tagged 14-3-3σ expression vector. Equal amounts of protein from cell lysates were immunoblotted with anti-p53, anti-Flag, and antiactin. Actin served as a loading control. (B) Immunofluorescence studies of p53 stability. R1B/L17 cells were transfected with either plasmid pCMV expressing p19ARF, p53, or 14-3-3σ or with an empty cytomegalovirus vector to examine the immunostaining of endogenous p53 as detected by immunofluorescence. The cells that received the indicated plasmids were seeded at 2 × 103 cells per chamber slide. The cells were fixed, and p53 was immunodetected with anti-p53 antibody (FL293; Santa Cruz), followed by indocarbocyanine-conjugated anti-rabbit immunoglobulin (B, D, F, and H). DAPI staining was used to show the localization of nuclei (A, C, E, and G). (C) p53 binding activity of 14-3-3σ and p19ARF. R1B/L17 cells were transfected with the indicated expression vectors. Cell lysates were immunoprecipitated (i.p.) with anti-p53 antibody (DO-1) and immunoblotted (I.B.) with anti-Flag antibody (M2) to observe the association between p53 and 14-3-3σ or p19ARF. Immunoprecipitation with normal rabbit serum (NRB) was used as a negative control. Equal amounts of protein from cell lysates were immunoblotted with anti-p53 to indicate the expression of p53. Actin served as a loading control.
To further confirm the observation that 14-3-3σ affects the stability of p53, we determined whether the endogenous level of p53 was changed when 14-3-3σ was present with an immunofluorescence study. R1B/L17 cells (highly transfectable) were transfected with either pCMV plasmid containing p19ARF, p53, or 14-3-3σ or with an empty vector pCMV, and the expression of endogenous p53 was detected by immunofluorescence study. A second plasmid directing the expression of the green fluorescent protein (GFP) was cotransfected to monitor equal transfection efficiency (data not shown). Due to the short half-life of p53, pCMV5-transfected cells showed weak staining of endogenous p53 (Fig. 2B, panel B). Significantly, the normally weak staining of endogenous p53 was dramatically increased in the 14-3-3σ-transfected cells (Fig. 2B, panel F) compared with pCMV5-transfected cells, suggesting that 14-3-3σ increased the stability of endogenous p53.
Interestingly, the level of p53 staining in 14-3-3σ-transfected cells was almost equal to the level of staining of p53-transfected cells (Fig. 2B, panel H). p19ARF, a well-characterized protein that stabilizes p53 by blocking Mdm2-mediated degradation (32, 46), was transfected to provide a positive control (Fig. 2B, panel D) for p53 stabilization. To compare the stabilizing activity of 14-3-3σ and p19ARF, we performed coimmunoprecipitation experiments and found that 14-3-3σ coimmunoprecipitated with p53 (Fig. 2C), whereas p19ARF, previously shown to stabilize p53 (32), did not directly bind to p53 (Fig. 2C). Taken together, these results demonstrated that 14-3-3σ increased p53's stability.
14-3-3σ blocked Mdm2 activity toward p53.
On the basis of the observation that 14-3-3σ increased p53's stability, we wondered whether 14-3-3σ interfered with p53-negative regulator Mdm2. To determine whether 14-3-3σ blocks Mdm2 activity for increasing p53's stability, we performed a cotransfection experiment. Cells were cotransfected with Mdm2, pCMV-p53, and increasing amounts of 14-3-3σ into H1299 cells. The normally decreased level of p53 due to the expression of Mdm2 was reversed in conditions where 14-3-3σ was increased (Fig. 3A), suggesting that 14-3-3σ blocked Mdm2-mediated p53 degradation.
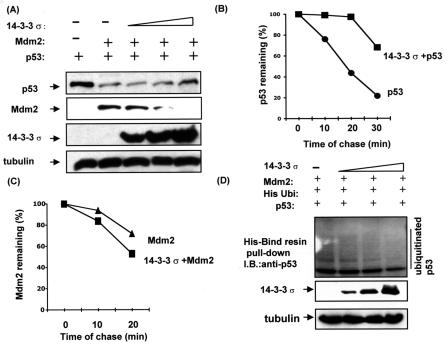
FIG. 3.
14-3-3σ inhibited Mdm2-mediated p53 ubiquitination. (A) 14-3-3σ stabilized p53 in the presence of Mdm2. H1299 cells were cotransfected with indicated pCMV-Mdm2, pCMV-p53, and increasing amounts of pCMV-Flag-14-3-3σ (0 μg, 0 μg, 1 μg, 4 μg, and 8 μg). Equal amounts of protein from cell lysates were immunoblotted with anti-Mdm2, anti-Flag, and antiactin. Levels of tubulin are shown as equal loading controls. (B) 14-3-3σ inhibited p53 turnover. 293T cells transiently transfected with pCMV-14-3-3σ and vector/pCMV-p53 were pulse-labeled with [35S]methionine for 30 min and chased for the indicated time. (C) 14-3-3σ accelerated Mdm2 degradation. 293T cells transiently transfected with pCMV-14-3-3σ and vector/pCMV-Mdm2 were pulse-labeled with [35S]methionine for 30 min and chased for the indicated time. Cells were harvested, and the amount of labeled Mdm2 or p53 protein immunoprecipitated at each time point was detected by phosphoimager for quantitation. p53 or Mdm2 remaining is indicated graphically. (D) 14-3-3σ inhibited Mdm2-mediated p53 ubiquitination. H1299 cells were cotransfected with equal amount of Mdm2 (3 μg), pCMV-p53 (1 μg), histidine-tagged ubiquitin-expressing vector (1 μg), and increasing amount of pCMV-Flag-14-3-3σ (0 μg, 1 μg, 4 μg, and 8 μg). The cell lysates were harvested, and the histidine-tagged ubiquitin-containing protein complexes were pulled down with His-Bind resin (Novagen). The protein complexes were then resolved by SDS-12% polyacrylamide gel and probed with anti-p53 antibody to observe the histidine-tagged ubiquitinated p53.
To explore whether 14-3-3σ overexpression can modulate p53 turnover, we performed pulse and chase experiments in transfected 293T cells. In the 14-3-3σ-transfected cells, 35S-labeled p53 immunoprecipitated by anti-p53 remained at more than 50% after 30 min, whereas the degradation rate of p53 was faster in vector-transfected cells (less than 30% remained after 30 min) (Fig. 3B). Thus, the 14-3-3σ-mediated p53 stabilization was resulted from increasing the half-life of p53 (Fig. 3B). Interestingly, the expression of 14-3-3σ seemed to accelerate the degradation of Mdm2 (Fig. 3A). Again, we performed pulse and chase experiments in transfected 293T cells to determine the turnover of Mdm2 in the presence of 14-3-3σ. The 35S-labeled Mdm2 immunoprecipitated from 14-3-3σ-transfected cells had a faster turnover than that immunoprecipitated from vector-transfected cells (Fig. 3C). This indicates that elevated levels of 14-3-3σ decrease the half-life of Mdm2.
Because Mdm2 is an E3 ubiquitin ligase for p53 and is involved in regulating the stability and half-life of p53 through the ubiquitin-proteasome pathway, we determined whether 14-3-3σ could inhibit Mdm2-mediated p53 ubiquitination. Cells were cotransfected with Mdm2, pCMV-p53, pCMV expressing histidine-tagged ubiquitin, and increasing amounts of 14-3-3σ into H1299 cells. Cells were harvested and analyzed by His-Bind resin binding for the presence of histidine-tagged ubiquitin-containing p53. Mdm2 can act as a ubiquitin ligase and will add histidine-tagged ubiquitin to its target, p53. Thus, the histidine-tagged ubiquitinated form of p53 can be detected in the histidine-tagged ubiquitin-containing protein complexes bound to His-Bind resin by immunoblotting with anti-p53. Under conditions where 14-3-3σ is creased, the amounts of the histidine-tagged ubiquitinated form of p53 decreased, suggesting that 14-3-3σ inhibited Mdm2-mediated p53 ubiquitination (Fig. 3D).
Because Mdm2 binds to p53 and facilitates its nuclear export and degradation by the cytoplasmic proteasome, we wondered whether 14-3-3σ interfered with Mdm2 to stabilize and retain p53 in the nucleus. Because Rat1-akt cells have high Akt activity that maintains Mdm2 in the nucleus (48), we used this cell line to study the subcellular localization of Mdm2 in the presence of 14-3-3σ. We found that Mdm2 was located in the nucleus of cells when cells were infected with the control β-galactosidase-expressing virus Ad-β-gal (Fig. 4A, panel D, red) with the immunostaining assay. The location of 14-3-3σ was found in the cytoplasm of cells infected with Ad-HA-14-3-3σ (Fig. 4A, panel B, green) were stained with anti-HA antibody, which is consistent with the previous observation due to a nuclear export signal in 14-3-3σ (21). No signals were detected in Ad-β-gal-infected cells with anti-HA antibody (Fig. 4A, panel E). DAPI signals indicated the location of the nuclei (Fig. 4A, panels C and F). Importantly, Mdm2's subcellular location, originally only in the nucleus, was also detected in the cytoplasm when cells were infected with adenovirus Ad-14-3-3σ (Fig. 4A, panel A, red), suggesting that 14-3-3σ affected the location of Mdm2 from the nucleus to the cytoplasm.
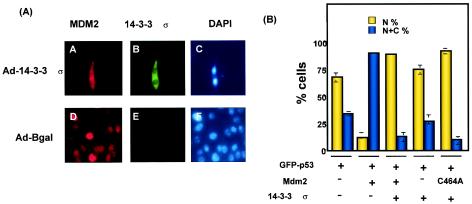
FIG. 4.
14-3-3σ blocked Mdm2-mediated p53 nuclear export. (A) Subcellular localization of Mdm2 was affected by overexpression of 14-3-3σ. The Rat1-akt cell lines were infected with Ad-HA-14-3-3σ or Ad-β-gal. After 48 h, cells were fixed and stained with anti-Mdm2 to detect the location of Mdm2 (A, D) or stained with anti-HA antibodies to observe the location of 14-3-3σ (B, E). Nuclei were stained with DAPI dye. (B) Activity of 14-3-3σ inhibited Mdm2-mediated p53 nuclear export. R1B/L17 cells were transfected with the indicated constructs to examine the subcellular localization of GFP-p53 as detected by green fluorescence. The cells that received the indicated plasmids were seeded at 2 × 103 in a chamber slide. The location of GFP-p53 was observed under a fluorescence microscope. A total of 300 cells were counted for the location of GFP-p53 in each condition. The percentages of nuclear-positive cells (N) and nuclear and cytoplasmic-positive cells (N+C) are presented. Data shown were from a typical experiment conducted in triplicate. Bars represent standard deviations.
Because Mdm2 has p53 nuclear-exporting activity (2, 11), we wondered whether 14-3-3σ was able to mediate Mdm2 mislocation and in turn affect p53's subcellular location. To determine such a link, we used GFP-p53 (36) to indicate the location of p53 in a cotransfection assay. Cells were transfected with GFP-p53 (36) alone or in combination with Mdm2, nuclear export mutant Mdm2 (C464A), 14-3-3σ, a mixture of Mdm2 and 14-3-3σ, or a mixture of Mdm2 (C464A) and 14-3-3σ. The location of p53 was detected by green fluorescence from GFP-p53 with a fluorescence microscope.
In a cotransfection assay, wild-type Mdm2's nuclear exporting activity resulted in a low percentage (about 10%) of cells with nuclear GFP-p53 fluorescence (Fig. 4B). Nuclear export mutant Mdm2 (C464A) did not mediate p53 nuclear export (11). Importantly, we found that the low percentage of cells with nuclear GFP-p53 fluorescence resulting from Mdm2 expression was dramatically reversed when 14-3-3σ was expressed in the transfection, suggesting that 14-3-3σ blocked Mdm2-mediated p53 nuclear export. Taken together, these results indicated that 14-3-3σ blocked Mdm2-mediated p53 degradation by inhibiting Mdm2's ubiquitin ligase activity and by interfering with the nuclear exporting activity of Mdm2 toward p53.
14-3-3σ enhanced p53 transcriptional activity.
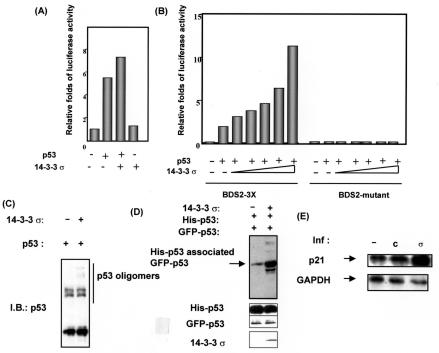
Because 14-3-3σ stabilized the p53 protein level and antagonized Mdm2 activity, it is possible that 14-3-3σ can enhance p53 transcriptional activity. Thus, we used a p53-responsive luciferase reporter gene assay to determine whether 14-3-3σ had any effect on the transcriptional activity of p53. We performed a reporter gene assay by cotransfecting the luciferase reporter gene BDS2-3x-luc, containing p53 response elements in the 14-3-3σ promoter (2), 14-3-3σ, and p53 into R1B/L17 cells. We found that 14-3-3σ enhanced p53 transcriptional activity in cells as determined by luciferase activity (Fig. 5A).
FIG. 5.
14-3-3σ enhanced p53 transcriptional activity. (A) 14-3-3σ activated the p53 luciferase reporter gene. The BDS2-3X-luc reporter containing a p53-responsive element was transfected with the indicated p53- or 14-3-3σ-expressing vectors into R1B/L17 cells. Relative luciferase activity is shown. (B) 14-3-3σ potentiated p53 transcriptional activity in a dose-dependent manner. The BDS2-3X-luc reporter was transfected with increasing amounts of the 14-3-3σ expression vector into p53-null MEF cells. The BDS2-mutant-luc reporter with a mutated p53-responsive element was used as a negative control. Relative luciferase activity is shown. (C) Oligomerization assay. H1299 cells were transfected with the indicated plasmids. Total cell extracts were incubated with 0.01% glutaraldehyde to determine the levels of the oligomerization of p53. p53 oligomers were immunoblotted (I.B.) with anti-p53 antibodies. (D) 14-3-3σ increased the interaction between p53 molecules. H1299 cells were transfected with pCMV-His-tagged-p53 and pCMV-GFP-p53 in the presence or absence of 14-3-3σ. His-tagged p53 was bound with His-Bind beads, and His-tagged p53-associated GFP-p53 was observed with the anti-GFP antibody. (E) Transcriptional activation of p53 target gene p21. R1B/L17 cells were infected (inf) with Ad-14-3-3σ (σ) or Ad-β-gal (C) or not infected (−). After 48 h, total RNAs were prepared. Northern blot analysis was performed on total RNAs to examine the expression of the p21 gene. Signals for GAPDH are shown to indicate the integrity and quantity of the RNA.
We confirmed this observation in a cell background without endogenous p53. We repeated the experiment by cotransfecting p53, the reporter gene, and increasing amounts of 14-3-3σ into p53-null mouse embryonic fibroblast (MEF) cells. Again, 14-3-3σ facilitated the p53 transcriptional activity under these conditions in a dose-dependent manner (Fig. 5B). As expected, 14-3-3σ had no effect on the negative control luciferase reporter gene (BDS2-mutant that has mutant p53 binding sites) (Fig. 5B), demonstrating that 14-3-3σ enhanced p53's transcriptional activity through p53 binding site.
The tetramerization domain of p53 is important for DNA binding and thus facilitates transcriptional activity (25). One of the important functions of the 14-3-3 family is to act as adaptors for signal mediators (38). Because we have demonstrated that 14-3-3σ bound to p53 (Fig. 1D), we wondered whether 14-3-3σ helped the cooperative p53 dimer-dimer interaction to occur so as to stabilize p53 DNA binding and in turn to potentiate p53's transcriptional activity. Cells were cotransfected with p53 and 14-3-3σ. Total cell extracts were incubated with glutaraldehyde, which cross-linked the oligomers, to determine the levels of p53 oligomerization.
As shown in Fig. 5C, we found that 14-3-3σ facilitated the p53 oligomerization process, as demonstrated by the appearance of slow-migrating trimers or tetramers in the SDS-PAGE. In addition, we cotransfected equal amounts of two different tagged p53 constructs into the cells to determine whether two different tagged p53 proteins interacted with each other more efficiently when 14-3-3σ was present. Cells were cotransfected with pCMV-His-p53 and pCMV-GFP-p53 in the presence or absence of 14-3-3σ into H1299 cells. Cells were harvested and analyzed by His-Bind resin binding. Due to the oligomerization process, levels of GFP-p53 associated with His-p53 bound to His-Bind resin can be detected by immunoblotting with anti-GFP. Under condition where 14-3-3σ was present, levels of GFP-p53 associated His-p53 were increased (Fig. 5D), suggesting that 14-3-3σ increased the intermolecular interaction of p53 and in turn facilitated the oligomerization process.
On the basis of the observation that 14-3-3σ enhanced p53 transcriptional activity, we wondered whether 14-3-3σ enhanced p53 transcriptional activity toward an endogenous target gene such as p21. Cells were infected with Ad-14-3-3σ, and gene expression of p21 was determined by mRNA expression. As shown in Fig. 5E, p21 expression was highly activated by 14-3-3σ, about fivefold, as determined by Northern blot analysis and quantitated by phosphorimager (data not shown). Taken together, these results demonstrated that 14-3-3σ facilitated p53 oligomerization and enhanced p53's transcriptional activity.
Inducible expression of 14-3-3σ suppressed tumor growth in nude mice.
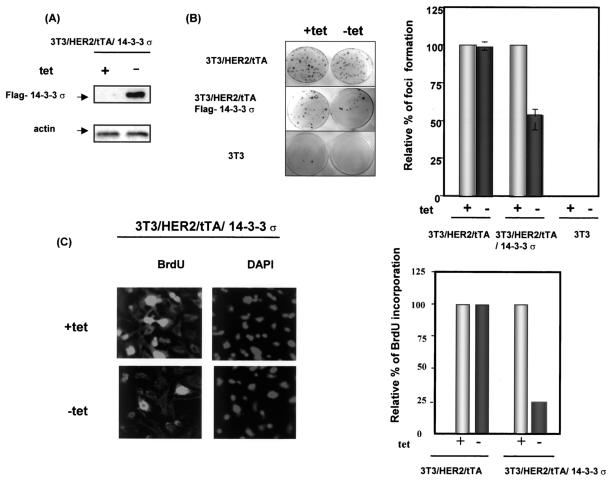
The most important tumor suppressor, p53, has been successfully employed as a molecular target for cancer gene therapy (4). Because we found that 14-3-3σ potentiates p53's stability and activity, we hypothesize that overexpression of 14-3-3σ counteracts mitogenic growth and tumorigenicity. To explore the tumor-suppressive effect of 14-3-3σ, we employed a cell system that had been transformed by the HER2/neu (HER2) oncogene and could be tetracycline regulated to control the expression of the gene of interest (44).
We successfully constructed and expressed 14-3-3σ in HER2-transformed NIH 3T3 cells (3T3/HER2 cells) (47) that expressed the tTA (tetracycline transcription activator) (3T3/HER2/tTA) (44) in the absence of tetracycline (Fig. 6A). The 14-3-3σ-expressing cells (3T3/HER2/tTA/14-3-3σ in the absence of tetracycline) reduced the number of HER2-induced microfoci about 50% compared with 3T3/HER2/tTA cells (Fig. 6B, bar graph), indicating that overexpression of 14-3-3σ inhibited transformed cell growth. In addition, bromodeoxyuridine incorporation (Fig. 6C) was dramatically reduced in the 14-3-3σ-expressing cells (NIH 3T3/HER2/tTA-14-3-3σ in the absence of tetracycline) compared with non-14-3-3σ-expressing cells (NIH 3T3/HER2/tTA-14-3-3σ in the presence of tetracycline). Thus, these data strongly suggested that overexpression of 14-3-3σ can function as a growth inhibitor in HER2/neu-overexpressing cells.
FIG. 6.
Tetracycline-regulated 14-3-3σ inhibited transformation phenotype and mitogenic signals of HER2/neu-overexpressing cells. (A) Tetracycline-inducible expression of 14-3-3σ. To examine the expression of 14-3-3σ, the culture medium of 3T3/HER2/tTA/14-3-3σ was switched to medium alone or medium containing 2 μg of tetracycline (tet) per ml. The Flag-14-3-3σ expressed in the absence of tetracycline was probed by immunoblotting with anti-Flag antibody. The levels of actin were used as an equal loading control. (B) Focus formation assay. NIH 3T3 (3T3), 3T3/HER2/tTA, and 3T3/HER2/tTA/Flag-14-3-3σ cells were subjected to a microfocus formation assay in the presence (+) or absence (−) of 2 μg of tetracycline per ml. Foci were counted in the presence and absence of tetracycline. The number of foci from 3T3/HER2/tTA and 3T3/HER2//tTA/14-3-3σ cells treated with tetracycline was set at 100%. The relative focus formation from cells cultured in the absence of tetracycline is shown as a bar graph. Bars represent standard deviations. (C) Tetracycline-regulated 14-3-3σ expression blocked cell cycle entry into S phase in HER2/neu-overexpressing cells. Bromodeoxyuridine (BrdU) incorporation assays were conducted after cells were cultured in the presence or absence of tetracycline for 20 h. Incorporation of bromodeoxyuridine was examined under a fluorescence microscope with fluorescein isothiocyanate-conjugated antibromodeoxyuridine. DAPI staining indicated the location of nuclei. A total of 300 cells were counted for bromodeoxyuridine staining in each condition. The number of bromodeoxyuridine-positive cells from 3T3/HER2/tTA and 3T3/HER2//tTA/14-3-3σ cells cultured in tetracycline-containing medium was set at 100%. The relative percentage of bromodeoxyuridine-positive cells in cells cultured in the absence of tetracycline is presented. Data shown are from a typical experiment conducted in triplicate.
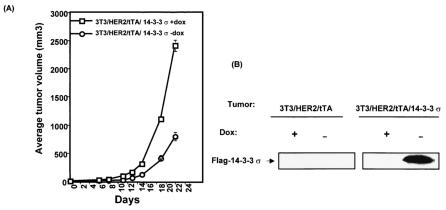
Since 14-3-3σ blocked HER2-induced cell growth and transformation in vitro, we then assessed its effect on HER2-mediated tumorigenicity in vivo. To verify the antitumor activity of 14-3-3σ, we performed a tumorigenicity assay in immunodeficient strains of mice. The 3T3/HER2/tTA/14-3-3σ cells were treated with or without doxycycline (a derivative of tetracycline) for 18 h, with the drug subcutaneously injected into the flanks of nude mice. Tumor formation was then assayed. There was dramatically less HER2-mediated tumorigenesis in the 14-3-3σ-expressing cells (3T3/HER2/tTA/14-3-3σ in the absence of doxycycline) than in the control cells (3T3/HER2/tTA-14-3-3σ in the presence of doxycycline) (Fig. 7A). The same results were obtained when two other independent clones were used for the tumorigenesis assay (data not shown).
FIG. 7.
Doxycycline-regulated expression of 14-3-3σ inhibits the tumorigenesis of HER2/neu-overexpressing cells in nude mice. (A) Tumorigenicity. 3T3/HER2/tTA/14-3-3σ cells were cultured in the presence (+) or absence (−) of doxycycline (dox) for 24 h. Then 106 cells were harvested and injected subcutaneously into the flank region of female nude mice. The mice were fed water with (+) or without (−) 200 μg of doxycycline per ml. Tumor volumes were monitored for 22 days. The change in tumor volume over a 22-day period is shown in the graph. Bars represent standard deviations. (B) Gene expression. Tumor tissues from the sites of implantation were assessed for expressed Flag-14-3-3σ levels by immunoblotting with anti-Flag antibody. Representative tumors are shown.
Tumor tissues from the sites of implantation were assessed for 14-3-3σ levels by immunoblotting (Fig. 7B). As expected, transduced 14-3-3σ (Flag tagged) was absent in tumors of 3T3/HER2/tTA-injected control mice regardless of the presence or absence of doxycycline in their drinking water. 14-3-3σ was also not detected in tumors of NIH 3T3/HER2/tTA-14-3-3σ-injected mice given drinking water containing doxycycline, whereas transduced Flag-14-3-3σ was detected in the small tumors of NIH 3T3/HER2/tTA-14-3-3σ-injected mice given drinking water without doxycycline. The data suggested that the expression of 14-3-3σ is directly involved in inhibiting tumor growth. These results demonstrated that overexpression of 14-3-3σ was sufficient to abolish tumorigenicity in cells transformed by the HER2 oncogene.
DISCUSSION
As described above, expression of 14-3-3σ is frequently reduced in human breast cancers, suggesting that loss of 14-3-3σ 's function is crucial in the development of cancer. Therefore, it is important to characterize the biological function of 14-3-3σ. In this study, we investigated the molecular mechanism of p53 and 14-3-3σ interaction. First, using cell lines that received DNA damage signals to investigate the interaction between 14-3-3σ and p53, we found that 14-3-3σ interacted with endogenous p53 when p53 was stabilized by treatment with the DNA-damaging agent adriamycin, which is a type II topoisomerase inhibitor. Other 14-3-3 isoforms (ɛ, τ, and γ) were found to interact with p53 (42), but the detailed mechanism remains unclear.
As for 14-3-3σ, the interaction between 14-3-3σ and p53 was difficult to characterize in a previous study (35), possibly due to different assay conditions. It is not clear why it was difficult to determine binding. However, several lines of evidence from our study indicate that this interaction does exist (Fig. 1). Because only 14-3-3σ is the target gene of p53 in the 14-3-3 family, our study of the interaction between p53 and 14-3-3σ provides important information for a positive-feedback loop driving p53 transactivation. Interestingly, when we used transient transfection to determine the domain responsible for p53 binding, the different domains bound p53 differently. The C-terminal region bound to p53 with better efficiency, which was consistent with the structures of 14-3-3 family members, which generally use the C-terminal region to interact with their target proteins (10, 14). Indeed, we also confirmed that the C-terminal region (amino acids 153 to 248) of 14-3-3σ bound to p53 in a GST-p53 pull-down assay (Fig. 1E).
Structural studies have demonstrated that 14-3-3 proteins form a dimer (18, 43) as an open-ended, cup-shaped structure, and it is possible that the C-terminal region of is 14-3-3σ is located at the open-ended structure to pull in the 14-3-3σ-interacting protein p53. The domains of 14-3-3σ (amino acids 1 to 90) and 14-3-3σ (amino acids 1 to 161) still interacted with p53, albeit to a lesser degree. It is possible that these domains formed a dimer with endogenous 14-3-3σ, possibly through dimerization domain (Fig. 1C), thus retaining some binding activity toward p53.
Second, we found that 14-3-3σ stabilized p53, which may occur through direct binding, and at least this characteristic was not shared by p19ARF. It remains to be determined whether p19ARF and 14-3-3σ are functionally coordinated. Third, we found that 14-3-3σ blocked Mdm2-mediated p53 degradation. Clearly, we showed that 14-3-3σ blocked Mdm2-mediated p53 ubiquitination, suggesting that 14-3-3σ acts as a negative regulator of Mdm2.
One interesting observation is that 14-3-3σ caused the downregulation of Mdm2 through enhancing the turnover of Mdm2 (Fig. 3C). Because p19ARF can promote Mdm2 degradation and stabilize p53 (5, 32, 46), again it will be interesting to investigate whether 14-3-3σ regulates Mdm2 via p19ARF. In addition, MdmX protein, a p53-binding protein with homology to Mdm2, was shown to bind to Arf and affect Mdm2 stability and p53 transactivation (16, 17), it will be important to know whether MdmX has any role in 14-3-3σ 's activity toward p53. In addition, we found that 14-3-3σ caused the mislocation of Mdm2: a well-characterized nuclear Mdm2 protein became cytoplasmic in the presence of 14-3-3σ overexpression. Consequently, we then found that 14-3-3σ blocked Mdm2-mediated p53 nuclear export, a process required for p53 degradation.
It remains to be determined whether 14-3-3σ causes the nuclear export of Mdm2 through its potential leucine-rich nuclear export signal sequence (202-STLIMQLLRDNLTLW-212) (20, 21, 23), thus preventing Mdm2's nuclear export of p53 and adding another p53-stabilizing effect. Fourth, using a reporter gene assay, we showed that 14-3-3σ enhanced the transcriptional activity of p53. The ability to enhance p53 transcriptional activity may result not only from stabilizing p53 protein level but also from enhancing the oligomerization of p53, a process important for DNA binding (36).
Because 14-3-3 protein can act as a possible molecular scaffold or chaperone (38), 14-3-3σ may help p53 form a dimer through direct binding, facilitating the cooperative p53 dimer-dimer interaction needed for the stabilization of p53 DNA binding. Alternatively, 14-3-3σ may act as a transcriptional coactivator of p53. Such a coactivator function was demonstrated in the observation that some 14-3-3 proteins participate in transcriptional regulation via interacting with TATA-binding protein and TFIIB (31). It remains to be determined whether 14-3-3σ interacts with the transcription machinery to potentiate p53 transcriptional activity.
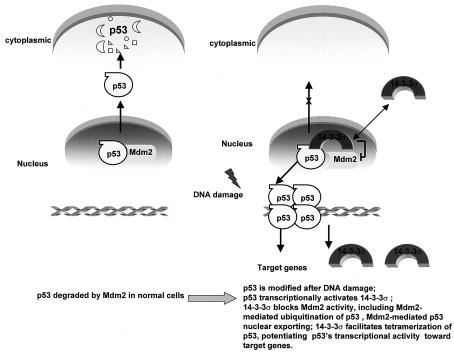
We present a model in Fig. 8 to highlight the effects of 14-3-3σ in facilitating the activity of p53. Finally, we found that 14-3-3σ had tumor-suppressive activity. Because 14-3-3σ is downregulated in primary breast cancers (9) and cell lines (33) and is a negative regulator of the cell cycle (21), overexpression of 14-3-3σ may counteract the mitogenic growth and tumorigenicity of cancer cells. Here, our studies indicated that 14-3-3σ inhibited the tumorigenicity of HER2-transformed cells, highlighting the tumor-suppressive role of 14-3-3σ. We previously demonstrated that overexpression of 14-3-3σ can inhibit the transformation and cell growth of several breast cancer cell lines (21). No studies have yet examined the tumor-suppressive activity of 14-3-3σ in a cancer xenograft mouse model, a prerequisite for developing cancer gene therapy. Therefore, our studies with inducible expression of 14-3-3σ to investigate 14-3-3σ as a possible anticancer agent in a tetracycline-based cancer model is a critical step for further therapeutic applications.
FIG. 8.
Model for effects of 14-3-3σ in regulating p53's activity.
In conclusion, our studies suggested that 14-3-3σ has a positive feedback role in regulating the activity of p53. Importantly, overexpression of 14-3-3σ counteracted the mitogenic growth and tumorigenicity of cells transformed by the oncogene. Our results may lead to the use of 14-3-3σ in therapeutic applications for rational cancer therapy.
Acknowledgments
We thank B. Vogelstein, M. Hung, and J. Wahl for valuable reagents.
This work was supported by the William McGowan Charitable foundation. H.-Y. Yang is a recipient of a postdoctoral fellowship from DOD Army Breast Cancer Research program (DAMD17-98-1-8243). M.-H. Lee is a recipient of a Flemin and Davenport research award and Susan G. Koman Breast Cancer Foundation research award.
REFERENCES
- 1.Aitken, A. 1995. 14-3-3 proteins on the MAP. Trends Biochem. Sci. 20:95-97. [DOI] [PubMed] [Google Scholar]
- 2.Boyd, S. D., K. Y. Tsai, and T. Jacks. 2000. An intact HDM2 RING-finger domain is required for nuclear exclusion of p53. Nat. Cell Biol. 2:563-568. [DOI] [PubMed] [Google Scholar]
- 3.Chan, T. A., H. Hermeking, C. Lengauer, K. W. Kinzler, and B. Vogelstein. 1999. 14-3-3σ is required to prevent mitotic catastrophe after DNA damage. Nature 401:616-620. [DOI] [PubMed] [Google Scholar]
- 4.Chene, P. 2001. p53 as a drug target in cancer therapy. Expert Opin. Ther. Patents 11:923-935. [Google Scholar]
- 5.Clark, P. A., S. Llanos, and G. Peters. 2002. Multiple interacting domains contribute to p14ARF mediated inhibition of Mdm2. Oncogene 21:4498-4507. [DOI] [PubMed] [Google Scholar]
- 6.Dellambra, E., O. Golisano, S. Bondanza, E. Siviero, P. Lacal, M. Molinari, S. D'Atri, and M. De Luca. 2000. Downregulation of 14-3-3σ prevents clonal evolution and leads to immortalization of primary human keratinocytes. J. Cell Biol. 149:1117-1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dellambra, E., M. Patrone, B. Sparatore, A. Negri, F. Ceciliani, S. Bondanza, F. Molina, F. D. Cancedda, and M. De Luca. 1995. Stratifin, a keratinocyte specific 14-3-3 protein, harbors a pleckstrin homology (PH) domain and enhances protein kinase C activity. J. Cell Sci. 108:3569-3579. [DOI] [PubMed] [Google Scholar]
- 8.Dhar, S., J. A. Squire, M. P. Hande, R. J. Wellinger, and T. K. Pandita. 2000. Inactivation of 14-3-3σ influences telomere behavior and ionizing radiation-induced chromosomal instability. Mol. Cell Biol. 20:7764-7772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferguson, A. T., E. Evron, C. B. Umbricht, T. K. Pandita, T. A. Chan, H. Hermeking, J. R. Marks, A. R. Lambers, P. A. Futreal, M. R. Stampfer, and S. Sukumar. 2000. High frequency of hypermethylation at the 14-3-3σ locus leads to gene silencing in breast cancer. Proc. Natl. Acad. Sci. USA 97:6049-6054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fu, H., R. R. Subramanian, and S. C. Masters. 2000. 14-3-3 proteins: structure, function, and regulation. Annu. Rev. Pharmacol. Toxicol. 40:617-647. [DOI] [PubMed] [Google Scholar]
- 11.Geyer, R. K., Z. K. Yu, and C. G. Maki. 2000. The Mdm2 RING-finger domain is required to promote p53 nuclear export. Nat. Cell Biol. 2:569-573. [DOI] [PubMed] [Google Scholar]
- 12.Gossen, M., and H. Bujard. 1992. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc. Natl. Acad. Sci. USA 89:5547-5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hermeking, H., C. Lengauer, K. Polyak, T. C. He, L. Zhang, S. Thiagalingam, K. W. Kinzler, and B. Vogelstein. 1997. 14-3-3σ is a p53-regulated inhibitor of G2/M progression. Mol. Cell 1:3-11. [DOI] [PubMed] [Google Scholar]
- 14.Ichimura, T., M. Ito, C. Itagaki, M. Takahashi, T. Horigome, S. Omata, S. Ohno, and T. Isobe. 1997. The 14-3-3 protein binds its target proteins with a common site located towards the C-terminus. FEBS Lett. 413:273-276. [DOI] [PubMed] [Google Scholar]
- 15.Iwata, N., H. Yamamoto, S. Sasaki, F. Itoh, H. Suzuki, T. Kikuchi, H. Kaneto, S. Iku, I. Ozeki, Y. Karino, T. Satoh, J. Toyota, M. Satoh, T. Endo, and K. Imai. 2000. Frequent hypermethylation of CpG islands and loss of expression of the 14-3-3σ gene in human hepatocellular carcinoma. Oncogene 19:5298-5302. [DOI] [PubMed] [Google Scholar]
- 16.Jackson, M. W., and S. J. Berberich. 2000. MdmX protects p53 from Mdm2-mediated degradation. Mol. Cell. Biol. 20:1001-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jackson, M. W., M. S. Lindstrom, and S. J. Berberich. 2001. MdmX binding to ARF affects Mdm2 protein stability and p53 transactivation. J. Biol. Chem. 276:25336-25341. [DOI] [PubMed] [Google Scholar]
- 18.Jones, D. H., S. Ley, and A. Aitken. 1995. Isoforms of 14-3-3 protein can form homo- and heterodimers in vivo and in vitro: implications for function as adapter proteins. FEBS Lett. 368:55-58. [DOI] [PubMed] [Google Scholar]
- 19.Kistner, A., M. Gossen, F. Zimmermann, J. Jerecic, C. Ullmer, H. Lubbert, and H. Bujard. 1996. Doxycycline-mediated quantitative and tissue-specific control of gene expression in transgenic mice. Proc. Natl. Acad. Sci. USA 93:10933-10938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kumagai, A., and W. G. Dunphy. 1999. Binding of 14-3-3 proteins and nuclear export control the intracellular localization of the mitotic inducer Cdc25. Genes Dev. 13:1067-1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laronga, C., H. Y. Yang, C. Neal, and M. H. Lee. 2000. Association of the cyclin-dependent kinases and 14-3-3σ negatively regulates cell cycle progression. J. Biol. Chem. 275:23106-23112. [DOI] [PubMed] [Google Scholar]
- 22.Lee, M. H., I. Reynisdottir, and J. Massague. 1995. Cloning of p57KIP2, a cyclin-dependent kinase inhibitor with unique domain structure and tissue distribution. Genes Dev. 9:639-649. [DOI] [PubMed] [Google Scholar]
- 23.Lopez-Girona, A., B. Furnari, O. Mondesert, and P. Russell. 1999. Nuclear localization of Cdc25 is regulated by DNA damage and a 14-3-3 protein. Nature 397:172-175. [DOI] [PubMed] [Google Scholar]
- 24.Lozano, G., and R. Montes de Oca Luna. 1998. Mdm2 function. Biochim. Biophys. Acta 1377:M55-M59. [DOI] [PubMed] [Google Scholar]
- 25.McLure, K. G., and P. W. Lee. 1998. How p53 binds DNA as a tetramer. EMBO J. 17:3342-3350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Melis, R., and R. White. 1999. Characterization of colonic polyps by two-dimensional gel electrophoresis. Electrophoresis 20:1055-1064. [DOI] [PubMed] [Google Scholar]
- 27.Midgley, C. A., J. M. Desterro, M. K. Saville, S. Howard, A. Sparks, R. T. Hay, and D. P. Lane. 2000. An N-terminal p14ARF peptide blocks Mdm2-dependent ubiquitination in vitro and can activate p53 in vivo. Oncogene 19:2312-2323. [DOI] [PubMed] [Google Scholar]
- 28.Nigro, J. M., S. J. Baker, A. C. Preisinger, J. M. Jessup, R. Hostetter, K. Cleary, S. H. Bigner, N. Davidson, S. Baylin, P. Devilee, et al. 1989. Mutations in the p53 gene occur in diverse human tumour types. Nature 342:705-708. [DOI] [PubMed] [Google Scholar]
- 29.Ostergaard, M., H. H. Rasmussen, H. V. Nielsen, H. Vorum, T. F. Orntoft, H. Wolf, and J. E. Celis. 1997. Proteome profiling of bladder squamous cell carcinomas: identification of markers that define their degree of differentiation. Cancer Res. 57:4111-4117. [PubMed] [Google Scholar]
- 30.Ouchi, T., A. N. Monteiro, A. August, S. A. Aaronson, and H. Hanafusa. 1998. BRCA1 regulates p53-dependent gene expression. Proc. Natl. Acad. Sci. USA 95:2302-2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pan, S., P. C. Sehnke, R. J. Ferl, and W. B. Gurley. 1999. Specific interactions with TBP and TFIIB in vitro suggest that 14-3-3 proteins may participate in the regulation of transcription when part of a DNA binding complex. Plant Cell 11:1591-1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pomerantz, J., N. Schreiber-Agus, N. J. Liegeois, A. Silverman, L. Alland, L. Chin, J. Potes, K. Chen, I. Orlow, H. W. Lee, C. Cordon-Cardo, and R. A. DePinho. 1998. The Ink4a tumor suppressor gene product, p19Arf, interacts with Mdm2 and neutralizes Mdm2's inhibition of p53. Cell 92:713-723. [DOI] [PubMed] [Google Scholar]
- 33.Prasad, G. L., E. M. Valverius, E. McDuffie, and H. L. Cooper. 1992. Complementary DNA cloning of a novel epithelial cell marker protein, Hme1, that may be down-regulated in neoplastic mammary cells. Cell Growth Differ. 3:507-513. [PubMed] [Google Scholar]
- 34.Reynisdottir, I., K. Polyak, A. Iavarone, and J. Massague. 1995. Kip/Cip and Ink4 Cdk inhibitors cooperate to induce cell cycle arrest in response to TGF-beta. Genes Dev. 9:1831-1845. [DOI] [PubMed] [Google Scholar]
- 35.Stavridi, E. S., N. H. Chehab, A. Malikzay, and T. D. Halazonetis. 2001. Substitutions that compromise the ionizing radiation-induced association of p53 with 14-3-3 proteins also compromise the ability of p53 to induce cell cycle arrest. Cancer Res. 61:7030-7033. [PubMed] [Google Scholar]
- 36.Stommel, J. M., N. D. Marchenko, G. S. Jimenez, U. M. Moll, T. J. Hope, and G. M. Wahl. 1999. A leucine-rich nuclear export signal in the p53 tetramerization domain: regulation of subcellular localization and p53 activity by nuclear export signal masking. EMBO J. 18:1660-1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Suzuki, H., F. Itoh, M. Toyota, T. Kikuchi, H. Kakiuchi, and K. Imai. 2000. Inactivation of the 14-3-3σ gene is associated with 5′ CpG island hypermethylation in human cancers. Cancer Res. 60:4353-4357. [PubMed] [Google Scholar]
- 38.Tzivion, G., Y. H. Shen, and J. Zhu. 2001. 14-3-3 proteins; bringing new definitions to scaffolding. Oncogene 20:6331-6338. [DOI] [PubMed] [Google Scholar]
- 39.Vellucci, V. F., F. J. Germino, and M. Reiss. 1995. Cloning of putative growth regulatory genes from primary human keratinocytes by subtractive hybridization. Gene 166:213-220. [DOI] [PubMed] [Google Scholar]
- 40.Wang, J., H. M. Goodman, and H. Zhang. 1999. An arabidopsis 14-3-3 protein can act as a transcriptional activator in yeast. FEBS Lett. 443:282-284. [DOI] [PubMed] [Google Scholar]
- 41.Wang, W., and D. C. Shakes. 1996. Molecular evolution of the 14-3-3 protein family. J. Mol. Evol. 43:384-398. [DOI] [PubMed] [Google Scholar]
- 42.Waterman, M. J., E. S. Stavridi, J. L. Waterman, and T. D. Halazonetis. 1998. ATM-dependent activation of p53 involves dephosphorylation and association with 14-3-3 proteins. Nat. Genet. 19:175-178. [DOI] [PubMed] [Google Scholar]
- 43.Xiao, B., S. J. Smerdon, D. H. Jones, G. G. Dodson, Y. Soneji, A. Aitken, and S. J. Gamblin. 1995. Structure of a 14-3-3 protein and implications for coordination of multiple signalling pathways. Nature 376:188-191. [DOI] [PubMed] [Google Scholar]
- 44.Yang, H. Y., R. Shao, M. C. Hung, and M. H. Lee. 2001. p27 Kip1 inhibits HER2/neu-mediated cell growth and tumorigenesis. Oncogene 20:3695-3702. [DOI] [PubMed] [Google Scholar]
- 45.Yang, H. Y., B. P. Zhou, M. C. Hung, and M. H. Lee. 2000. Oncogenic signals of HER-2/neu in regulating the stability of the cyclin-dependent kinase inhibitor p27. J. Biol. Chem. 275:24735-24739. [DOI] [PubMed] [Google Scholar]
- 46.Zhang, Y., Y. Xiong, and W. G. Yarbrough. 1998. ARF promotes Mdm2 degradation and stabilizes p53: ARF-INK4a locus deletion impairs both the Rb and p53 tumor suppression pathways. Cell 92:725-734. [DOI] [PubMed] [Google Scholar]
- 47.Zhou, B. P., M. C. Hu, S. A. Miller, Z. Yu, W. Xia, S. Y. Lin, and M. C. Hung. 2000. HER-2/neu blocks tumor necrosis factor-induced apoptosis via the Akt/NF-kappaB pathway. J. Biol. Chem. 275:8027-8031. [DOI] [PubMed] [Google Scholar]
- 48.Zhou, B. P., Y. Liao, W. Xia, Y. Zou, B. Spohn, and M. C. Hung. 2001. HER-2/neu induces p53 ubiquitination via Akt-mediated Mdm2 phosphorylation. Nat. Cell Biol. 3:973-982. [DOI] [PubMed] [Google Scholar]