Abstract
Prior ubiquitinylation of the unstable c-Fos proto-oncoprotein is thought to be required for recognition and degradation by the proteasome. Contradicting this view, we report that, although c-Fos can form conjugates with ubiquitin in vivo, nonubiquitinylatable c-Fos mutants show regulated degradation identical to that of the wild-type protein in living cells under two classical conditions of study: transient c-fos gene expression during the G0/G1 phase transition upon stimulation by mitogens and constitutive expression during asynchronous growth. Moreover, c-Fos destruction during the G0/G1 phase transition is unusual because it depends on two distinct but cumulative mechanisms. We report here that one mechanism involves a C-terminal destabilizer which does not need an active ubiquitin cycle, whereas the other involves an N-terminal destabilizer dependent on ubiquitinylation of an upstream c-Fos breakdown effector. In addition to providing new insights into the mechanisms of c-Fos protein destruction, an important consequence of our work is that ubiquitinylation-dependent proteasomal degradation claimed for a number of proteins should be reassessed on a new experimental basis.
The proteasome is the proteolytic machinery responsible for the degradation of a large number of cellular proteins. It is formed by a proteolytic core, the 20S proteasome, which associates with various regulatory complexes instrumental for the selection and processing of substrates. It is widely thought that most, if not nearly all, of its substrates must previously be modified by covalent attachment of ubiquitin chains, usually on the ɛ-amino group of internal lysines and, occasionally, on their N-terminal amino group, to be recognized (11). For many proteins, this assumption is based on the observation that they can be polyubiquitinylated in vivo and/or in vitro in cell extracts or in the presence of purified ubiquitinylating enzymes. Most often, additional indirect criteria of analysis are also considered. One such criterion is protein stabilization, either at the nonpermissive temperature in cell lines harboring a thermosensitive mutant of the ubiquitin-activating enzyme (E1), the first enzyme of the ubiquitin cycle (11), or upon ectopic expression of nonpolymerizable mutants of ubiquitin. Another criterion is the alteration of protein turnover in cells which express transdominant mutants of ubiquitinylating enzymes putatively operating on the protein of interest. In practice, unambiguous cellular and biochemical demonstration of the requirement for prior ubiquitinylation is, so far, available for only a relatively limited number of proteasome substrates (see Discussion). In particular, whether the inhibition of ubiquitinylation at the level of the substrate itself, through, for example, the use of nonubiquitinylatable mutants, may lead to protein stabilization has rarely been studied. Besides this, a few proteins, such as ornithine decarboxylase (ODC) (8) and the cyclin-dependent kinase inhibitor p21 (29), have been shown to be degraded physiologically by the proteasome in a ubiquitin-independent manner; although in the case of ODC, an ancillary protein, antizyme, is necessary. They are, however, considered exceptions to the ubiquitin-targeting hypothesis. Interestingly, whereas multiubiquitinylated forms of ODC have never been described in vivo, some have for p21. This led Verma and Deshaies to wonder about the actual number of unusual exceptions and to propose that a class of previously underappreciated proteasome substrates may have been missed (36). Moreover, growing evidence indicates that conjugation of multiubiquitin chains to proteins may serve purposes other than protein destruction, including regulation of transcription factor and enzyme activity as well as intracellular trafficking (21).
The c-Fos proto-oncoprotein is a key cell regulator whose improper, or dysregulated, expression is oncogenic in various lineages both in cultured cells and in living organisms (15). It is a basic DNA-binding domain-leucine zipper (LZ) transcription factor which heterodimerizes with different protein partners, such as the Jun family members, via LZ-LZ interactions to stimulate target genes through binding to the so-called AP-1 or CRE DNA motifs (7). It is constitutively expressed in certain embryonic and adult tissues in vivo (22) as well as in a variety of proliferating tumors and cell lines (for references, see reference 10). Its expression can be induced, in general rapidly and transiently, from nondetectable or low levels, in virtually all other cells to integrate many types of stimuli (13, 28). c-Fos is a short-lived protein. Its instability is essential for controlling its accumulation levels during the physiological periods of its expression as well as for rapid gene activity shutoff at the end of the latter and, thereby, avoidance of deleterious effects on the cell biology. c-Fos has been shown to be mainly, if not exclusively, degraded by the proteasome in vivo under two classical conditions of study: constitutive expression in asynchronously growing cells and transient induction in cells undergoing a G0/G1 phase transition upon stimulation by growth factors (2, 26). Its recognition by the proteasome significantly differs in the two situations (2, 10). In asynchronous cells, c-Fos destruction is under the control of a unique C-terminal destabilizer, whereas in G0/G1 cells, c-Fos is regulated by both C- and N-terminal destabilizers. Ubiquitinylation of c-Fos itself has been claimed to be necessary for proteasomal degradation (14, 31, 35). This conclusion was drawn from the observations that (i) c-Fos can be ubiquitinylated in vitro by semipurified ubiquitinylating enzymes (14, 31), (ii) c-Fos hydrolysis is promoted by semipurified ubiquitinylation enzymes in vitro (35), and (iii) c-Fos is stabilized to some extent in serum-stimulated E1 thermosensitive hamster E36-ts20 cells cultured at the nonpermissive temperature (31). Using nonubiquitinylatable c-Fos mutants, we have revisited this issue and shown that ubiquitinylation of c-Fos per se is not necessary for rapid and regulated proteasomal degradation. We also report that the mechanisms by which the two c-Fos destabilizers operate during the G0/G1 transition are largely different. Finally, together with those obtained with p21, our results show that great care should be taken when analyzing the dependence on ubiquitin for protein degradation and support the notion that the number of proteasome substrates recognized and hydrolyzed in a ubiquitin-independent manner might have been underestimated.
MATERIALS AND METHODS
Plasmids, mutagenesis, and expression vectors.
The rat c-Fos open reading frame was PCR amplified from a synthetic cDNA (1). For constitutive expression, it was cloned downstream of the cytomegalovirus promoter into the pcDNA3 plasmid (Invitrogen), and for transient serum-induced expression, it was cloned in the PM302 vector (2). Mutagenesis of c-Fos was done by PCR. The Myc6 tag (MEQKLISEEDLNE6) was PCR amplified from the pCS2+MT plasmid (5). Myc6K/R was assembled by using a set of synthetic oligonucleotides. The pcDNA3-Ha-Mos plasmid expressing a human c-mos cDNA is a gift from S. Leibovitch. The His6-tagged ubiquitin expression vector was described by Treier et al. (34).
Cell culture and transfection conditions.
Cells were grown in Dulbecco's modified Eagle medium containing 10% fetal calf serum. HeLa, E36, E36-ts20 (17), and f10 (6) cells were transfected by using classical procedures (10). For protein stability analysis in asynchronously growing cells, 1.5 × 106 HeLa cells were transiently transfected with 10 μg of pcDNA3-based expression plasmids, and 36 h later, either pulse-chase experiments were conducted or cells were collected for immunoblotting analysis. In the latter case, cells were also cotransfected with 1 μg of the enhanced green fluorescent protein (EGFP)-expressing pEGFPC1 plasmid (Clontech) as an internal standard. EGFPs were assayed by fluorescence or by immunoblotting with the anti-EGFP antiserum from Roche. For synchronization in G0/G1, cells were serum deprived for 36 h to arrest them in G0 and then stimulated by the addition of fresh medium containing 20% serum. E36 and E36-ts20 cells were amplified at 32°C. For E1 inactivation, they were incubated at 43°C for 30 min and then cultured at 39°C. MG132, lactacystin, and cycloheximide were used at final concentrations of 10, 10, and 50 μM, respectively. In cotransfection experiments, 5 μg of c-Fos and 10 μg of c-Mos expression vector was used.
Pulse-chase and immunoblotting experiments.
Protein extracts were prepared and half-life measurements were carried out as described in references 2 and 26: approximately 3 × 106 cells were radiolabeled for 1 h in the presence of 150 μCi of a [35S]methionine-cysteine mixture (NEG-772; NEN-Life Science) per ml of culture medium either at 36 h posttransfection, in the case of constitutive c-Fos expression, or at 30 min post-serum stimulation in the case of transient expression in G0/G1 cells before chases were started. In the case of E36-ts20 cells, E1 was inactivated by the incubation of cells at 43°C for 30 min during the labeling period, as described by Stancovksi et al. (31), a treatment which does not interfere with transient c-fos gene induction. Immunoprecipitations were carried out with either the sc52 anti-c-Fos rabbit antiserum (Santa Cruz Biotechnology), the 9E10 anti-Myc tag antibody, or the anti-T7 phage epitope tag monoclonal antibody (Novagen), depending on the experiment. Immunocomplexes were recovered by centrifugation with bovine serum albumin-preblocked protein A-agarose (sc2002; Santa-Cruz Biotechnology) and directly resuspended in electrophoresis loading buffer (2, 26). Proteins were fractionated through sodium dodecyl sulfate-containing 12% polyacrylamide gels. Dried gels were analyzed by using the 445SI PhosphorImager (Molecular Dynamics). For immunoblotting analysis, proteins were electrotransferred on polyvinylidene difluoride membranes and immunodetection was carried out with the same antibodies as described above or the anti-EGFP antiserum by using the Renaissance chemiluminescence kit (New England Nuclear) (2).
In vivo ubiquitinylation assay.
Analysis of ubiquitinylated species formed in vivo was carried out as described by Treier et al. (34) with 5 × 106 transfected HeLa cells per sample. One percent of the cell lysate was saved for immunoblotting analysis. Ubiquitin conjugates were affinity chromatography purified from the rest of the lysate with Ni2+-nitrilotriacetic acid-agarose beads (Qiagen). Ubiquitinylated proteins were eluted in electrophoresis loading sample buffer for immunoblotting analysis.
Gel shift assay.
Proteins used for electrophoretic mobility shift assays (EMSA) were in vitro translated in a volume of 25 μl by using the TNT kit (Promega). For each reaction mixture, 1 μl of each protein was incubated at room temperature for 20 min in binding buffer (5% glycerol, 1 mM EDTA, 10 mM HEPES [pH 7.9], 20 mM NaCl, 4 mM MgCl2, 10 mM dithiothreitol, 2 mM spermidine) in the presence of 2 μg of poly(dI-dC). Reaction mixtures were then mixed with 20,000 cpm of [32P] end-labeled oligonucleotide containing an AP-1 site (TTCCGGCTGACTCATCAAGCG). Incubation was continued at room temperature for 10 min, and DNA-protein complexes were resolved by electrophoresis through a nondenaturing 5% polyacrylamide gel at room temperature. The gel was dried, and DNA-protein complexes were visualized by autoradiography.
In vivo transcription assays.
HeLa cells were transfected with Fugene (Roche) by using the reporter plasmid pColl517luc, which contained a firefly luciferase gene under the control of the AP-1-responsive mouse collagenase promoter (4). Luciferase assays were performed 24 h later with the Reportalight Biowhittaker kit. The pEGFP-C1 plasmid was cotransfected as an internal control for transfection efficiency. Fluorescence was measured directly on living cells by spectrofluorometry.
Immunolocalization experiments.
Indirect immunofluorescence assays were carried out as described by Roux et al. (25) with the sc52 and H125 (Santa Cruz Biotechnology) anti-c-Fos antisera.
RESULTS
c-Fos can be ubiquitinylated in vivo.
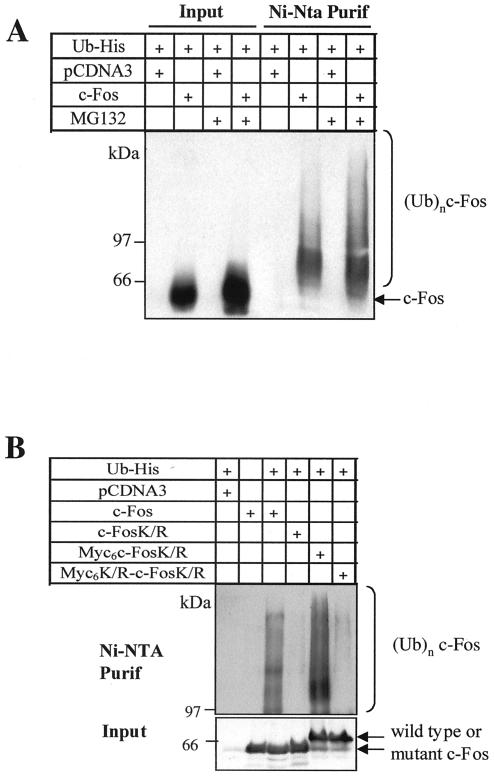
We first asked whether c-Fos could be ubiquitinylated in vivo in asynchronous human HeLa cells. For this, we used the transfection assay of Treier et al. (34), which permits purification and analysis of specific ubiquitin-protein conjugates under denaturing conditions. This offers the advantage of inhibiting isopeptidases that could possibly remove ubiquitin chains from studied proteins during sample manipulation. A small fraction of c-Fos (lower than 1%) was found to be ubiquitinylated (Fig. 1A). The same was observed in simian Cos-7 cells and mouse BALB/c embryo fibroblasts (data not shown). Moreover, the addition of a proteasome inhibitor (MG132) stabilized both nonconjugated and ubiquitin-conjugated c-Fos. However, the ratio of ubiquitinylated versus nonubiquitinylated c-Fos, as determined from densitometer scanning of luminograms (Fig. 1A), was identical in MG132-treated and nontreated cells. This differs from what is generally observed for substrates of the ubiquitin-proteasome pathway, the ubiquitinylated fraction of which increases upon inhibition of the proteasome.
FIG. 1.
c-Fos-ubiquitin conjugates in asynchronously growing HeLa cells. (A) Ubiquitinylation of c-Fos in HeLa cells. Asynchronously growing HeLa cells were transiently cotransfected with the His6-tagged ubiquitin expression plasmid and either the PM645 c-Fos expression vector or the void pcDNA3 parental vector. At 36 h posttransfection, the cells were incubated or not in the presence of MG132 for 8 h before cell protein extract preparation and affinity chromatography purification of ubiquitin conjugates on nickel agarose. Immunoblotting experiments were conducted with the specific sc52 anti-c-Fos antiserum. To compare the relative amounts of ubiquitinylated and nonubiquitinylated c-Fos, 1% of the total cell protein extracts was saved before nickel agarose chromatography and analyzed in parallel (Input) with the ubiquitin conjugates (Ni-Nta Purif). c-Fos and c-Fos-ubiquitin conjugates [(Ubn)-c-Fos] are indicated. (B) Ubiquitinylation of the c-FosK/R and its N-terminally tagged versions in asynchronous HeLa cells. Asynchronous HeLa cells were transiently cotransfected with constitutive expression vectors coding for His6-tagged ubiquitin and either c-Fos (PM645), c-FosK/R (PM695), Myc6-c-FosK/R (PM775), or Myc6K/R-c-FosK/R (PM841). Cotransfection of the parental pcDNA3 plasmid and the His6-tagged ubiquitin vector and transfection of the c-Fos expression vector alone were used as negative controls. Ubiquitin conjugates were prepared and analyzed as described for panel A.
c-Fos does not require ubiquitinylation on internal lysines to be degraded by the proteasome.
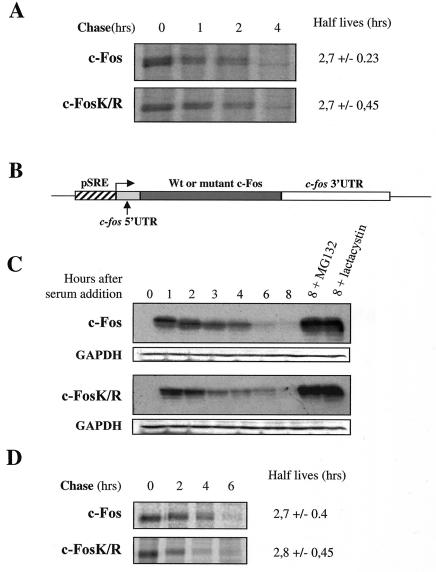
Next, we asked whether c-Fos ubiquitinylation is necessary for proteasomal degradation under conditions of constitutive expression in asynchronously growing cells and of transient expression upon serum stimulation of serum-deprived cells. In the latter case, c-Fos accumulation peaks by 1 to 2 h poststimulation and returns to basal levels a few hours latter. Because most proteins are ubiquitinylated on internal lysines, we first constructed a c-Fos mutant (c-FosK/R) in which all lysines were changed into arginines and we compared its turnover to that of wild-type c-Fos. No ubiquitinylated species of c-FosK/R could be detected in asynchronous HeLa cells (Fig. 1B), indicating that ubiquitin conjugation occurs essentially, if not exclusively, on internal lysines. However, both proteins showed similar half-lives (2.7 h) in pulse-chase experiments conducted with asynchronous HeLa cells (Fig. 2A) and were stabilized upon addition of MG132 (Fig. 3A). To study their degradation in serum-stimulated cells, c-fos−/− mouse embryo fibroblasts (f10 cells) (6) were stably transfected with expression plasmids reproducing transient c-fos gene expression due to a minimal serum-regulatable promoter and the presence of the 3′ c-fos mRNA untranslated region carrying the main mRNA destabilizer (Fig. 2B) (2). Cells arrested in the G0 phase by serum deprivation were stimulated by serum, and c-Fos and c-FosK/R abundances were monitored by immunoblotting as a function of time. Both proteins peaked 1 h after induction and decayed with comparable kinetics thereafter (Fig. 2C). Pulse-chase experiments confirmed similar half-lives (approximately 2.7 h) (Fig. 2D). Importantly, the levels of ectopic proteins in transfected f10 cells were comparable to those of endogenous c-Fos in serum-stimulated mouse BALB/c 3T3 fibroblasts, as assayed by immunofluorescence and immunoblotting (data not shown), ruling out any possible bias due to aberrant protein levels. Finally, the addition of MG132 or lactacystin, another proteasome inhibitor, stabilized both proteins (Fig. 2C). Taken together, these data indicate that c-Fos proteasomal degradation is independent of ubiquitinylation on the internal lysines.
FIG. 2.
Ubiquitinylation of c-Fos on internal lysine residues is not required for proteasomal degradation. (A) Half-lives of c-Fos and c-FosK/R in asynchronous HeLa cells. Asynchronous HeLa cells were transfected in parallel with either PM645 or PM695 for pulse-chase experiments as described in Materials and Methods. Values are the averages (± standard deviations) of the results of four independent experiments. (B) Structure of expression plasmids reproducing transient c-fos gene expression. Wild-type and mutant c-Fos protein open reading frames are under the transcriptional control of a minimal mouse c-fos serum-regulatable promoter. The 5′ and 3′ untranslated regions are those of the c-fos mRNA. The former allows efficient translation, and the latter confers instability to mRNAs. This plasmid reproduces transient mRNA accumulation similar to that of the endogenous c-fos mRNA (2). (C) Accumulation kinetics of c-Fos and c-FosK/R in serum-stimulated f10 cells. A population of f10 cells was stably transfected with PM763 and PM826 expression vectors to express c-Fos and c-FosK/R, respectively, in a serum-inducible manner. They were arrested in G0 upon serum deprivation and then stimulated by serum in the presence or absence of lactacystin or MG132. Cell extracts prepared at various time points poststimulation were analyzed by immunoblotting for the presence of c-Fos. Immunoblots were reprobed with an antiserum directed against glyceraldehyde-3-phosphate dehydrogenase (GAPDH) to verify that the same amount of protein was loaded in all lanes. Previous work has shown that transient c-fos mRNA accumulation in G0/G1 is not affected by the presence of proteasome inhibitors (2, 26). (D) Half-lives of c-Fos and c-FosK/R in serum-stimulated f10 cells. Pulse-chase experiments at the peak of induction were conducted as described in Materials and Methods. Values are the averages (± standard deviations) of the results of at least three independent experiments.
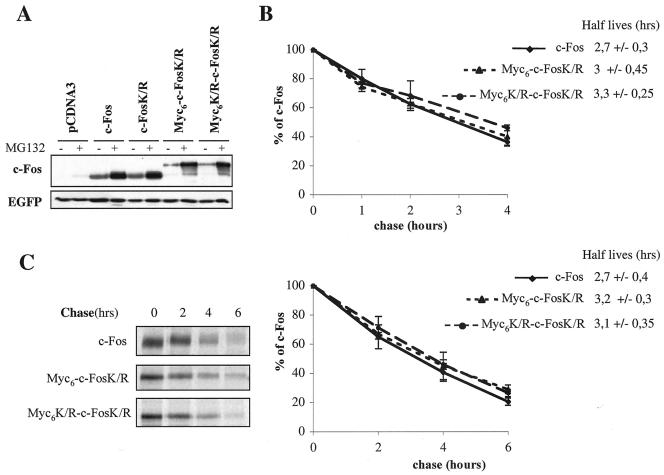
FIG. 3.
N-terminal ubiquitinylation of c-Fos is not required for proteasomal degradation. (A) Accumulation of wild-type and mutant c-Fos in asynchronous HeLa cells in the presence of MG132. HeLa cells transiently transfected with PM645, PM695, PM775, or PM841 as well as with the pEGFP-C1 vector expressing EGFP, which was used as an internal invariant transfection control. At 36 h posttransfection, cells were incubated or not in the presence of MG132 for 8 h. Immunoblot analyses were carried out with both anti-c-Fos and anti-EGFP antisera. Note the retarded migration of Myc6-c-FosK/R and Myc6K/R-c-FosK/R because of the length of the tag (90 amino acids). MG132 treatment does not affect the abundance of mRNAs expressed from transfected vectors, which implies that higher wild-type or mutant c-Fos accumulation reflects protein stabilization. (B) Quantitative analysis of pulse-chase experiments in asynchronous HeLa cells. At least three independent pulse-chase experiments per construct were conducted as described in the legend to Fig. 2A. Half-life values are the averages (± standard deviations) of the results of at least three independent experiments, such as the one presented in panel C. (C) Pulse-chase experiments in serum-stimulated f10 cells. Experiments were conducted as described in the legend to Fig. 2D. +, present; −, absent.
N-terminal ubiquitinylation is not required for c-Fos degradation by the proteasome.
Although no c-FosK/R ubiquitinylated species were detected in transfection assays (Fig. 1B), we wanted to more formally rule out a possible contribution of N-terminal ubiquitin conjugation to c-Fos instability. Thus, we modified the c-FosK/R N terminus to prevent any conjugation of ubiquitin by grafting two long tags (90 amino acids): (i) a Myc6 tag, which contains potentially ubiquitinylatable lysines, but which was previously shown to be capable of inhibiting N-terminal ubiquitinylation-dependent degradation of MyoD (5), E7 of human papilloma virus 16 (24), and LMP1 (3) proteins, and (ii) a mutated nonubiquitinylatable version of it (Myc6K/R) in which all lysines were changed into arginines. No ubiquitin conjugates could be seen in the case of Myc6K/R-c-FosK/R,whereas some were easily detected in the case of Myc6-c-FosK/R (Fig. 1B). Although the latter observation does not question the conclusion drawn from the three above-cited studies, it nevertheless indicates that Myc6 must be used with caution because it can lead to ubiquitinylation where it is supposed to suppress it. We, then, compared the half-lives of c-Fos, c-FosK/R, and the N-terminally tagged versions in asynchronous HeLa cells. All proteins accumulated to the same steady-state levels in standardized transient transfection experiments (Fig. 3A) and decayed with similar rates in pulse-chase experiments (Fig. 3B), indicating similar turnovers. Moreover, both c-Fos and the tagged mutants accumulated in the presence of MG132, suggesting an involvement of the proteasome in their degradation (Fig. 3A).
The half-lives of c-Fos, Myc6-c-FosK/R, and Myc6K/R-c-FosK/R were then compared in serum-stimulated f10 cells stably transfected with the serum-inducible expression vector presented in Fig. 2B. Pulse-chase experiments revealed similar half-lives for the three proteins at the G0/G1 phase transition (Fig. 3C), supporting the idea that a possible N-terminal ubiquitinylation of c-Fos does not have a major role in its rapid degradation.
Myc6-c-FosK/R and Myc6K/R-c-FosK/R nevertheless showed slightly longer half-lives (less than 20% longer) than c-Fos and c-FosK/R both in asynchronous and G0/G1 cells, raising the possibility of a minor contribution of N-terminal ubiquitinylation to c-Fos instability. We, however, do not favor this hypothesis for various reasons: (i) Myc6K/R-c-FosK/R does not show detectable ubiquitinylation in living cells (Fig. 1B), (ii) tagging with a nonubiquitinylatable T7 phage epitope stabilizes c-FosK/R neither in asynchronous HeLa cells nor in serum-stimulated f10 cells (data not shown) and hamster fibroblasts (see Fig. 6), and (iii) artificially increased ubiquitinylation of Myc6-c-FosK/R in its N-terminal region, due to the presence of the ubiquitinylatable Myc6 tag, does not accelerate breakdown. Moreover, it is interesting that c-Fos begins with a doublet of methionines and that methionine is one of the most acetylated N-terminal amino acids in eucaryotic cells (23). Although the point has not been experimentally addressed due to the low amounts of the protein in vivo, the probability that the c-Fos N terminus is blocked by an acetyl group is high, which is in line with the fact that we detected no N-terminal ubiquitinylation of c-Fos in our experiments.
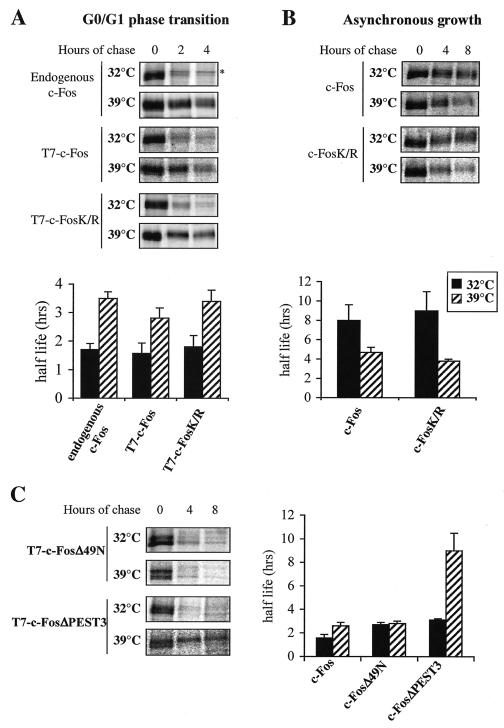
FIG. 6.
Involvement of the ubiquitin pathway in c-Fos degradation. (A) Degradation of c-Fos and c-FosK/R in serum-stimulated E36-ts20 cells. E36-ts20 cells were transfected with PM302 and PM694 to express T7-tagged versions of c-Fos and c-FosK/R in a serum-inducible manner. Either nontransfected cells (endogenous c-Fos) or transfected cell populations (T7-c-Fos and T7-c-FosK/R) were cultured at 32°C and serum starved for 36 h before stimulation by serum. E1 inactivation and pulse-chase experiments were conducted as described in Materials and Methods. The presented half-lives are the averages (± standard deviations) of the results of at least three independent experiments. The star indicates a background band immunoprecipitated by the sc52 antiserum. (B) Degradation of c-Fos and c-FosK/R in asynchronous E36-ts20 cells. E36-ts20 cells were transfected with constitutive expression vectors PM645 or PM695 coding for wild-type c-Fos and c-FosK/R, respectively. Pulse-chase experiments were carried out 36 h later, after inactivation or not of E1. The presented half-lives are the averages (± standard deviations) of the results of at least three independent experiments. (C) Stabilities of N- and C-terminally truncated c-Fos in E36-ts20 cells. Experiments were conducted as described for panel A, with the PM698 and PM327 vectors coding for T7-tagged c-FosΔ49N and c-FosΔPEST3 proteins. The presented half-lives correspond to the averages (± standard deviations) of the results of three independent experiments.
c-FosK/R is functionally active, and its degradation is regulated like that of wild-type c-Fos.
Abnormal proteins may undergo proteasomal degradation (11), some of which most probably through ubiquitin-independent mechanisms (30, 32). It was thus important to establish that c-FosK/R was still biologically active and not addressed to the proteasome via a pathway not operating on the wild-type protein.
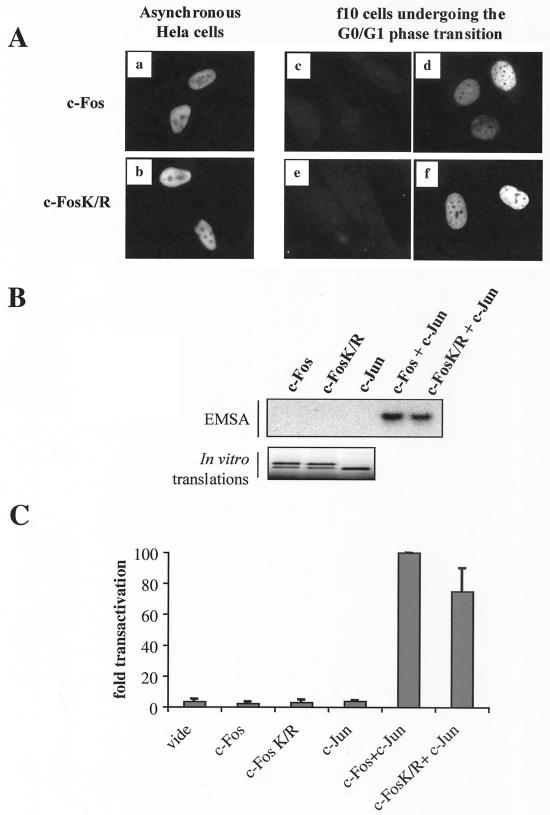
Immunolocalization experiments showed that c-FosK/R localized primarily within the nucleus of both asynchronous and serum-stimulated cells as did the wild-type protein (Fig. 4A), excluding that gross intracellular redistribution might account for aberrant degradation. We then verified that c-FosK/R could heterodimerize with c-Jun (7) and bind to an AP-1 DNA motif as efficiently as c-Fos in EMSAs (Fig. 4B). Next, we compared c-FosK/R and c-Fos transactivations in vivo by cotransfecting a luciferase reporter gene under the control of an AP-1-responsive collagenase promoter with either c-Fos or c-FosK/R expression plasmid as well as a vector for c-Jun. Neither c-Fos nor c-FosK/R alone was able to transactivate the reporter gene. However, both of them were capable of transactivation in the presence of c-Jun, although c-FosK/R was slightly less (30%) efficient than wild-type c-Fos (Fig. 4C). This is consistent with the idea that the overall structure of the protein is not profoundly affected by the mutation of the lysines into arginines.
FIG. 4.
Intracellular localization, DNA binding, and transactivation activity of c-FosK/R. (A) Intracellular localization of c-Fos and c-FosK/R. Asynchronously growing HeLa cells were transiently transfected with PM645 or PM695 to express c-Fos or c-FosK/R, respectively. Immunofluorescence assays were performed with an anti-c-Fos antiserum 16 h later (a and b). f10 cells stably transfected with PM763 and PM826 to express c-Fos and c-FosK/R in a serum-inducible manner were arrested in G0 (c and e) and stimulated by serum for 1 h (d and f) before immunofluorescence analysis. (B) DNA-binding activity of c-Fos and c-FosK/R. In vitro-translated c-Fos, c-FosK/R, and c-Jun (lower panel) were incubatedalone or in combination with a radiolabeled AP-1 probe and used for EMSAs (upper panel) as described in Materials and Methods. (C) Transactivation activity of c-Fos and c-FosK/R. HeLa cells were transfected under standardized conditions with the pcDNA3 control plasmid (mock), PM645 c-Fos, PM695 c-FosK/R, and the CD330 c-Jun expression vectors as indicated on the figure in the presence of an AP-1-inducible luciferase expression vector (pColl517luc). Luciferase activity was assayed at 24 h posttransfection. A plasmid p-EGFP-C1 carrying an EGFP gene under the transcriptional control of the cytomegalovirus promoter was included in each transfection for normalization of data. The presented data are the averages of the results of three independent experiments. The transactivation efficiencies were calculated according to the c-Fos plus c-Jun transactivation efficiency, which was set to 100.
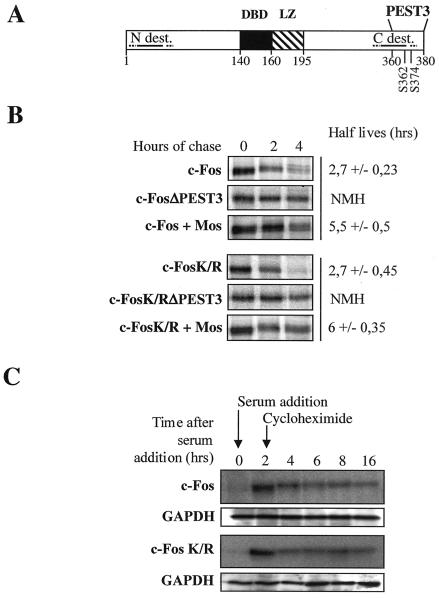
Then we investigated whether c-Fos and c-FosK/R degradations were similarly regulated. In asynchronous cells, we took advantage of two observations: (i) c-Fos degradation is under the control of a unique C-terminal destabilizer containing a PEST motif (PEST3) whose deletion is sufficient for dramatic stabilization (Fig. 5A) and (ii) phosphorylation of serines 362 and 374 within PEST3 by Mos-induced mitogen-activated protein kinases stabilizes c-Fos, although to a limited extent (10, 20). As expected, deletion of PEST3 (c-FosΔPEST3 and c-FosK/RΔPEST3 mutants) led to strong stabilization of both c-Fos and c-FosK/R and coexpression with Mos led to a comparable twofold stabilization in the two cases, as shown in pulse-chase experiments (Fig. 5B). The phosphorylation of serines 362 and 374 is associated with retarded electrophoretic migration (10, 20). It is thus of note that both c-Fos and c-FosK/R both showed the same retardation in the presence of Mos, since this provides further support for the idea of a lack of c-Fos C terminus structural alteration. Because degradation of c-Fos is regulated by significantly different mechanisms in serum-stimulated cells (2, 10), we had to turn to other criteria for analysis. It was previously shown that blocking protein synthesis selectively leads to c-Fos stabilization in a hyperphosphorylated form which migrates with a higher apparent molecular weight (26). Thus, cycloheximide was added 2 h after serum stimulation to c-fos−/− f10 cells stably transfected with serum-inducible expression vectors for c-Fos and c-FosK/R. Both proteins were strongly stabilized and showed slowed down migration (Fig. 5C). In summary, our data strongly suggest that changing all lysines to arginines does not affect the physiological mechanisms addressing c-Fos to the proteasome in exponentially growing and serum-stimulated cells (also see below).
FIG. 5.
Regulation of c-Fos and c-FosK/R degradation in asynchronously growing and serum-synchronized cells. (A) c-Fos-destabilizing elements. The positions of the DNA-binding domain (DBD), LZ, N- (N-dest.) and C-terminal (C-dest.) destabilizers, and PEST3 motif as well as that of serines 362 and 374 are indicated. (B) Degradation of c-Fos and c-FosK/R with deletions of PEST3 or coexpressed with Mos in asynchronous cells. HeLa cells were transfected in parallel with either PM645, PM695, PM551, or PM790 to express c-Fos, c-FosK/R, c-FosΔPEST3, or c-FosK/RΔPEST3, respectively, or with PM645 and PM695 in the presence of the Mos expression vector pcDNA3-Ha-Mos. Half-lives are the averages (± standard deviations) of the results of three independent experiments. NMH, not measurable half-life. (C) Stabilization of c-Fos and c-FosK/R in serum-synchronized cells. f10 cells stably transfected with PM763 and PM826 to express serum-inducible c-Fos and c-FosK/R, respectively, were stimulated by serum, and cycloheximide was added 2 h poststimulation. This figure is to be compared with Fig. 2C. Note the retarded migration of both proteins in the presence of cycloheximide, which is due to phosphorylation (26). The differences in abundances between time points 2 and 4 h are due to the degradation of c-Fos before cycloheximide exerts its stabilizing effect. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an internal marker to verify that the same amount of protein was analyzed in each lane.
The ubiquitin pathway is indirectly involved in the control of c-Fos degradation during the G0/G1 phase transition but not in asynchronously growing cells.
Contrasting with our data, other authors have argued that ubiquitinylation of c-Fos is required for proteasomal degradation in particular because c-Fos was partially stabilized in E1 thermosensitive hamster E36-ts20 cells stimulated by serum at the nonpermissive temperature (31). One possibility for reconciling these apparently contradictory observations would be an indirect involvement of the ubiquitin pathway in c-Fos degradation. To test this hypothesis, we compared the behavior of wild-type c-Fos to that of a nonubiquitinylatable mutant in E36-ts20 cells at the restrictive temperature.
First, we confirmed the observation described in the work by Stancovski et al. E36-ts20 cells were stimulated by serum after arrest in G0, and half-life measurements of endogenous c-Fos were carried out during the peak of induction under permissive (32°C) and nonpermissive (39°C) conditions for the ubiquitin pathway (Fig. 6A). No alteration in c-fos gene induction was observed under the nonpermissive conditions because the shift of temperature blocks the cell cycle only in the G2 phase. As expected, inhibition of E1 activity at 39°C caused an approximately twofold stabilization of c-Fos. Then, using the expression vector described in the legend to Fig. 2B, we analyzed the stabilities of c-Fos and c-FosK/R proteins N-terminally tagged with a T7 phage epitope to permit discrimination from endogenous c-Fos in G0/G1 cells. Both were as unstable as endogenous c-Fos at 32°C and were stabilized with the same efficiency at 39°C (Fig. 6A). In contrast, in keeping with Stancovski et al. (31), the temperature shift from 32 to 39°C did not increase the half-life of both endogenous and transfected c-Fos in parental E36 cells containing a functional E1 (data not shown). Given the fact that a nonubiquitinylatable c-Fos mutant is still stabilized when E1 is inactivated, we can conclude that the ubiquitin pathway is involved only indirectly in c-Fos degradation at the G0/G1 cells.
As the regulation of c-Fos degradation is different according to the expression conditions, we wondered whether a ubiquitinylation-sensitive effector was also required for the degradation of c-Fos and c-FosK/R in asynchronously growing cells. At 32°C, the half-lives of c-Fos and c-FosK/R were longer than in serum-stimulated cells, in accordance with previous observations with mouse embryo fibroblasts (10). However, in contrast to G0/G1 cells, both proteins were not stabilized at 39°C. In fact, they were degraded significantly faster than at 32°C (Fig. 6B), which is most probably due to temperature-linked kinetic differences already observed for a number of enzymatic reactions in thermosensitive cell lines (12, 17). Thus, the ubiquitin pathway is dispensable for c-Fos destruction in asynchronous cells.
The activity of the N-terminal, but not that of the C-terminal, c-Fos destabilizer is regulated by the ubiquitin pathway during the G0/G1 transition.
To determine whether a functional ubiquitin cycle was necessary for the activity of the N- and/or the C-terminal c-Fos destabilizers in G0/G1 cells, we used two mutants of c-Fos with deletions of either the first 49 amino acids of the protein (c-FosΔ49N) or of the last 20 amino acids (c-FosΔPEST3). Since only one destabilizer was altered in each one of these mutants, both of them were moderately stabilized in mouse embryo fibroblasts with half-lives approximately twofold longer than that of c-Fos (2, 10). This is different from what is observed in asynchronously growing cells (Fig. 5B), where deletion of PEST3 inactivates the C-terminal destabilizer, which is the only one to function under this condition (10), and leads to a strong stabilization. The two mutants were also stabilized about twofold in E36-ts20 cells stimulated by serum at 32°C, confirming that each one of the two destabilizers was active in G0/G1 in this cellular context as well (Fig. 6C). When the cells were shifted to the restrictive temperature, the half-life of c-FosΔ49N was not modified, suggesting that the C-terminal destabilizer did not need ubiquitin to be still active at 39°C. In contrast, c-FosΔPEST3 was strongly stabilized, indicating that an active E1 enzyme is necessary for the activity of the N-terminal destabilizer.
DISCUSSION
c-Fos is degraded by the proteasome independently of its own ubiquitinylation.
The comparison of nonubiquitinylatable mutants to wild-type c-Fos showed that neither internal nor N-terminal ubiquitinylation of the protein itself is necessary for proteasomal degradation, both in asynchronously growing cells and during the G0/G1 phase transition in different cell types. This conclusion differs from those drawn by other authors on the basis of indirect experiments carried out in vitro or in vivo with the E36-ts20 E1 thermosensitive cell line (14, 31, 35). We, nevertheless, reconcile these two sets of results by showing that the ubiquitin pathway is indeed indirectly required, but only during the G0-to-G1 phase transition, for maximal degradation of c-Fos. Our results, moreover, give insight into the mechanisms of efficient c-Fos destruction by the action of two independent destabilizers during the G0-to-G1 phase transition to avoid sustained activity of the protein, which could lead to deregulated activation of target genes and transformation.
Importantly, the c-FosK/R mutant used here retained most of the c-Fos transcriptional activity, and its degradation was regulated like that of the wild-type protein in different assays. This makes it very unlikely that strong conformation alterations might entail destruction via nonphysiological mechanisms or via pathways responsible for the elimination of abnormal proteins. We cannot formally exclude the possibility that structural effects related to the lysine-to-arginine change may account for the slightly diminished transcriptional activity of c-FosK/R. It is, however, worth considering that lysines are targets, not only for ubiquitin attachment but also for a variety of other posttranslational modifications, such as acetylation, methylation, or conjugation of ubiquitin-like polypeptides. Whether one, or several, of them is necessary for optimal c-Fos activity deserves further investigation.
How is c-Fos targeted to the proteasome?
If prior ubiquitinylation of c-Fos itself is not necessary for degradation, then how is this protein recognized by the proteasome? We have previously reported that c-Fos degradation depends on a unique C-terminal destabilizer in asynchronous cells and on both an N-terminal and a C-terminal destabilizer, which is probably the same as the one used in asynchronous cells but whose activity is limited because of the phosphorylation of serines 362 and 374, in cells undergoing a G0/G1 phase transition (10). We stress here that the mechanisms underlying the action of the N- and C-terminal destabilizers are clearly different since the N-terminal destabilizer requires an active ubiquitin cycle, whereas the C-terminal destabilizer does not. It is also worth underlining that the alterations of the N- or C-terminal destabilizer each led to no more than a twofold stabilization (10), which is consistent with the idea that both destabilizers function independently, and that individual c-Fos molecules are recognized either via the N- or via the C-terminal destabilizer.
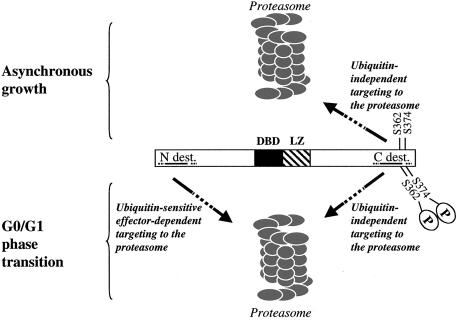
How could the N-terminal destabilizing element permit the addressing of c-Fos to the proteasome? Since c-Fos does not need to be ubiquitinylated to be degraded, inactivation of this destabilizer upon E1 inactivation suggests that at least one ubiquitinylation-sensitive upstream effector of c-Fos controls its action. One obvious possibility would be the trans-targeting of c-Fos to the proteasome by a ubiquitinylated dimerization partner of the Jun family. Although it is not possible to eliminate this possibility for a minor fraction of c-Fos, this mechanism does not concern the bulk of the protein because, as reported elsewhere (10), (i) mutation or deletion of the LZ does not eliminate c-Fos instability and (ii) grafting of an N-terminal fragment carrying the destabilizer but lacking the LZ is sufficient to destabilize the stable EGFP reporter protein. The identification of the proteins interacting with the c-Fos N terminus is being tested to determine whether the ubiquitinylation event(s) is responsible for the inactivation of an inhibitor (either through functional inactivation or induction of destruction) or the activation of a positive effector of c-Fos destruction. How could c-Fos be addressed to the proteasome via its C terminus? One obvious possibility is direct recognition, as has been documented for the p21 cyclin-dependent kinase inhibitor, which directly binds the C8 subunit of the 20S proteasome and whose instability correlates with the ability to bind to C8 (33). Alternatively, a cofactor(s) might be necessary, as was shown for ODC, whose proteolysis by the purified 26S proteasome in vitro is dependent upon the presence of its antizyme cofactor (19). Whatever the case, two important points must be stressed here with respect to C-terminal domain-mediated degradation of c-Fos. First, even if c-Fos were recognized directly by the proteasome, this would not exclude the possibility that ancillary molecules may promote or favor this interaction in vivo. Second, the ubiquitinylation-independent destruction mechanism does not just correspond to a basal and slow process which can be dramatically accelerated upon ubiquitinylation of the protein, as has been reported in the case of IκΒα after cytokine treatment of cells (16). Rather, it actually constitutes an efficient destruction pathway, which is the only mechanism accounting for c-Fos instability in asynchronous cells and which contributes to approximately half of the c-Fos elimination in G0/G1 cells (Fig. 7).
FIG. 7.
Degradation of c-Fos in asynchronous cells and cells undergoing a G0/G1transition. c-Fos shows unusual properties with regard to its destruction. First, in spite of being a ubiquitinylatable protein in vivo, its prior ubiquitinylation is dispensable for proteasomal degradation. Second, it possesses at least two destabilizing elements which are used differently depending on the situation. One destabilizer (C-dest.) is located within the C-terminal part of the protein. It constitutes the only active destabilizer of c-Fos in asynchronous cells. It is also active during the G0/G1transition, although phosphorylation of serines 362 and 374 limits its activity. It does not necessitate an active ubiquitin pathway for functioning in asynchronous or in serum-stimulated cells. The other destabilizer (N-dest.) is located at the N terminus of the molecule. It is conditionally active during the G0/G1transition, and its activity is dependent on a functional ubiquitin cycle, presumably through ubiquitinylation of an upstream effector of c-Fos degradation. Discontinuous lines indicate that the precise limits of the N- and C-terminal destabilizers are not known. In G0/G1, c-Fos molecules are probably recognized via either the C or N terminus. Whether cooperation between N- and C-terminal destabilizers occurs after recognition for further processing of c-Fos by the proteasome cannot be ruled out. Discontinuous arrows indicate that upstream effectors are (N-terminal destabilizer-mediated degradation) or may be (C-terminal destabilizer-mediated degradation) involved in c-Fos being addressed to the proteasome.
Biological relevance of c-Fos ubiquitinylation.
Because degradation of the bulk of c-Fos does not require prior ubiquitinylation of the protein, the observation of c-Fos-ubiquitin conjugates is intriguing. The fraction of ubiquitinylated species is, however, very small. A simple explanation for this would be that these conjugates correspond to a small proportion of misfolded c-Fos molecules eliminated following a ubiquitinylation-dependent clearance mechanism, as has been reported for a number of abnormal or in-excess proteins (27, 32). We also do not exclude that they could just reflect a nonspecific ubiquitinylation background occurring or tolerated in the cell. It is, however, worth considering that, on one hand, it is presently unknown what fraction of c-Fos is biologically active in a cell at a given time and whether all c-Fos molecules have similar fates. On the other hand, it is clear that ubiquitinylation is involved in the regulation of a variety of biological processes, including the control of the activity of several transcription factors, independent of proteasomal degradation (9). It thus cannot be ruled out that ubiquitinylation may govern the activity of a subset of c-Fos protein. Finally, even though we clearly demonstrated that ubiquitinylation is not necessary for the degradation of the bulk of c-Fos in asynchronously growing and G0/G1 phase transitioning fibroblasts, we do not exclude the possibility that c-Fos ubiquitinylation might be used for the degradation of a minor fraction of the protein or under conditions that have not been investigated here. More-extensive analysis of this point in other cell types and other conditions of c-Fos expression is thus required to answer this question.
Demonstration that prior ubiquitinylation of a protein is required for its proteasomal degradation.
Our data show that visualization of ubiquitinylated species of a given protein in vivo is not sufficient to conclude that this ubiquitinylation is instrumental for proteasomal degradation even though this protein is stabilized upon E1 inactivation in a thermosensitive cell line. They also suggest that rigorous addressing of this issue requires inhibition of ubiquitinylation at the level of the protein itself without affecting other ubiquitinylation events occurring in the cell, because the ubiquitin pathway might be indirectly involved in the control of protein turnover. Thus, our results have important general consequences. First, they suggest that the degradation of certain proteins, whose degradation is thought to be ubiquitinylation dependent based on indirect evidence, should be reexamined on a novel experimental basis. Second, they provide a novel support for the idea, initially proposed by Verma and Deshaies (36), that ubiquitinylation-independent substrates of the proteasome may constitute an underappreciated group of proteins. These authors have already stressed that proteasomes are present in bacteria and archaea while ubiquitin and ubiquitin-conjugating systems are not (18, 37). It is thus tempting to speculate that, while acquiring the ability to recognize and process ubiquitinylated proteins through evolution, higher eucaryote proteasomes have also kept the ability of primitive proteasomes to degrade numerous nonubiquitinylated proteins.
At present, the lysine-to-arginine mutation is most probably the only approach available for the inhibition of internal protein ubiquitinylation, and the modification of the N terminus, through deletion and/or grafting of tags, is the only one to inhibit N-terminal ubiquitinylation. Since mutagenesis can induce deleterious conformational changes, criteria of confidence, such as conservation of the protein functions and of its turnover regulation, are necessary to eliminate the possibility of nonphysiological degradation of such mutants. Absence or partial stabilization of a protein in E1 thermosensitive cell lines also deserves a comment. On one hand, absence of stabilization does not necessarily mean ubiquitin-independent degradation because a number of these cell lines show a leaky phenotype. This characteristic most probably explains why c-Fos stabilization is not detectable in serum-stimulated mouse A31N-ts20 cells (26), whereas it is detectable in E36-ts20 cells (31) upon a shift at 39°C. On the other hand, limited stabilization does not necessarily imply a leaky phenotype of the cell line used but may hide a complex degradation mechanism. This situation is most probably exemplified here for the first time by c-Fos, whose N-terminal destabilizer is efficiently inactivated in serum-stimulated E36-ts20 cells at 39°C while its C-terminal destabilizer is still active.
Acknowledgments
We are indebted to E. Andermarcher, O. Coux, R. Hipskind, and M. Scheffner for critical reading of the manuscript.
This work was supported by grants from the CNRS, the ARC, the Ligue Nationale contre le Cancer, and the European Commission contract QLG1-CT-2001-02026.
REFERENCES
- 1.Abate, C., D. Luk, and T. Curran. 1990. A ubiquitous nuclear protein stimulates the DNA-binding activity of fos and jun indirectly. Cell Growth Differ. 1:455-462. [PubMed] [Google Scholar]
- 2.Acquaviva, C., F. Brockly, P. Ferrara, G. Bossis, C. Salvat, I. Jariel-Encontre, and M. Piechaczyk. 2001. Identification of a C-terminal tripeptide motif involved in the control of rapid proteasomal degradation of c-Fos proto-oncoprotein during the G0-to-S phase transition. Oncogene 20:7563-7572. [DOI] [PubMed] [Google Scholar]
- 3.Aviel, S., G. Winberg, M. Massucci, and A. Ciechanover. 2000. Degradation of the epstein-barr virus latent membrane protein 1 (LMP1) by the ubiquitin-proteasome pathway. Targeting via ubiquitination of the N-terminal residue. J. Biol. Chem. 275:23491-23499. [DOI] [PubMed] [Google Scholar]
- 4.Bakiri, L., D. Lallemand, E. Bossy-Wetzel, and M. Yaniv. 2000. Cell cycle-dependent variations in c-Jun and JunB phosphorylation: a role in the control of cyclin D1 expression. EMBO J. 19:2056-2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Breitschopf, K., E. Bengal, T. Ziv, A. Admon, and A. Ciechanover. 1998. A novel site for ubiquitination: the N-terminal residue, and not internal lysines of MyoD, is essential for conjugation and degradation of the protein. EMBO J. 17:5964-5973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brusselbach, S., U. Mohle-Steinlein, Z. Q. Wang, M. Schreiber, F. C. Lucibello, R. Muller, and E. F. Wagner. 1995. Cell proliferation and cell cycle progression are not impaired in fibroblasts and ES cells lacking c-Fos. Oncogene 10:79-86. [PubMed] [Google Scholar]
- 7.Chinenov, Y., and T. K. Kerppola. 2001. Close encounters of many kinds: Fos-Jun interactions that mediate transcription regulatory specificity. Oncogene 20:2438-2452. [DOI] [PubMed] [Google Scholar]
- 8.Coffino, P. 1998. Degradation of ornithine decarboxylase, p. 411-428. In J.-M. Peters, J. R. Robins, and D. Finley (ed.), Ubiquitin and the biology of the cell. Plenum Press, New York, N.Y.
- 9.Conaway, R., C. Browner, and J. Conaway. 2002. Emerging roles of ubiquitin in transcription regulation. Science 296:1254-1258. [DOI] [PubMed] [Google Scholar]
- 10.Ferrara, P., E. Andermarcher, G. Bossis, C. Acquaviva, F. Brockly, I. Jariel-Encontre, and M. Piechaczyk. 2003. The structural determinants responsible for c-Fos protein proteasomal degradation differ according to the conditions of expression. Oncogene 22:1461-1474. [DOI] [PubMed] [Google Scholar]
- 11.Glickman, M. H., and A. Ciechanover. 2002. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol. Rev. 82:373-428. [DOI] [PubMed] [Google Scholar]
- 12.Gropper, R., R. Brandt, S. Elias, C. Beaver, A. Mayer, A. Schwartz, and A. Ciechanover. 1991. The ubiquitin-activating enzyme, E1, is required for stress-induced lysosomal degradation of cellular proteins. J. Biol. Chem. 266:3602-3610. [PubMed] [Google Scholar]
- 13.Herdegen, T., and V. Waetzig. 2001. AP-1 proteins in the adult brain: facts and fiction about effectors of neuroprotection and neurodegeneration. Oncogene 20:2424-2437. [DOI] [PubMed] [Google Scholar]
- 14.Hermida-Matsumoto, M. L., P. B. Chock, T. Curran, and D. C. Yang. 1996. Ubiquitinylation of transcription factors c-Jun and c-Fos using reconstituted ubiquitinylating enzymes. J. Biol. Chem. 271:4930-4936. [DOI] [PubMed] [Google Scholar]
- 15.Jochum, W., E. Passegue, and E. F. Wagner. 2001. AP-1 in mouse development and tumorigenesis. Oncogene 20:2401-2412. [DOI] [PubMed] [Google Scholar]
- 16.Krappmann, D., F. G. Wulczyn, and C. Scheidereit. 1996. Different mechanisms control signal-induced degradation and basal turnover of the NF-kappaB inhibitor IkappaB alpha in vivo. EMBO J. 15:6716-6726. [PMC free article] [PubMed] [Google Scholar]
- 17.Kulka, R. G., B. Raboy, R. Schuster, H. A. Parag, A. Diamond, A. Ciechanover, and M. Marcus. 1988. A Chinese hamster cell cycle mutant arrested at G2 phase has a temperature-sensitive ubiquitin-activating enzyme E1. J Biol. Chem. 263:15726-15731. [PubMed] [Google Scholar]
- 18.Maupin-Furlow, J. A., S. J. Kaczowka, M. S. Ou, and H. L. Wilson. 2001. Archaeal proteasomes: proteolytic nanocompartments of the cell. Adv. Appl. Microbiol. 50:279-338. [DOI] [PubMed] [Google Scholar]
- 19.Murakami, Y., S. Matsufuji, T. Kameji, S. Hayashi, K. Igarashi, T. Tamura, K. Tanaka, and A. Ichihara. 1992. Ornithine decarboxylase is degraded by the 26S proteasome without ubiquitination. Nature 360:597-599. [DOI] [PubMed] [Google Scholar]
- 20.Okazaki, K., and N. Sagata. 1995. The Mos/MAP kinase pathway stabilizes c-Fos by phosphorylation and augments its transforming activity in NIH 3T3 cells. EMBO J. 14:5048-5059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pickart, C. M. 2001. Ubiquitin enters the new millennium. Mol. Cell 8:499-504. [DOI] [PubMed] [Google Scholar]
- 22.Piechaczyk, M., and J. M. Blanchard. 1994. c-fos proto-oncogene regulation and function. Crit. Rev. Oncol. Hematol. 17:93-131. [DOI] [PubMed] [Google Scholar]
- 23.Polevoda, B., and F. Sherman. 2003. N-terminal acetyltransferases and sequence requirements for N-terminal acetylation of eukaryotic proteins. J. Mol. Biol. 325:595-622. [DOI] [PubMed] [Google Scholar]
- 24.Reinstein, E., M. Scheffner, M. Oren, A. Ciechanover, and A. Schwartz. 2000. Degradation of the E7 human papillomavirus oncoprotein by the ubiquitin-proteasome system: targeting via ubiquitination of the N-terminal residue. Oncogene 19:5944-5950. [DOI] [PubMed] [Google Scholar]
- 25.Roux, P., J. M. Blanchard, A. Fernandez, N. Lamb, P. Jeanteur, and M. Piechaczyk. 1990. Nuclear localization of c-Fos, but not v-Fos proteins, is controlled by extracellular signals. Cell 63:341-351. [DOI] [PubMed] [Google Scholar]
- 26.Salvat, C., I. Jariel-Encontre, C. Acquaviva, S. Omura, and M. Piechaczyk. 1998. Differential directing of c-Fos and c-Jun proteins to the proteasome in serum-stimulated mouse embryo fibroblasts. Oncogene 17:327-337. [DOI] [PubMed] [Google Scholar]
- 27.Schubert, U., L. Anton, J. Gibbs, C. Norbury, J. Yewdell, and J. Bennink. 2000. Rapid degradation of a large fraction of newly synthesized proteins by proteasomes. Nature 404:770-774. [DOI] [PubMed] [Google Scholar]
- 28.Shaulian, E., and M. Karin. 2001. AP-1 in cell proliferation and survival. Oncogene 20:2390-2400. [DOI] [PubMed] [Google Scholar]
- 29.Sheaff, R. J., J. D. Singer, J. Swanger, M. Smitherman, J. M. Roberts, and B. E. Clurman. 2000. Proteasomal turnover of p21Cip1 does not require p21Cip1 ubiquitination. Mol. Cell 5:403-410. [DOI] [PubMed] [Google Scholar]
- 30.Shringarpure, R., T. Grune, J. Mehlhase, and K. J. Davies. 2003. Ubiquitin-conjugation is not required for the degradation of oxidized proteins by the proteasome. J. Biol. Chem. 278:311-318. [DOI] [PubMed] [Google Scholar]
- 31.Stancovski, I., H. Gonen, A. Orian, A. L. Schwartz, and A. Ciechanover. 1995. Degradation of the proto-oncogene product c-Fos by the ubiquitin proteolytic system in vivo and in vitro: identification and characterization of the conjugating enzymes. Mol. Cell. Biol. 15:7106-7116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tarcsa, E., G. Szymanska, S. H. Lecker, C. M. O'Connor, and A. L. Goldberg. 2000. Ca2+-free calmodulin and calmodulin damaged by in vitro aging are selectively degraded by 26S proteasomes without ubiquitination. J. Biol. Chem. 275:20295-20301. [DOI] [PubMed] [Google Scholar]
- 33.Touitou, R., J. Richardson, S. Bose, M. Nakanishi, J. Rivett, and M. J. Allday. 2001. A degradation signal located in the C-terminus of p21WAF1/CIP1 is a binding site for the C8 alpha-subunit of the 20S proteasome. EMBO J. 20:2367-2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Treier, M., L. M. Staszewski, and D. Bohmann. 1994. Ubiquitin-dependent c-Jun degradation in vivo is mediated by the delta domain. Cell 78:787-798. [DOI] [PubMed] [Google Scholar]
- 35.Tsurumi, C., N. Ishida, T. Tamura, A. Kakizuka, E. Nishida, E. Okumura, T. Kishimoto, M. Inagaki, K. Okazaki, N. Sagata, et al. 1995. Degradation of c-Fos by the 26S proteasome is accelerated by c-Jun and multiple protein kinases. Mol. Cell. Biol 15:5682-5687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Verma, R., and R. J. Deshaies. 2000. A proteasome howdunit: the case of the missing signal. Cell 101:341-344. [DOI] [PubMed] [Google Scholar]
- 37.Zwickl, P., W. Baumeister, and A. Steven. 2000. Dis-assembly lines: the proteasome and related ATPase-assisted proteases. Curr. Opin. Struct. Biol. 10:242-250. [DOI] [PubMed] [Google Scholar]