Abstract
Gene-specific transcriptional activation is a multistep process that requires numerous protein factors and DNA elements, including enhancers and the core promoter. To investigate the roles of core promoter elements in transcriptional activation in vertebrates, we examined expression and factor occupancy on representative promoters in chicken DT40 cells containing a conditional TATA binding protein (TBP)-associated factor 9 allele (TAF9). Characterized core elements, including TATA box-flanking regions and the downstream promoter element, were found to play significant roles in determining promoter strength, response to activators, and factor occupancy and recruitment. The requirement for TAF9 was found to be highly promoter specific, and TAF9 dependence and promoter occupancy were not always correlated. We also describe contrasting examples of factor recruitment and activation mechanisms at different promoters, highlighted by the nearly opposite mechanisms utilized by the simian virus 40 enhancer and p53. With the core promoters analyzed, the former functions by facilitating RNA polymerase II (RNAP II) recruitment to a preassembled TBP/TFIIB-containing scaffold and p53 strongly recruits TBP and TFIIB while RNAP II levels remain modest. Taken together, our results illustrate both the important roles of core promoter elements and the remarkable diversity that characterizes transcriptional activation mechanisms in vertebrates.
Initiation of transcription by RNA polymerase II (RNAP II) requires cis-acting DNA elements including core promoters and enhancers. Core promoters, usually situated between −40 to +40 bp from the transcription start site (+1), define the docking sites for the general transcription machinery and are sufficient for directing authentic transcription initiation (11, 57, 59). On the other hand, enhancers are bound by cognate transcriptional activators to mediate activated mRNA synthesis (7, 47). Four major core promoter elements have been identified: the TATA box, initiator (Inr), downstream promoter element (DPE), and TFIIB recognition element (BRE) (11, 59). These elements not only influence core promoter strength through direct contacts with the transcriptional machinery but can also play significant roles in determining activation potential, i.e., response to enhancers (8, 18, 19, 20, 69). GC-rich sequences flanking the adenovirus major late promoter (MLP) TATA box, including the upstream BRE and a downstream G track, have been shown to modulate core promoter strength and activation potential (20, 35, 69, 72). Furthermore, enhancers specific for the TATA-Inr or Inr-DPE core promoter combination have been identified in Drosophila melanogaster (10). Taken together, these and other studies have provided important clues into the dynamic functions of the core promoter elements, including the DPE and TATA-flanking sequences, although precisely how they are recognized and contribute to promoter strength are not well understood.
Transcription initiation by RNAP II requires many proteins, including a number of general transcriptional factors (GTFs) (55). TATA-binding protein (TBP) and TFIIB are the only GTFs shown to have sequence-specific promoter-binding activity. Recognition of the TATA box by TBP, most frequently as a component of TFIID, constitutes the first step toward preinitiation complex (PIC) formation (27, 55). A close correlation between promoter activity and TBP occupancy has been observed in Saccharomyces cerevisiae cells, and many activators seem to function directly or indirectly by enhancing TBP binding to promoters (33, 40). Given this central role of TBP in initiating transcription, it is not surprising that TBP-TATA interactions are subject to many levels of regulation by numerous factors (26, 37, 73). Following TBP recruitment, TFIIB enters the complex to form a more stable ternary complex, which in turn recruits the RNAP II-TFIIF complex. While TFIIB binding was shown to be rate-limiting on an adenovirus E4 TATA promoter (42), some other promoters, most notably the MLP, contain a BRE upstream of the TATA box that confers high-affinity binding by TFIIB (35). Paradoxically, this avid BRE-TFIIB interaction seems to be repressive to promoter activity in transfected cells (72) and in cell extracts, where it appears to represent a possible target of activators (20). TFIIB not only interacts with sequences upstream of TATA but also makes base-specific contacts immediately downstream of TATA (67, 70). How these interactions affect TFIIB recruitment and promoter activity is not understood.
Apart from TBP and TFIIB, specific core promoter elements can also be recognized by certain TBP-associated factors (TAFs) present in TFIID (9, 68). Several TAFs can make sequence-specific contacts apparently independently of TBP. Two notable examples are the TAF2/TAF1-Inr and TAF6/TAF9-DPE interactions detected in vitro (8, 13; see reference 65 for nomenclature). In agreement with these promoter interactions, TAFs were shown to be required for in vitro transcription from promoters containing Inr and/or DPEs (30, 56). In both cases, the absence of a TATA box in the promoters tested seems to necessitate direct interactions between TAFs and promoter sequences other than TATA. On TATA-containing promoters, TATA-flanking sequences, as well as the Inr, can play important roles in TAF-dependent transcription. For example, the dependence of the yeast RPS5 promoter on a specific TAF, TAF1, was mapped to sequences flanking the noncanonical TATA box (58). Additionally, TFIID subunits were shown to interact extensively in vitro with the GC-rich core MLP from upstream of the TATA box to downstream of the Inr (16, 51), whereas the non-GC-rich E4 and human Hsp70 TATA promoters displayed a limited TFIID footprint centered on the TATA box (16, 50). Consistent with these differential TFIID-promoter binding patterns, the MLP was shown to utilize TFIID to form PICs much more efficiently than the E4 and Hsp70 promoters in a single-round in vitro transcription assay (52). Alternatively, TAFs can be recruited to core promoters lacking any strict sequence consensus through direct interactions with specific activators. In yeast, promoter occupancy by TAFs at such promoters was, surprisingly, relatively unaffected by the absence of TBP and other GTFs but had a stringent requirement for activators (32, 39, 46). These observations are consistent with the previously proposed “coactivator” functions of TAFs, whereby TAFs function to bridge activators to the general transcription machinery via protein-protein interactions (2, 9, 25). These studies and others have suggested a variety of roles for TAFs in vitro and in yeast cells, but how they function in promoter recognition and activation in living vertebrate cells is largely unknown.
Previously we constructed a conditional TAF9 knockout cell line from chicken DT40 cells (DT40-TAF9) (15). TAF9 is a histone fold-containing TAF present not only in TFIID but also in other complexes analogous to the SAGA complex in yeast. We provided evidence that TAF9, although essential for viability, was not generally required for RNAP II-mediated transcription in vivo. While these findings are on the one hand consistent with generally selective roles of TAFs in yeast (25), on the other they contrast with the apparently more general requirement for yeast TAF9 (3, 28, 48, 49), highlighting the fact that the nature of gene-specific requirements of TAFs remains poorly defined in vivo, especially in vertebrates. To address this issue and the function of core promoter elements more generally, we employed DT40-TAF9 cells to analyze promoters with different core element combinations and to investigate the roles of TAFs on these promoters under physiological conditions. Combining stable-transfection assays and chromatin immunoprecipitation (ChIP) analysis, we show that core promoter elements, including TATA-flanking regions and the DPE, play active but distinct roles in determining promoter activity and occupancy. A potent enhancer element, the simian virus 40 (SV40) 72-bp repeats, as expected greatly increased transcription, but the levels achieved were significantly influenced by core promoter structure. Strikingly, TBP and TFIIB promoter occupancy levels were not increased by the SV40 enhancer but RNAP II recruitment was, indicating that the enhancer activates transcription by influencing a step(s) subsequent to TBP/TFIIB binding. Consistent with our previous studies, many promoters were only minimally affected by TAF9 depletion, and, as expected from studies of yeast, TAF9 dependence and recruitment were frequently correlated. However, this was not always observed, as two TAF9-dependent promoters actually displayed very low TAF9 occupancy. Finally, an activator that interacts with and requires TAF9 in vitro, p53, is shown to be TAF9 independent in vivo and to employ an activation mechanism distinct from that employed by the SV40 enhancer. Taken together, these results illustrate the diverse roles of core promoter/enhancer elements and TAFs in transcriptional control.
MATERIALS AND METHODS
Plasmid constructs.
The luciferase reporter construct pGL3-promoter (Promega) was digested with XhoI/HindIII to remove the SV40 promoter, and the blasticidin selection gene (Brs; see the DT40 website: http://swallow.gsf.de/dt40.html) was inserted into the BamHI site downstream of the luciferase sequence to make the GL3-Brs vector. Adenovirus MLP (−50 to +31 relative to the +1 start site) was inserted into pKS(+) (Stratagene) to serve as template for generation of triple point mutants mBRE and mGT by inverse PCR. These core promoters, as well as the c-fos core promoter (−55 to +40), were blunt end ligated into the SmaI site of GL3-Brs to make core MLP and fos reporter vectors. To construct SV40 72-bp enhancer repeat (SVE)-containing plasmids, the SVEs were amplified by PCR from pcDNA3.1 (Invitrogen) and inserted in the KpnI site upstream of the core promoter-luciferase expression cassette. The human IRF-1 full promoter (−1312 to +39), as well as the DPE-scrambled mutant promoter (8), was ligated into SmaI-cut GL3-Brs as described above. These plasmid DNAs were purified by CsCl centrifugation and linearized by SalI prior to transfection. SalI generates 3- and 5-kb arms flanking the target promoters to minimize the genomic position effect.
Gal4 (amino acids [aa] 1 to 147)-p53 (aa 1 to 82)-encoding sequences were excised from pAB-Galp53 (kindly provided by S.Y. Sheih and C. Prives; derived from pABGal as described in reference 4) and inserted into expression vector pAPSV-zeo containing the chicken β-actin promoter, the SV40 late poly(A) signal, and the zeocin resistance gene (zeo; see the DT40 website). The DNA fragment encoding the L22Q W23S double-mutant p53 activation domain was amplified by PCR from pCP53-22/23 (41) and used to replace the wild-type (WT) counterpart in pAB-Galp53 and similarly cloned into pAPSV-zeo. KpnI was used for linearization before transfection. The reporter vector RE-luc was constructed as follows. The responsive element (RE) containing five tandem copies of the Gal4 binding site and a short E1b TATA box was cut out of G5E1b TATA-CAT (18) and inserted into pKS(+). Subsequently, the luciferase reporter gene and the Brs were placed downstream and upstream of the promoter, respectively. An EcoRI site between the Brs cassette and the RE was used for linearization.
To construct the expression vector encoding TAF9 fused to three copies of Flag tags at the C terminus, human TAF9 cDNA (31) was amplified by PCR and inserted in frame into the EcoRI/EcoRV-digested 3XFlag-CMV-Neo vector (Sigma).
Cell culture and transfections.
Chicken DT40-TAF9 cells were maintained and transfected by electroporation as previously described (15). Human HeLa cells were grown in Dulbecco's modified Eagle medium (Invitrogen) supplemented with 10% fetal bovine serum (HyClone) at 37°C and 5% CO2. Transient transfections of HeLa cells were carried out with CaPO4. For stable transfections of HeLa cells, 10 μg of linearized plasmid DNA was incubated with cells from one 100-mm-diameter dish at 70% confluence in 0.7 ml of 1× HEPES buffered saline and the mixture was electroporated at 250 V and 950 μF. Two days later, the cells were plated out to eight 100-mm dishes in selective media. Blasticidin (Calbiochem), zeocin (Invitrogen), and neomycin (G418; Invitrogen) were used at 10, 200, and 400 μg/ml, respectively.
RNA and protein analyses.
RNase protection and Western blot analyses were carried out as previously described (15). RNA probes for start site mapping experiments were transcribed from PCR fragments containing the T7 promoter. Primer extension experiments for start site mapping were carried out essentially as described previously (12) with the following modifications: labeled primers were purified from 20% denaturing polyacrylamide gel electrophoresis gels and, after annealing at 65°C, reaction mixtures were left at room temperature (RT) to cool slowly to below 35°C.
Luciferase reporter analysis.
Clonal analysis of DT40-TAF9 stable transfections was carried out as follows. Typically 15 to 40 individual clones were selected after each electroporation and were grown in parallel with or without tetracycline (TET) for 72 h. Protein extracts were subjected to luciferase analysis as recommended by Promega using a luminometer (Berthold). Median expression levels of normal growing cells (TET−) were calculated for individual transfections to indicate promoter strength. To measure TAF9 dependence, reductions of luciferase expression for individual clones were calculated as ratios of expression in TET− and TET+ samples and a median factor of reduction was derived for each transfection. For each promoter, two to five, typically three or more, independent transfections were carried out. The above parameters were averaged among these transfections, and standard deviations were calculated. Statistical significance was evaluated in all experiments, and in all cases P values were less that 0.05, typically less than 0.01.
ChIP analysis.
ChIP assays were carried out as described by Orlando and Paro (54) with several modifications. Briefly, for each sample, 8 × 107 DT40-TAF9 cells were grown in the presence or absence of TET for 72 h. For HeLa cells, six 100-mm dishes of cells were cultured. Cells were fixed with 1% formaldehyde for 45 min at 37°C and then quenched with glycine. After being washed three times with phosphate-buffered saline, cells were lysed in 10 ml of lysis buffer (0.25% Triton X-100, 0.5% NP-40, 10 mM EDTA, 0.5 mM EGTA, 10 mM Tris [pH 8.0], 1 mM phenylmethylsulfonyl fluoride [PMSF]) and set on ice for 10 min. Pellets were resuspended in 10 ml of presonication buffer (0.2 M NaCl, 1 mM EDTA, 0.5 mM EGTA, 10 mM Tris [pH 8.0], 1 mM PMSF) for 10 min at RT. Nuclei were then resuspended in 3.5 ml of sonication buffer (1 mM EDTA, 0.5 mM EGTA, 10 mM Tris [pH 8.0], 1 mM PMSF) and sonicated 25 times for 20 s each on ice with a Branson 250 sonifier. The median size of DNAs at this point was usually 0.8 kb. After 10 min of incubation at RT in 0.5% Sarkosyl, supernatants were adjusted to a final density of 1.42 g/ml with CsCl and centrifuged in an SW55 rotor (Beckman) at 40,000 rpm for 48 h. Fractions enriched in chromatin, usually just beneath the lipid layer, were collected and dialyzed. Equal amounts of chromatin from TET− and TET+ samples, determined by measuring optical density at 260 nm in 2 M NaCl, were adjusted to 1× radioimmunoprecipitation assay (RIPA) buffer (20 mM Tris [pH 8.0], 1 mM EDTA, 1% Triton X-100, 0.1% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS], 140 mM NaCl, 1 mM PMSF). After a preclearing step, equal aliquots of chromatin were rocked with antibodies or no antibody (mock) overnight at 4°C. In the meantime, protein A/G-Sepharose beads (20 μl/reaction) were blocked in RIPA buffer with 100 μg of bovine serum albumin/ml and single-stranded DNA overnight. The blocked beads were briefly washed and incubated with the chromatin-antibody mixtures for 3 to 4 h at 4°C. The beads were washed with RIPA buffer twice, with RIPA buffer-275 mM NaCl twice, with RIPA buffer-500 mM NaCl four times, and with Tris-EDTA (TE), pH 8.0, twice. The washed beads and 2% inputs were resuspended in 100 μl of TE (pH 8.0)-0.5% SDS-200 μg of proteinase K/ml, and reversal of cross-links was carried out at 55°C for 3 h followed by 65°C overnight. After extraction and precipitation, DNAs were resuspended in 10 mM Tris (pH 8.0), with 40 μl for inputs and 20 μl for immunoprecipitation (IP) samples.
PCRs were carried out with the Qiagen Taq PCR system containing 10 μM promoter-specific primers and 0.05 μCi of [α-32P]dCTP/μl in 10 μl. Equal aliquots of PCR products were run on 6% denaturing polyacrylamide gel electrophoresis gels. To ensure linear amplification, a series of dilutions of DNA templates were routinely used. PCR products were typically 150 to 300 bp. Occupancy units represent percentages (whole numbers) of the IP samples (normalized against mock IP) compared with 1% input. All quantitations were done with a PhosphorImager (Molecular Dynamics). Copy numbers of transfected plasmids among independent clones were analyzed by genomic Southern blot analysis, and promoter occupancy was normalized for copy number.
Antibodies used in this study include anti-TBP sc-273 (Santa Cruz Biotechnology; 3 μg/reaction); anti-TFIIB sc-274 (Santa Cruz Biotechnology; 3 μg/reaction); anti-Flu 12CA5 monoclonal antibody (MAb; Roche; 2 μg/reaction); anti-Flag M2 MAb (Sigma; 4 μg/reaction); anti-RNAP II 8WG16 (Covance; 5 μl of ascites fluid); and polyclonal antibodies against human TAF6 and TAF7 (kind gifts from R. Roeder and C. M. Chiang, respectively) and chicken TAF9 (15).
Several observations strongly suggest that promoter occupancy levels reflect amounts of protein factors on the promoter and were not limited by cross-linking efficiency. Relative occupancy levels of TAFs TAF9, TAF6, and TAF7 were tightly correlated on several promoters, including SV-MLP, IRF-1, and histone 2B (H2B) promoters (see Fig. 3; data not shown), indicating that promoter DNA sequence variations similarly affected these TAFs. However, these TAFs have been shown to contact promoter DNA differently (8, 51), suggesting that TAF-TAF (and/or TAF-TBP), rather than TAF-promoter, cross-links were primarily responsible for TAF occupancy levels (see similar arguments in reference 32). In addition, in contrast to other studies of vertebrates, where cross-linking was usually carried out at RT for 10 to 15 min (5, 17), our conditions (37°C for 45 min) likely induced extensive protein-DNA and protein-protein cross-links (53), which were then enriched in the chromatin fraction after CsCl centrifugation.
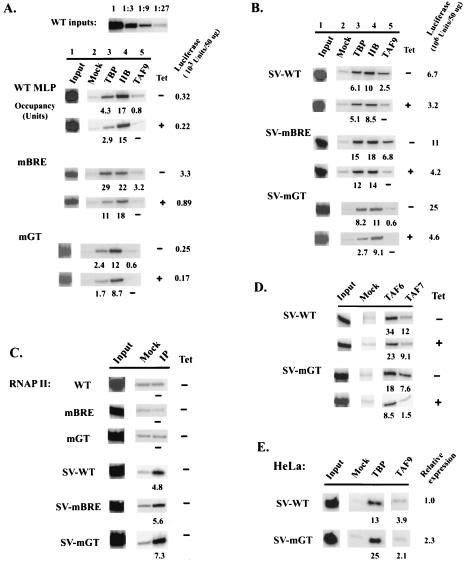
FIG. 3.
ChIP analysis of factor occupancy on MLP and SV-MLP promoters. Cells were grown in the absence (−) or presence (+) of TET for 72 h. Following formaldehyde cross-linking, soluble chromatin was prepared. After IP with antibodies against the indicated proteins, precipitated DNAs were used in PCR analysis. Occupancy units, as percentages of IP samples (normalized against mock IP) relative to 1% input, are indicated. Reporter expression levels are also shown. (A and B) ChIP analysis of TBP, TFIIB, and TAF9 occupancy on core (A) and SVE-containing (B) MLPs. PCRs using a series of dilutions of WT input samples are shown on top. (C) ChIP analysis of RNAP II occupancy in normally growing cells. (D) ChIP analysis of TAF6 and TAF7 occupancy on SV-WT and SV-mGT2 promoters. (E) ChIP analysis using a HeLa cell line that stably expressed Flag-TAF9. Cells were transiently transfected with core MLP and SV-MLP constructs (5 μg of DNA/100-mm plate). After 48 h of incubation, cells were collected for ChIP analysis as described above, except that an anti-Flag M2 MAb (Sigma) was used to immunoprecipitate tagged TAF9. Relative expression levels are shown.
RESULTS
Experimental design.
The aim of this study was to analyze how core promoter elements contribute to promoter activity in metazoans and how this is affected by the presence or absence of TAFs. To examine the roles of core promoter elements in determining promoter strength and response to activators, we first wished to analyze core promoters with combinations of defined, characterized promoter elements, in the presence or absence of activators and TAFs. To this end, we constructed a series of luciferase reporter constructs containing combinations of promoter elements, with or without an upstream enhancer element (Fig. 1). The first group consisted of the MLP and two TATA-flanking-region mutant promoters (Fig. 1A). The GC-rich TATA-flanking regions of the MLP are capable of interacting with both TFIIB and TAFs and can have a significant impact on promoter properties (see above). The SVEs were used to construct corresponding chimeric full promoters to examine the role of core promoter elements in modulating response to activators. A TATA-containing core promoter lacking GC-rich elements, c-fos, was also analyzed (Fig. 1B). To examine the role of the DPE in vertebrates, the DPE-containing human IRF-1 promoter was analyzed along with a DPE-scrambled mutant promoter (8) (Fig. 1C). These plasmids were all confirmed to initiate transcription accurately in transiently transfected human 293 and HeLa cells by primer extension and RNase protection assays; for SVE-containing promoters, which were expressed strongly in DT40 cells, representative DT40 stable transfectants were also analyzed, which confirmed accurate initiation (data not shown). Luciferase has been shown to have a short half-life (∼3 to 4 h) in mammalian cells (36), and start site mapping also revealed close correlations between luciferase activities and transcript levels, indicating that luciferase expression is a valid representation of transcription. For analysis of promoter activity, stable transfectants in DT40-TAF9 cells were isolated. Both TAF9 alleles are disrupted in these cells, and TAF9 is expressed from a transgene driven by a TET-regulated promoter, allowing in vivo depletion of the protein (15). Reporter expression profiles in normally growing and TAF9-depleted DT40-TAF9 cells were analyzed for core promoter strength, activation potential, and TAF9 dependence in multiple isolated clones (see Materials and Methods). Representative clones for each promoter were then analyzed for factor occupancy by ChIP (54).
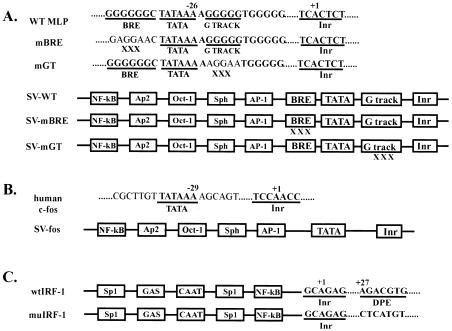
FIG. 1.
Diagram of core and full promoters used in reporter vectors. (A) GC-rich adenovirus MLP derivatives. Sequences of the WT core promoter and two triple point mutants (mBRE and mGT) are shown, with intact promoter elements underlined. The numbers correspond to positions relative to the +1 start site. Boxes, representative enhancer and core promoter elements of SV-MLPs containing the SVE. (B) c-fos core and SVE-containing promoters. Core promoter and enhancer elements are denoted as for panel A. (C) Human DPE-containing IRF-1 promoter and DPE-scrambled mutant promoter. Sequences of the Inr and DPE are shown. Boxes, representative enhancer elements on the IRF-1 promoters.
The BRE and downstream G track play antagonistic roles in MLP activity and also modulate TAF9 dependence.
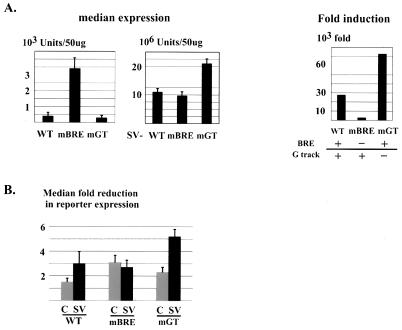
The transcriptional functions of the TATA box and the initiator element have been extensively studied (11, 57), whereas the roles of TATA flanking sequences are not as well characterized, especially in vivo. The MLP is a prototypical GC-rich TATA promoter with the TATA box and Inr as the only AT-rich sequences embedded in a generally GC-rich environment, conforming well to a “majority” consensus sequence defined by Wolner and Gralla (69), with preferred nucleotides at each position. To investigate the function in transcription of the MLP GC-rich TATA-flanking sequences, we transfected DT40-TAF9 cells with plasmids containing wild-type or mutant MLPs in the presence or absence of the SVE (MLPs and SV-MLPs; Fig. 1A), and multiple clones were isolated and analyzed for reporter expression (see Materials and Methods). Consistent with its lack of enhancer elements, the intact (WT) MLP was barely active when stably integrated into the DT40 genome: the median expression (∼400 luciferase units) was only fivefold higher than background (Fig. 2A, left). The BRE mutation (mBRE) significantly increased median expression levels, to ∼40-fold above background. In contrast, the downstream G-track mutation (mGT) showed a modest repressive effect, reducing expression levels to less than threefold above background.
FIG. 2.
Expression profile and TAF9 dependence of MLP and SV-MLP derivatives. Stable transfections of reporter constructs into DT40-TAF9 cells were carried out, and transfectants were analyzed individually after growing in media in the presence or absence of TET for 72 h (see Materials and Methods). (A) Promoter strength and activation potential of MLPs. Median expression levels under normal growing conditions, measured as luciferase units per 50 μg of extracts, were calculated for individual transfections and averaged over several independent experiments for each promoter. Averages are shown, with error bars indicating standard deviations. To the right, levels of induction (ratios of median expression between full and core promoters) are shown. (B) TAF9 dependence of promoter activity. Median reductions of reporter expression were calculated for individual transfections and averaged over independent experiments. C and SV, core and SVE-containing MLPs, respectively. Error bars represent standard deviations.
We next examined the above promoters in the presence of the SVE. Median expression levels of SVE-containing promoters were determined as described above (Fig. 2A, middle), and induction due to the SVE, calculated as the ratio of activated to core promoter expression, was also determined (Fig. 2A, right). Although strong activation was observed in all cases and the absolute levels differed by no more than twofold, significant differences were also detected. mBRE, with the highest core promoter activity, was the least responsive to the enhancer (2,800-fold induction compared with 27,000-fold for WT), while mGT, despite its very weak core promoter activity, displayed both the highest absolute levels and also the greatest activation potential (73,000-fold). These expression profiles were not specific to DT40 cells, as parallel stable transfections in HeLa cells produced similar relative patterns of core and activated transcription (data not shown). Taken together, these results indicate that the BRE and G track make distinct contributions to modulating core promoter strength and activation potential.
The above experiments were carried out with DT40-TAF9 cells growing under normal conditions. To gain insight into possible promoter-specific functions of TAF9, we examined how core promoter sequences affect TAF9 dependence. Individual DT40-TAF9 transfectants were analyzed for reporter expression after growing with or without TET for 72 h. At this time, TAF9 was undetectable, but total RNAP II transcription, based on pulse-labeling of poly(A)+ RNA, was almost unaffected (15). The median reductions of reporter expression following TAF9 depletion were calculated for both core and SVE-containing MLPs (Fig. 2B). As shown in Fig. 2B, the WT core MLP displayed modest TAF9 dependence, 1.5-fold overall reduction in reporter expression (Fig. 2B, lane C in WT lanes). Both mBRE and mGT displayed higher TAF9 dependence. The presence of the SVE in general conferred higher TAF9 sensitivity, except for mBRE, which already possessed rather significant TAF9 dependence (compare C and SV lanes for mBRE). The most active promoter (SV-mGT; Fig. 2A) was also the most TAF9 dependent (more than a factor of five reduction; Fig. 2B). However, it is notable that all the SVE promoters, including SV-mGT, retained significant activity in the absence of TAF9.
Promoter activation by the SVE significantly increases promoter occupancy by RNAP II, but not TBP and TFIIB.
We next sought to gain insight into the molecular mechanism(s) underlying the transcriptional effects described above. TBP, TFIIB, and TAFs have been shown to interact in vitro with the MLP in footprinting and photo-cross-linking analysis (16, 51, 74), and alterations of TATA-flanking regions have been shown to affect promoter binding by TBP and TFIIB in gel shift assays (69). We therefore performed ChIP analyses to examine TBP and TFIIB promoter occupancy. For each promoter, cells with typical (usually within 1.5-fold of median) expression and TAF9 dependence were grown and lysed; solubilized chromatin was immunoprecipitated, initially with antibodies against TBP or TFIIB, and recovered DNAs were amplified by PCR using promoter-specific primers. To ensure reproducibility and allow comparisons between promoters, two to four clones with typical (usually within 1.5-fold of median) expression and TAF9 dependence were analyzed for each promoter. After normalization for copy number (which displayed only modest variation [one- to threefold]), similar occupancy patterns were detected in all cases, and representative results are shown. A series of dilutions of input samples (1% and 1:3 dilutions; one example is shown in Fig. 3A) were also analyzed to ensure linear PCR amplification in each case. Occupancy units were calculated as percentages of IP samples (normalized against mock IP) relative to 1% input (see Materials and Methods). As a negative control, factor occupancy, reflecting the antibody background, on several nonpromoter regions was examined (e.g., the TAF1 3′ coding region) and was shown to be less than one-half of that on the weakest core promoter, mGT (data not shown). In addition, the promoter for the drug selection gene (drug promoter) also had minimal influence on factor occupancy on the test promoters. The drug promoter was separated from the test promoter by ∼2.5 kb, a distance much larger than the median size of chromatin fragments after sonication, ∼0.8 kb. The drug promoter also displayed only modest factor occupancy (data not shown), which was essentially constant among all clones examined. Although we have optimized the procedure for chromatin preparation to minimize effects due to differences in cross-linking efficiencies (see Materials and Methods), as with all ChIP experiments, it is conceivable that such differences exist and contributed to our results.
Significant factor occupancy was detected on the MLP core promoters, despite their very low transcription activities (Fig. 3A). Consistent with previous findings for yeast (33, 40), there was a direct correlation between TBP occupancy and core promoter activity, with mBRE promoter occupancy nearly 10-fold higher than WT and mGT (TET−; Fig. 3A). TFIIB occupancy levels also correlated with activity, but not as strongly (Fig. 3A, lane 4). mBRE displayed the highest occupancy, which is consistent with its activity but, interestingly, not with the idea that the BRE is a necessary element for TFIIB recruitment. TFIIB levels on the WT and mGT core promoters were reduced but higher than expected relative to TBP levels. This supports the idea that the BRE can enhance the TFIIB-promoter interaction but indicates that this interaction is not important, and may in fact be detrimental, for promoter activation. TAF9 levels (Fig. 3A, lane 5) also correlated with activity (see also below).
Next we examined TBP and TFIIB recruitment to promoters activated by the SVE. Strikingly, and in sharp contrast to the close correlations between factor occupancy and transcriptional activation found in yeast, the SVE did not significantly increase overall factor occupancy (compare Fig. 3A and B). The SVE increased TBP occupancy on the weakest core promoter, mGT, by approximately fourfold and effected less than a twofold increase in occupancy on the WT promoter. The most active core promoter, mBRE, actually displayed at least twofold-higher TBP occupancy than any of the SVE-containing promoters. TFIIB levels were also equivalent to those detected on the corresponding core promoters. These results both provide a mechanistic explanation for MLP core activity, known to be especially strong in in vitro assays, and indicate that the SVE functions to activate transcription at a step(s) subsequent to core promoter recognition.
To extend these results, we examined RNAP II occupancy on core and SVE-containing promoters. Importantly, significant RNAP II occupancy was detected on the SVE-containing promoters, but all three core promoters displayed RNAP II levels indistinguishable from background (Fig. 3C). Additionally, relative RNAP II levels correlated with promoter activity for all three SVE promoters. These results indicate that the SVE exerts its activating effect at least in part by directly or indirectly facilitating RNAP II recruitment to a preassembled core scaffold.
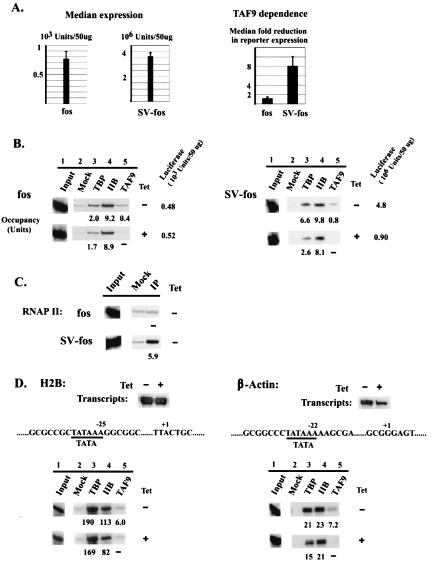
We next wished to investigate whether another type of core promoter recruits significant levels of TBP/TFIIB and how the SVE influences factor occupancy. To this end, we examined a non-GC-rich core promoter, derived from the c-fos proto-oncogene, which is devoid of a BRE and G track. As shown in Fig. 4A, the c-fos core promoter displayed weak basal promoter activity (∼9-fold above background) and the SVE induced activation ∼4,700-fold. TBP and TFIIB (and TAF9) occupancy on the core promoter was comparable to that observed with the relatively weak WT and mGT MLPs (Fig. 4B). Importantly, the SVE increased TBP occupancy only modestly and did not significantly enhance TFIIB levels. However, a greater increase in RNAP II occupancy was again detected (Fig. 4C). These results indicate that both the MLP and c-fos core promoters are capable of recruiting significant levels of TBP and TFIIB and that the strong activation induced by the SVE occurs primarily at a subsequent step(s).
FIG. 4.
Expression and factor occupancy on distinct TATA-containing promoters. (A to C) Analysis of c-fos core and SVE-containing promoters was carried out as described for Fig. 2 and 3. (A) Expression profile and TAF9 dependence. (B) ChIP analysis of TBP, TFIIB, and TAF9 occupancy. (C) ChIP analysis of RNAP II occupancy. (D) Analysis of two highly expressed endogenous promoters, the H2B and β-actin promoters. Transcript levels for cells grown in the absence or presence of TET for 72 h were analyzed by RNase protection and are shown at the top. Partial core promoter sequences and ChIP analysis are shown below.
TAF9 dependence and promoter occupancy can be uncoupled.
We next examined TAF occupancy on these promoters and specifically how TAF occupancy correlated with TAF9 dependence. In yeast, most TAF-dependent promoters were shown to recruit higher levels of TAFs than TAF-independent promoters (32, 39), but this has not been examined in metazoan organisms. To address this, ChIP assays were performed to measure TAF9 promoter occupancy in DT40-TAF9 cells by using first an anti-Flu epitope antibody. Detectable levels of TAF9 were found on all three MLP core promoters, with the more active mBRE derivative showing the highest levels, analogous to TBP occupancy (Fig. 3A). Growing cells in the presence of TET effectively removed TAF9 from the promoters (lane 5, TET+ samples), consistent with the efficient TAF9 depletion that occurs in these cells following TET addition (15).
We then examined TAF occupancy patterns on the SVE-activated promoters. SV-WT and SV-mBRE, promoters with medium TAF9 dependence (two- to threefold reduction in reporter expression; Fig. 2B) showed moderate-to-high TAF9 content, roughly correlating with activity and TBP occupancy (Fig. 3B, lane 5). Surprisingly, however, SV-mGT, which was both the strongest and the most TAF9-dependent promoter (Fig. 2B), showed minimal TAF9 occupancy (Fig. 3B), suggesting an “uncoupling” of TAF dependence and TAF promoter occupancy. In all cases, TET treatment reduced TAF9 to background levels. To rule out the possibility that the differential TAF9 IP efficiency was due to a conformational constraint on the accessibility of the Flu tag epitope, ChIP assays were performed with an antibody raised against chicken TAF9 (15), which gave identical results (results not shown).
We also examined interactions of additional TAFs with SVE-MLP derivatives in the presence or absence of TAF9. Previously we showed that depletion of TAF9 also resulted in significant but less-complete reductions in the levels of TFIID-specific TAF6 and TAF7, likely reflecting turnover following disruption of TFIID (15, 15a). Consistent with the observed relative TAF9 occupancy pattern (Fig. 3B), lower levels of TAF6 and TAF7 were detected on the stronger SV-mGT promoter than on SV-WT (Fig. 3D). Furthermore, TAF6 and TAF7 levels on SV-mGT were also significantly reduced by TET treatment, while depletion of these TAFs from SV-WT was much more modest, similar to that of TBP and TFIIB (Fig. 3B). These results extend those observed with TAF9, indicating that SV-mGT, despite its strong activity and TAF9 dependence, interacts weakly with several TAFs, suggesting that less-stable promoter binding by TAFs can actually be conducive to high transcriptional activity. Consistent with this, when SV-MLP derivatives were transiently transfected into HeLa cells that stably expressed Flag-TAF9, SV-mGT also showed the highest expression but lowest Flag-TAF9 occupancy (Fig. 3E and data not shown).
We next examined TAF9 dependence and promoter occupancy on several additional promoters. To investigate core promoter specificity, we again examined the c-fos core promoter derivatives. The c-fos core promoter is TAF9 independent (Fig. 4A, fos lanes), and TAF9 depletion had little, if any, effect on TBP or TFIIB occupancy (Fig. 4B). However, activation by the SVE again revealed an inverse correlation between TAF dependence and occupancy: while strongly TAF9 dependent (eightfold median reduction; Fig. 4A), SV-fos displayed very low TAF9 occupancy (Fig. 4B). It is also noteworthy that, in contrast to SV-fos, the endogenous c-fos promoter in DT40 cells is TAF9 independent (15), indicating that, as in yeast (39a), TAF dependence can be influenced by activator identity. We also tested the relationship between TAF9 dependence and occupancy with two strong endogenous promoters, the H2B and β-actin promoters. These two promoters possess GC-rich TATA flanking sequences (Fig. 4D, middle) and, despite distinct levels of factor occupancy, produced similarly large amounts of transcripts (estimated by RNase protection to be about fivefold higher than that from SV-mGT; data not shown). In both cases, TAF9 dependence and occupancy correlated well. H2B transcript levels were essentially unchanged by TAF9 depletion (Fig. 4D, top) (15); concomitantly, TAF9 occupancy was very low, which contrasts with the very high levels of TBP (and TFIIB) on this promoter (Fig. 4D, bottom). In contrast, β-actin appeared moderately TAF9 dependent (approximately threefold reduction in mRNA levels), and indeed there was significantly higher TAF9 occupancy (compared to the much more modest TBP/TFIIB levels). Together, our results indicate that, although in yeast TAF dependence and TAF promoter occupancy can be directly correlated, in vertebrate cells this is not necessarily the case. Indeed, the two most TAF9-dependent promoter derivatives examined displayed the lowest levels of bound TAF9.
The DPE constitutes a TAF9 response element.
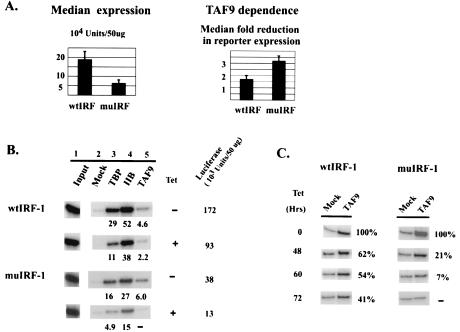
TAFs can also function as direct core promoter recognition factors (2, 25). Previously, TAF6 and TAF9 were shown to cross-link specifically to the DPE in vitro (8). To investigate the potential role of TAF9 in DPE recognition and function in vivo, we stably transfected DT40-TAF9 cells with reporter constructs containing the human IRF-1 promoter or a DPE-scrambled mutant promoter (wtIRF-1 and muIRF-1, respectively, both with an intact Inr; Fig. 1B) (8). Reporter expression in the resulting clones was analyzed as for Fig. 2. Although we analyzed uninduced transcription, significant levels of expression were detected. Consistent with previous results (8), mutation of the DPE decreased IRF-1 promoter strength, by three- to fourfold (Fig. 5A, left). We then examined expression following TAF9 depletion. Intriguingly, while expression from both promoters was modestly TAF9 dependent, depletion had about a twofold-greater effect on the DPE mutant promoter (Fig. 5A, right).
FIG. 5.
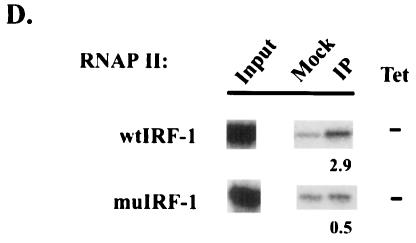
Expression and factor occupancy on WT and DPE-scrambled human IRF-1 promoters. (A) Expression profile (left) and TAF9 dependence (right). (B) ChIP analysis of TBP, TFIIB, and TAF9. (C) Time course ChIP analysis of TAF9 occupancy. Cells were grown in TET for 0, 48, 60, and 72 h and subjected to ChIP analysis. The levels of relative occupancy compared with that at time zero are shown. (D) ChIP analysis of RNAP II occupancy.
We next wished to examine further the link between TAF9 dependence and the possible TAF9-DPE interaction documented in vitro. To this end, ChIP analysis was performed on the WT and mutant IRF-1 promoters after 72 h treatment with TET, which resulted in ∼98% depletion of total TAF9, based on quantitative Western blotting (15) (data not shown), and the results are shown in Fig. 5B. muIRF-1 showed a typical, essentially complete, loss of TAF9, which was confirmed by using the polyclonal anti-TAF9 antibody (data not shown). Strikingly, however, and unlike what we observed with any other promoters, significant amounts of TAF9 (∼45%) remained associated with the wtIRF-1 promoter following depletion. Similar results were obtained with two other representative clones for each promoter (data not shown). ChIP analyses were also performed at different times following TET addition to document more completely the ability of the DPE to retain TAF9 (Fig. 5C). TAF9 was essentially depleted from the muIRF-1 promoter by 60 h (∼7% of time-zero level), while significant retention was observed at the wtIRF-1 promoter throughout the time course. These results support a role for the DPE in augmenting promoter activity and suggest that TAF9 dissociates more slowly from a DPE-containing promoter, providing functional support for the TAF9-DPE interaction detected in vitro. These findings also provide an explanation for the greater sensitivity of the mutant IRF-1 promoter to TAF9 depletion.
The requirements for TBP and TFIIB recruitment to TATA-less promoters remain largely undefined. To provide insights into this problem, we first examined TBP and TFIIB occupancy on the IRF-1 promoter and determined how this correlated with expression and how it changed following TAF9 depletion. Strikingly, both TBP and especially TFIIB levels were very high (Fig. 5B), in fact as high or higher than those of any promoters examined in this study with the sole exception of the H2B promoter. This is especially noteworthy given that the uninduced IRF-1 promoter is weak, nearly 50-fold weaker than any of the SV promoters. These values also indicate that TBP can be recruited effectively to a TATA-less promoter, likely through either bound activators or TAFs that contact promoter DNA. However, in contrast to results for TATA-containing promoters, where TBP occupancy either correlated with or decreased less than reporter expression following TAF9 depletion (Fig. 3 and 4), TBP promoter association at both WT and mutant IRF-1 promoters displayed significantly higher sensitivity to TAF9 depletion than did expression in multiple clones tested (Fig. 5B). TFIIB, on the other hand, showed lower dissociation upon TAF9 depletion, which correlated better with promoter activity. This differential pattern of TAF-dependent TBP and TFIIB promoter association was also seen on the endogenous TATA-less ASF/SF2 promoter, which does not contain an Inr or DPE (data not shown). Together our results indicate that TAF(s) play a more significant role in TBP recruitment to TATA-less promoters than in recruitment to TATA-containing promoters, although the DPE, despite its functional interaction with TAF9, is not essential for this.
Recruitment of TBP and TFIIB to the IRF-1 promoter was unexpectedly high. We next asked whether RNAP II levels were correspondingly high or whether they were weaker, in keeping with the reduced activity of this promoter. ChIP analysis (Fig. 5D) indicates that the RNAP II levels at both WT and mutant promoters were relatively low, reflecting transcriptional activity and not TBP/TFIIB promoter occupancy (compare with RNAP II levels on the stronger SVE promoters; Fig. 3C and 4C). Additionally, RNAP II occupancy on wtIRF-1 was significantly higher than on muIRF-1, also correlating with promoter activity. Together, these results suggest that RNAP II, not TFIID, recruitment limits expression of the TATA-less IRF-1 promoter.
The p53 activation domain recruits TBP and TFIIB but is TAF9 independent.
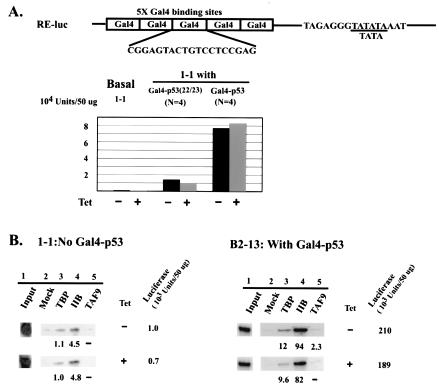
Aside from promoter recognition, TAFs were originally proposed to function as coactivators to bridge activators and the general transcription machinery (2, 25). Several activators, notably p53 and VP16, have been shown to interact directly with TAF9 and to be dependent on TAF9 for function in vitro (24, 45, 64). Since DT40 cells lack endogenous p53 expression (62), Gal4-p53 fusion proteins were used to examine the TAF9 dependence of a minimal promoter activated by the p53 activation domain, allowing a direct comparison with previous in vitro studies (64),. A reporter plasmid containing five tandem copies of the Gal4 binding site linked to the adenovirus E1B TATA box was constructed (RE-luc; Fig. 6A) and stably transfected into DT40-TAF9 cells. Several individual clones were isolated and analyzed for luciferase expression. This basal promoter, like the previously examined non-GC-rich TATA core promoters, was not TAF9 dependent (data not shown). One basal clone, 1-1, was transfected with vectors encoding Gal4 fusion proteins containing either the WT or double-mutant (aa 22 and 23) p53 activation domain, and several individual clones for each activator were isolated. Reporter expression in the presence or absence of TAF9 was analyzed, and median expression levels were determined. Consistent with previous results, the WT fusion protein was much more potent than the double mutant (Fig. 6A) (41). Most importantly, however, both sets of activated clones were TAF9 independent, as depletion of TAF9 caused essentially no change in reporter expression. These results indicate that TAF9 can be dispensable for p53-mediated activation in vivo.
FIG. 6.
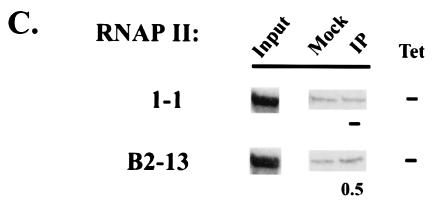
Mechanisms of Gal4-p53-mediated activation in vivo. (A) At the top is a diagram of the Gal4-p53-responsive promoter (RE-luc). Sequences of the Gal4 binding site and a short E1b TATA fragment are shown. Below is shown median levels of expression of a basal clone (1-1) and activated clones expressing Gal4 fusions with either mutant (aa 22 and 23) or WT p53 activation domains. The values were derived from three separate measurements for 1-1 and from two separate measurements for eight activated clones. (B) ChIP analysis of TBP, TFIIB, and TAF9 on basal and activated promoters. Three Gal4-p53-activated clones were analyzed, and representative results from clone B2-13 are shown. (C) ChIP analysis of RNAP II occupancy.
We next performed ChIP assays to examine transcription factor recruitment to the basal and Gal4-p53-activated promoters. While the basal promoter showed only near-background levels of TBP and TFIIB occupancy, TBP/TFIIB occupancy on the activated promoter (one example, from clone B2-13, is shown) was significantly increased and TFIIB levels were especially high (Fig. 6B). In contrast, RNAP II occupancy on both basal and activated promoters was very low (Fig. 6C). This contrasts, for example, with the wtIRF-1 promoter, which displayed very similar activity but significantly higher RNAP II occupancy (Fig. 5D). Additionally, there was no preferential TAF9 recruitment, relative to other promoters analyzed (Fig. 6B, lane 5), and TAF9 depletion had only minor effects on TBP and TFIIB promoter occupancy (lane 3 and 4). Taken together, these results indicate not only that Gal4-p53 functions independently of TAF9 but also that it preferentially recruits TBP and TFIIB, but not RNAP II, at least in the core promoter context examined here.
DISCUSSION
The experiments described here have provided a view of the unexpected plasticity evident in the mechanisms underlying transcriptional activation from vertebrate promoters. Our data have shown, for example, that core promoter elements can contribute in diverse ways to promoter activity, interactions with GTFs, and/or response to transcriptional activators. Reflecting this, as well as the ability of activators to function in distinct ways, is the diversity we uncovered in mechanisms of transcription factor recruitment, in which relative (and absolute) levels of TBP/TFIID, TFIIB, and even RNAP II can vary considerably but nonetheless lead to significant, and in some cases very similar, levels of activated transcription. In keeping with this variability in factor recruitment and activity, we also observed an unanticipated variability in TAF dependence and TAF promoter occupancy. Below we discuss these results, highlighting the striking diversity in promoter structure and function in vertebrate cells.
The BRE is the most recently described core element, but its functional significance has been somewhat controversial. The initial characterization of the element by Lagrange et al. (35), which suggested that the BRE stimulates transcription via its interaction with TFIIB, utilized in vitro assays with purified components. In contrast, much earlier studies analyzing the effects of single base substitutions in this region by transient-transfection assays are largely consistent with a negative function (71, 72), a conclusion supported by the more-recent study of Evans et al. (20), who utilized in vitro assays of unfractionated nuclear extract. However these later experiments also suggested that the inhibitory effect on basal transcription in fact facilitated activation by the acidic activator GAL4-VP16. Finally, Wolner and Gralla (69) analyzed a BRE substitution mutant element that had essentially the opposite effects, suggesting that the BRE modestly enhances basal transcription but reduces activation potential. Our results extend these previous studies and are significant because they are the first to examine BRE function in a chromosomal context. Most notably, our results show that disruption of the BRE resulted in a significant activation of basal, but not activated, transcription, and also increased both TBP and TFIIB association with the promoter. While different mutations were analyzed in all studies and while it may be that the BRE functions differently in other promoter contexts, our findings support the view that the role of the BRE in vivo is to negatively modulate basal promoter activity, thereby increasing activation potential.
The region just downstream of the TATA box, which we have called the G track, has not been considered a core promoter element, but our data and previous observations suggest that it can play a significant role in promoter function. For example, TFIIB can make base-specific contacts in this region in vitro (21, 67), and it has also been suggested that the G track affects the secondary structure of the promoter region (72). Our mutational data suggest that the G track has a role related to but distinct from that of the BRE. Mutation of the G track significantly enhanced activated transcription, but in this case repressed basal expression, allowing for the highest activation ratio of any promoter tested. Also unlike what was found for the BRE, TBP and TFIIB promoter association was not affected by the G-track mutation, while TAF occupancy was actually decreased. Together our results illustrate the ability of TATA-flanking sequences to modulate basal versus activated transcription.
Our studies on the DPE, and TATA-less promoters more generally, on the one hand, agree with expectations from previous analyses, while on the other provide new insights into how such promoters function. The initial studies of Kadanoga and colleagues indicated that the DPE is a conserved element in many TATA-lacking promoters in Drosophila and that it is required for optimal transcription in vitro and in transfected mammalian cells and, using photo-cross-linking with purified Drosophila TFIID, provided evidence that the DPE is recognized by TAF6 and to a lesser degree its partner TAF9 (8). Additional studies revealed that accumulation of a DPE-containing transcript in flies containing a mutant TAF9 allele was reduced (34, 60). Our results are significant because they establish that a DPE can contribute to optimal transcription in vertebrate cells in a normal chromosomal context. More importantly, our data indicate that the interaction between TAF9 in purified TFIID and the DPE is physiologically significant, reflecting a tighter interaction of TFIID with the TATA-less promoter that results in a slower dissociation of TAF9. This, together with the enhanced GTF and RNAP II occupancy levels in wtIRF-1 relative to those in the DPE-scrambled mutant promoter, provides a likely explanation for the DPE's ability to enhance transcription. But there are undoubtedly additional interactions required for TATA-less promoter function. For example, our data showed that TBP dissociated more rapidly from TATA-less than from TATA-containing promoters, but this was not affected by the DPE. Also, TFIIB dissociation following TAF9 depletion was not significantly affected by the presence or absence of a TATA box, nor were levels of transcription from the TATA-less promoters tested significantly more sensitive to TAF9 depletion than was transcription from TATA-containing promoters. These results together are indicative of a complex set of protein-DNA interactions at the core promoter that cumulatively lead to assembly of a productive PIC.
Recent studies have illustrated a surprising diversity in the mechanisms by which activators in metazoan cells can function to enhance PIC assembly and hence transcription, and our results have extended these possibilities in novel ways. In vitro experiments, supported by studies of yeast, have led to the classical model of an ordered, or in some cases concerted, pathway, beginning with recruitment of TFIID/TBP and culminating with RNAP II (38, 55). But, as ChIP assays have allowed more-detailed examination of PIC assembly during transcriptional activation in vivo, it is becoming apparent that alternate pathways exist (23). For example, RNAP II is already present at the beta interferon promoter prior to activation, and activators, at least in part through the activity of chromatin-remodeling complexes, facilitate the recruitment of TBP as a final step in activation (1). A nearly complete PIC appears to be preassembled on the α1-antitrypsin promoter, with activators, again through chromatin remodeling, allowing initiation to occur (61). Our results suggest additional variations on this theme, and contrasting examples are provided by the SVE and the p53 activation domain. Factors bound to the SVE, at least in the context of the core promoters we analyzed, bring about massive activation without significantly increasing the levels of TFIID/TBP and TFIIB detected. These findings are in agreement with recent results of Bertolino and Singh (6), who found that distal enhancers, including the SVE, were unable to activate transcription from a minimal core promoter, likely reflecting an inability to recruit TBP. However, the presence of a promoter-proximal activator capable of recruiting TBP allowed the enhancers to function. These findings are consistent with our conclusion that factors associated with the SVE function at a step(s) subsequent to TBP/TFIIB recruitment. In our case an additional activator was not required because the core promoters that we analyzed were able to bind sufficient levels of TBP and TFIIB and/or because of the promoter-proximal position of the SVE. Our results suggest a novel mechanism whereby activation occurs on a preassembled TBP/TFIIB core scaffold.
The p53 activation domain works by what appears to be almost the opposite mechanism: TBP and TFIIB levels were increased dramatically, by 10- and 20-fold, respectively, while the levels of RNAP II remained low. A considerable amount of early data suggested that a p53-TBP interaction might be important for activation. Numerous studies demonstrated a direct interaction between the p53 activation domain and TBP and correlated the requirements for the interaction with those for transcriptional activation (44, 66). TBP and p53 were also shown to bind DNA cooperatively (14) and to activate transcription synergistically in transient-cotransfection assays (22). While additional cotransfection experiments provided evidence that the TBP-p53 interaction is not necessary for activation in such assays (63), our data indicate that the p53 activation domain, directly or indirectly, enhances TBP promoter occupancy. Interactions between p53 and TFIIB are less well documented. However, it has been shown that inhibition of in vitro transcription by excess p53 (i.e., squelching) can be reversed specifically by TFIIB and TFIID, leading to the suggestion that p53 activation involves interactions with both these factors (43). Our data are consistent with this and suggest that a significant aspect of p53 activation function involves recruitment of TBP and TFIIB. It is possible that other interactions are also involved. For example, an interaction between p53 and the TRAP/Mediator component TRAP80 has been observed and has been shown to be important for p53 function in vitro (29). In any event, our data support a model for p53 activation in which p53 facilitates the assembly of a stable scaffold containing TBP/TFIID and TFIIB that is able to bring about significant levels of transcription in the absence of high levels of RNAP II.
Studies of yeast employing conditional mutants coupled with transcriptional measurements and ChIP assays have provided detailed insights into the mechanisms by which GTFs and other transcription factors function in vivo. For example, these methods led to the conclusion that most TAFs serve gene-specific functions in yeast and are generally present on TAF-dependent promoters but absent from TAF-independent promoters (references 32 and 39 and references therein). In higher organisms, this powerful type of analysis is much more difficult, and indeed to our knowledge ours is the first study to attempt this. While more work is certainly required, our results suggest a more complex picture of TAF dependence and promoter occupancy than the one apparently applicable to yeast. For example, the TAF9-independent p53-activated promoter was found to have a significantly higher TAF9 content, measured either as an absolute value or as a TBP/TAF9 ratio, than did the two most TAF9-dependent promoters, the SV-mGT and SV-fos derivatives, which had among the lowest TAF9 content. The presence of appreciable levels of TAF9 on nearly all promoters examined suggests that in vertebrates there may not be a significant fraction of TAF-free TBP, as has been suggested for yeast (32, 39). However, the fact that the TBP/TAF9 ratio was quite variable from promoter to promoter is consistent with the possibility that TFIID complexes with different TAF contents may assemble on different promoters, an idea supported by the presence of apparently distinct TFIID complexes on the α1-antitrypin promoter (61). But especially intriguing are the two promoters displaying high TAF9 dependence and low TAF occupancy. An interesting possibility is that these promoters are bound by a TFIID with low TAF content and that the TAF9 dependence results from the requirement for another TAF9-containing complex, such as PCAF, STAGA, or TFTC, which perhaps interacts more weakly or transiently with the promoter, limiting TAF9 cross-linking. Yeast TAF9 was also observed to cross-link weakly to certain yTAF9-dependent promoters, and a similar explanation was suggested (32). In any case, it is notable that the simple mutation in the MLP G track induced such striking effects: it reduced basal activity but increased activation potential and decreased TAF content but enhanced TAF dependence. These diverse effects illustrate the important and complex role of core promoter architecture in transcriptional activation.
Acknowledgments
We thank J. Kadonaga, A. Kutach, C. Prives, S. Y. Sheih, R. Prywes, J. Shen, J. M. Buerstedde, Y. Takagaki, T. Kashima, and J. Wang for kind gifts of plasmid DNAs. We are grateful to R. Roeder and C. M. Chiang for providing antibodies. We also thank S. Lomvardas, O. Calvo, and other members of Manley lab for helpful discussion. Inna Boluk is thanked for help preparing the manuscript.
This work was supported by NIH grant GM 37971.
REFERENCES
- 1.Agalioti, T., S. Lomvardas, B. Parekh, J. Yie, T. Maniatis, and D. Thanos. 2000. Ordered recruitment of chromatin modifying and general transcription factors to the IFN-beta promoter. Cell 103:667-678. [DOI] [PubMed] [Google Scholar]
- 2.Albright, S. R., and R. Tjian. 2000. TAFs revisited: more data reveal new twists and confirm old ideas. Gene 242:1-13. [DOI] [PubMed] [Google Scholar]
- 3.Apone, L. M., C. A. Virbasius, F. C. Holstege, J. Wang, R. A. Young, and M. R. Green. 1998. Broad, but not universal, transcriptional requirement for yTAFII17, a histone H3-like TAFII present in TFIID and SAGA. Mol. Cell 2:653-661. [DOI] [PubMed] [Google Scholar]
- 4.Baniahmad, A., A. C. Kohne, and R. Renkawitz. 1992. A transferable silencing domain is present in the thyroid hormone receptor, in the v-erbA oncogene product and in the retinoic acid receptor. EMBO J. 11:1015-1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bell, B., E. Scheer, and L. Tora. 2001. Identification of hTAF(II)80 delta links apoptotic signaling pathways to transcription factor TFIID function. Mol. Cell 8:591-600. [DOI] [PubMed] [Google Scholar]
- 6.Bertolino, E., and H. Singh. 2002. POU/TBP cooperativity: a mechanism for enhancer action from a distance. Mol. Cell 10:397-407. [DOI] [PubMed] [Google Scholar]
- 7.Blackwood, E. M., and J. T. Kadonaga. 1998. Going the distance: a current view of enhancer action. Science 281:61-63. [DOI] [PubMed] [Google Scholar]
- 8.Burke, T. W., and J. T. Kadonaga. 1997. The downstream core promoter element, DPE, is conserved from Drosophila to humans and is recognized by TAFII60 of Drosophila. Genes Dev. 11:3020-3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burley, S. K., and R. G. Roeder. 1996. Biochemistry and structural biology of transcription factor IID (TFIID). Annu. Rev. Biochem. 65:769-799. [DOI] [PubMed] [Google Scholar]
- 10.Butler, J. E., and J. T. Kadonaga. 2001. Enhancer-promoter specificity mediated by DPE or TATA core promoter motifs. Genes Dev. 15:2515-2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Butler, J. E., and J. T. Kadonaga. 2002. The RNA polymerase II core promoter: a key component in the regulation of gene expression. Genes Dev. 16:2583-2592. [DOI] [PubMed] [Google Scholar]
- 12.Carey, M., and S. T. Smale. 2000. Transcriptional regulation in eukaryotes: concepts, strategies and techniques. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 13.Chalkley, G. E., and C. P. Verrijzer. 1999. DNA binding site selection by RNA polymerase II TAFs: a TAF(II)250-TAF(II)150 complex recognizes the initiator. EMBO J. 18:4835-4845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen, X., G. Farmer, H. Zhu, R. Prywes, and C. Prives. 1993. Cooperative DNA binding of p53 with TFIID (TBP): a possible mechanism for transcriptional activation. Genes Dev. 7:1837-1849. [DOI] [PubMed] [Google Scholar]
- 15.Chen, Z., and J. L. Manley. 2000. Robust mRNA transcription in chicken DT40 cells depleted of TAFII31 suggests both functional degeneracy and evolutionary divergence. Mol. Cell. Biol. 20:5064-5076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15a.Chen, Z., and J. L. Manley. J. Biol. Chem., in press.
- 16.Chiang, C. M., H. Ge, Z. Wang, A. Hoffmann, and R. G. Roeder. 1993. Unique TATA-binding protein-containing complexes and cofactors involved in transcription by RNA polymerases II and III. EMBO J. 12:2749-2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Christova, R., and T. Oelgeschlager. 2002. Association of human TFIID-promoter complexes with silenced mitotic chromatin in vivo. Nat. Cell Biol. 4:79-82. [DOI] [PubMed] [Google Scholar]
- 18.Colgan, J., and J. L. Manley. 1995. Cooperation between core promoter elements influences transcriptional activity in vivo. Proc. Natl. Acad. Sci. USA 92:1955-1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Emami, K. H., W. W. Navarre, and S. T. Smale. 1995. Core promoter specificities of the Sp1 and VP16 transcriptional activation domains. Mol. Cell. Biol. 15:5906-5916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Evans, R., J. A. Fairley, and S. G. Roberts. 2001. Activator-mediated disruption of sequence-specific DNA contacts by the general transcription factor TFIIB. Genes Dev. 15:2945-2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fairley, J. A., R. Evans, N. A. Hawkes, and S. G. Roberts. 2002. Core promoter-dependent TFIIB conformation and a role for TFIIB conformation in transcription start site selection. Mol. Cell. Biol. 22:6697-6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Farmer, G., J. Colgan, Y. Nakatani, J. L. Manley, and C. Prives. 1996. Functional interaction between p53, the TATA-binding protein (TBP), and TBP-associated factors in vivo. Mol. Cell. Biol. 16:4295-4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fry, C. J., and C. L. Peterson. 2002. Transcription. Unlocking the gates to gene expression. Science 295:1847-1848. [DOI] [PubMed] [Google Scholar]
- 24.Goodrich, J. A., T. Hoey, C. J. Thut, A. Admon, and R. Tjian. 1993. Drosophila TAFII40 interacts with both a VP16 activation domain and the basal transcription factor TFIIB. Cell 75:519-530. [DOI] [PubMed] [Google Scholar]
- 25.Green, M. R. 2000. TBP-associated factors (TAFIIs): multiple, selective transcriptional mediators in common complexes. Trends Biochem. Sci. 25:59-63. [DOI] [PubMed] [Google Scholar]
- 26.Hampsey, M. 1998. Molecular genetics of the RNA polymerase II general transcriptional machinery. Microbiol. Mol. Biol. Rev. 62:465-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hernandez, N. 1993. TBP, a universal eukaryotic transcription factor? Genes Dev. 7:1291-1308. [DOI] [PubMed] [Google Scholar]
- 28.Holstege, F. C., E. G. Jennings, J. J. Wyrick, T. I. Lee, C. J. Hengartner, M. R. Green, T. R. Golub, E. S. Lander, and R. A. Young. 1998. Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95:717-728. [DOI] [PubMed] [Google Scholar]
- 29.Ito, M., C. X. Yuan, S. Malik, W. Gu, J. D. Fondell, S. Yamamura, Z. Y. Fu, X. Zhang, J. Qin, and R. G. Roeder. 1999. Identity between TRAP and SMCC complexes indicates novel pathways for the function of nuclear receptors and diverse mammalian activators. Mol. Cell 3:361-370. [DOI] [PubMed] [Google Scholar]
- 30.Kaufmann, J., K. Ahrens, R. Koop, S. T. Smale, and R. Muller. 1998. CIF150, a human cofactor for transcription factor IID-dependent initiator function. Mol. Cell. Biol. 18:233-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klemm, R. D., J. A. Goodrich, S. Zhou, and R. Tjian. 1995. Molecular cloning and expression of the 32-kDa subunit of human TFIID reveals interactions with VP16 and TFIIB that mediate transcriptional activation. Proc. Natl. Acad. Sci. USA 92:5788-5792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuras, L., P. Kosa, M. Mencia, and K. Struhl. 2000. TAF-Containing and TAF-independent forms of transcriptionally active TBP in vivo. Science 288:1244-1248. [DOI] [PubMed] [Google Scholar]
- 33.Kuras, L., and K. Struhl. 1999. Binding of TBP to promoters in vivo is stimulated by activators and requires Pol II holoenzyme. Nature 399:609-613. [DOI] [PubMed] [Google Scholar]
- 34.Kutach, A. K., and J. T. Kadonaga. 2000. The downstream promoter element DPE appears to be as widely used as the TATA box in Drosophila core promoters. Mol. Cell. Biol. 20:4754-4764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lagrange, T., A. N. Kapanidis, H. Tang, D. Reinberg, and R. H. Ebright. 1998. New core promoter element in RNA polymerase II-dependent transcription: sequence-specific DNA binding by transcription factor IIB. Genes Dev. 12:34-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leclerc, G. M., F. R. Boockfor, W. J. Faught, and L. S. Frawley. 2000. Development of a destabilized firefly luciferase enzyme for measurement of gene expression. BioTechniques 29:590-591, 594-596, 598. [DOI] [PubMed] [Google Scholar]
- 37.Lee, T. I., and R. A. Young. 1998. Regulation of gene expression by TBP-associated proteins. Genes Dev. 12:1398-1408. [DOI] [PubMed] [Google Scholar]
- 38.Lemon, B., and R. Tjian. 2000. Orchestrated response: a symphony of transcription factors for gene control. Genes Dev. 14:2551-2569. [DOI] [PubMed] [Google Scholar]
- 39.Li, X. Y., S. R. Bhaumik, and M. R. Green. 2000. Distinct classes of yeast promoters revealed by differential TAF recruitment. Science 288:1242-1244. [DOI] [PubMed] [Google Scholar]
- 39a.Li, X. Y., S. R. Bhaumik, X. Zhu, L. Li, W. C. Shen, B. L. Dixit, and M. R. Green. 2002. Selective recruitment of TAFs by yeast upstream activating sequences. Implications for eukaryotic promoter structure. Curr. Biol. 12:1240-1244. [DOI] [PubMed] [Google Scholar]
- 40.Li, X. Y., A. Virbasius, X. Zhu, and M. R. Green. 1999. Enhancement of TBP binding by activators and general transcription factors. Nature 399:605-609. [DOI] [PubMed] [Google Scholar]
- 41.Lin, J., J. Chen, B. Elenbaas, and A. J. Levine. 1994. Several hydrophobic amino acids in the p53 amino-terminal domain are required for transcriptional activation, binding to mdm-2 and the adenovirus 5 E1B 55-kD protein. Genes Dev. 8:1235-1246. [DOI] [PubMed] [Google Scholar]
- 42.Lin, Y. S., and M. R. Green. 1991. Mechanism of action of an acidic transcriptional activator in vitro. Cell 64:971-981. [DOI] [PubMed] [Google Scholar]
- 43.Liu, X., and A. J. Berk. 1995. Reversal of in vitro p53 squelching by both TFIIB and TFIID. Mol. Cell. Biol. 15:6474-6478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu, X., C. W. Miller, P. H. Koeffler, and A. J. Berk. 1993. The p53 activation domain binds the TATA box-binding polypeptide in Holo-TFIID, and a neighboring p53 domain inhibits transcription. Mol. Cell. Biol. 13:3291-3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lu, H., and A. J. Levine. 1995. Human TAFII31 protein is a transcriptional coactivator of the p53 protein. Proc. Natl. Acad. Sci. USA 92:5154-5158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mencia, M., Z. Moqtaderi, J. V. Geisberg, L. Kuras, and K. Struhl. 2002. Activator-specific recruitment of TFIID and regulation of ribosomal protein genes in yeast. Mol. Cell 9:823-833. [DOI] [PubMed] [Google Scholar]
- 47.Merika, M., and D. Thanos. 2001. Enhanceosomes. Curr. Opin. Genet. Dev. 11:205-208. [DOI] [PubMed] [Google Scholar]
- 48.Michel, B., P. Komarnitsky, and S. Buratowski. 1998. Histone-like TAFs are essential for transcription in vivo. Mol. Cell 2:663-673. [DOI] [PubMed] [Google Scholar]
- 49.Moqtaderi, Z., M. Keaveney, and K. Struhl. 1998. The histone H3-like TAF is broadly required for transcription in yeast. Mol. Cell 2:675-682. [DOI] [PubMed] [Google Scholar]
- 50.Nakajima, N., M. Horikoshi, and R. G. Roeder. 1988. Factors involved in specific transcription by mammalian RNA polymerase II: purification, genetic specificity, and TATA box-promoter interactions of TFIID. Mol. Cell. Biol. 8:4028-4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Oelgeschlager, T., C. M. Chiang, and R. G. Roeder. 1996. Topology and reorganization of a human TFIID-promoter complex. Nature 382:735-738. [DOI] [PubMed] [Google Scholar]
- 52.Oelgeschlager, T., Y. Tao, Y. K. Kang, and R. G. Roeder. 1998. Transcription activation via enhanced preinitiation complex assembly in a human cell-free system lacking TAFIIs. Mol. Cell 1:925-931. [DOI] [PubMed] [Google Scholar]
- 53.Orlando, V. 2000. Mapping chromosomal proteins in vivo by formaldehyde-crosslinked-chromatin immunoprecipitation. Trends Biochem. Sci. 25:99-104. [DOI] [PubMed] [Google Scholar]
- 54.Orlando, V., and R. Paro. 1993. Mapping Polycomb-repressed domains in the bithorax complex using in vivo formaldehyde cross-linked chromatin. Cell 75:1187-1198. [DOI] [PubMed] [Google Scholar]
- 55.Orphanides, G., T. Lagrange, and D. Reinberg. 1996. The general transcription factors of RNA polymerase II. Genes Dev. 10:2657-2683. [DOI] [PubMed] [Google Scholar]
- 56.Park, J. M., J. Werner, J. M. Kim, J. T. Lis, and Y. J. Kim. 2001. Mediator, not holoenzyme, is directly recruited to the heat shock promoter by HSF upon heat shock. Mol. Cell 8:9-19. [DOI] [PubMed] [Google Scholar]
- 57.Roeder, R. G. 1996. The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem. Sci. 21:327-335. [PubMed] [Google Scholar]
- 58.Shen, W. C., and M. R. Green. 1997. Yeast TAF(II)145 functions as a core promoter selectivity factor, not a general coactivator. Cell 90:615-624. [DOI] [PubMed] [Google Scholar]
- 59.Smale, S. T. 2001. Core promoters: active contributors to combinatorial gene regulation. Genes Dev. 15:2503-2508. [DOI] [PubMed] [Google Scholar]
- 60.Soldatov, A., E. Nabirochkina, S. Georgieva, T. Belenkaja, and P. Georgiev. 1999. TAFII40 protein is encoded by the e(y)1 gene: biological consequences of mutations. Mol. Cell. Biol. 19:3769-3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Soutoglou, E., and I. Talianidis. 2002. Coordination of PIC assembly and chromatin remodeling during differentiation-induced gene activation. Science 295:1901-1904. [DOI] [PubMed] [Google Scholar]
- 62.Takao, N., H. Kato, R. Mori, C. Morrison, E. Sonada, X. Sun, H. Shimizu, K. Yoshioka, S. Takeda, and K. Yamamoto. 1999. Disruption of ATM in p53-null cells causes multiple functional abnormalities in cellular response to ionizing radiation. Oncogene 18:7002-7009. [DOI] [PubMed] [Google Scholar]
- 63.Tansey, W. P., and W. Herr. 1995. The ability to associate with activation domains in vitro is not required for the TATA box-binding protein to support activated transcription in vivo. Proc. Natl. Acad. Sci. USA 92:10550-10554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Thut, C. J., J. L. Chen, R. Klemm, and R. Tjian. 1995. p53 transcriptional activation mediated by coactivators TAFII40 and TAFII60. Science 267:100-104. [DOI] [PubMed] [Google Scholar]
- 65.Tora, L. 2002. A unified nomenclature for TATA box binding protein (TBP)-associated factors (TAFs) involved in RNA polymerase II transcription. Genes Dev. 16:673-675. [DOI] [PubMed] [Google Scholar]
- 66.Truant, R., H. Xiao, C. J. Ingles, and J. Greenblatt. 1993. Direct interaction between the transcriptional activation domain of human p53 and the TATA box-binding protein. J. Biol. Chem. 268:2284-2287. [PubMed] [Google Scholar]
- 67.Tsai, F. T., and P. B. Sigler. 2000. Structural basis of preinitiation complex assembly on human pol II promoters. EMBO J. 19:25-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Verrijzer, C. P., and R. Tjian. 1996. TAFs mediate transcriptional activation and promoter selectivity. Trends Biochem. Sci. 21:338-342. [PubMed] [Google Scholar]
- 69.Wolner, B. S., and J. D. Gralla. 2000. Roles for non-TATA core promoter sequences in transcription and factor binding. Mol. Cell. Biol. 20:3608-3615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Woychik, N. A., and M. Hampsey. 2002. The RNA polymerase II machinery: structure illuminates function. Cell 108:453-463. [DOI] [PubMed] [Google Scholar]
- 71.Yu, Y. T., and J. L. Manley. 1984. Generation and functional analyses for base-substitution mutants of the adenovirus 2 major late promoter. Nucleic Acids Res. 12:9309-9321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yu, Y. T., and J. L. Manley. 1986. Structure and function of the S1 nuclease-sensitive site in the adenovirus late promoter. Cell 45:743-751. [DOI] [PubMed] [Google Scholar]
- 73.Zhao, X., and W. Herr. 2002. A regulated two-step mechanism of TBP binding to DNA: a solvent-exposed surface of TBP inhibits TATA box recognition. Cell 108:615-627. [DOI] [PubMed] [Google Scholar]
- 74.Zhou, Q., P. M. Lieberman, T. G. Boyer, and A. J. Berk. 1992. Holo-TFIID supports transcriptional stimulation by diverse activators and from a TATA-less promoter. Genes Dev. 6:1964-1974. [DOI] [PubMed] [Google Scholar]