Abstract
Alterations in MYC and p53 are hallmarks of cancer. p53 coordinates the response to gamma irradiation (γ-IR) by either triggering apoptosis or cell cycle arrest. c-Myc activates the p53 apoptotic checkpoint, and thus tumors overexpressing MYC often harbor p53 mutations. Nonetheless, many of these cancers are responsive to therapy, suggesting that Myc may sensitize cells to γ-IR independent of p53. In mouse embryo fibroblasts (MEFs) and in Eμ-myc transgenic B cells in vivo, c-Myc acts in synergy with γ-IR to trigger apoptosis, but alone, when cultured in growth medium, it does not induce a DNA damage response. Surprisingly, c-Myc also sensitizes p53-deficient MEFs to γ-IR-induced apoptosis. In normal cells, and in precancerous B cells of Eμ-myc transgenic mice, this apoptotic response is associated with the suppression of the antiapoptotic regulators Bcl-2 and Bcl-XL and with the concomitant induction of Puma, a proapoptotic BH3-only protein. However, in p53-null MEFs only Bcl-XL expression was suppressed, suggesting levels of Bcl-XL regulate the response to γ-IR. Indeed, Bcl-XL overexpression blocked this apoptotic response, whereas bcl-X-deficient MEFs were inherently and selectively sensitive to γ-IR-induced apoptosis. Therefore, MYC may sensitize tumor cells to DNA damage by suppressing Bcl-X.
Deregulated cell growth and the inhibition of apoptosis are hallmarks of cancer. The MYC family of oncogenes and the p53 tumor suppressor gene are pivotal players in tumorigenesis and are altered in most tumor types. c-Myc is the founding member of a family of structurally related basic helix-loop-helix-leucine zipper (bHLH-Zip) proteins that function as sequence-specific transcription factors and are aberrantly expressed in most cancers (46). c-Myc is a key regulator of cell proliferation and differentiation, and its expression is both necessary (14, 26) and sufficient (11, 16) to drive quiescent cells into S phase. Following mitogenic stimulation, c-Myc is rapidly induced, remains elevated throughout the cell cycle and, through dimerization with its bHLH-Zip partner Max, regulates the transcription of genes essential for cell growth and division (4). Conversely, c-myc expression is rapidly suppressed by growth inhibitory signals such as transforming growth factor β (12). However, these controls are lost in cancers by translocations, amplifications, and alterations in regulatory signaling pathways, resulting in abnormally high levels of MYC oncoproteins (9). The precise roles that Myc oncoproteins provide to provoke tumorigenesis are not fully resolved but do include the regulation of target genes that control cell division (6, 13), differentiation (33), cell size (27), and angiogenesis (5, 48).
Despite its role in promoting tumorigenesis, in normal cells Myc overexpression triggers apoptosis. For example, c-Myc triggers rapid cell death following the withdrawal of essential survival factors (2, 21) that are continuously required to suppress c-Myc-induced apoptosis (24). Moreover, in vivo studies using transgenic mice have underscored the concept that in most scenarios (but not all), Myc triggers the apoptotic program (19, 47, 48). Indeed, the molecular analysis of lymphomas arising in Eμ-myc transgenic mice established that the Arf-p53 and the Bcl-2 and Bcl-XL apoptotic pathways triggered by Myc are disabled during lymphomagenesis (19, 20). The Bcl-2 family of apoptotic regulators includes both antiapoptotic proteins, such as Bcl-2 and Bcl-XL, as well as proapoptotic proteins such as Bax, Bad, Bak, Puma, and Noxa (8), and those implicated as regulators of Myc-induced apoptosis include Bax (18, 30), Bcl-XL (20, 47), and Bcl-2 (17, 20).
Effective anticancer regimens trigger tumor cell apoptosis and, thus, therapy failures usually coincide with alterations in apoptotic pathways, in particular the inactivation of the p53 pathway (53) or with alterations in the Bcl-2 family (28). p53 is activated by a diverse number of signals that stress the cell, including hyperproliferation, hypoxia, and DNA damage (63). Normally p53 levels and function are harnessed by Mdm2, which is a p53 transcription target that blocks p53's transactivation functions (41) and programs p53 for destruction by initiating p53 ubiquitination and shuttling it to the cytosol for degradation by the proteasome (23, 51, 60). Signals that activate p53 disrupt this interaction by inducing phosphorylation of p53 (3, 10) and/or Mdm2 (39) or by inducing the ARF nucleolar tumor suppressor, which binds to and inhibits Mdm2 functions (50, 64). The net result is stabilization of p53 and activation of its transcription functions. In turn, p53 either induces G1 arrest by inducing transcription of the cyclin-dependent kinase (cdk) inhibitor p21Cip1, allowing time for DNA repair, or eliminates cells by triggering apoptosis. p53-induced apoptosis is thought to occur through its ability to induce proapoptotic Bcl-2 family members, including Bax, Noxa, and Puma (63).
Myc's propensity to provoke apoptosis presents a paradox as to why MYC genes are so often activated in human tumors. In part, this may reflect Myc's ability to function as the angiogenic switch (5, 48), but Myc also appears to antagonize checkpoints that harness the cell cycle. For example, in immortal and tumor-derived cell lines Myc overexpression antagonizes p53's ability to activate p21Cip1 following DNA damage (56). Here we report that in mouse embryonic fibroblasts (MEFs) and B cells derived from Eμ-myc mice, Myc activation acts in synergy with gamma irradiation (γ-IR) to trigger apoptosis. Surprisingly, Myc also sensitizes p53-deficient MEFs to γ-IR-induced apoptosis, and we pinpoint this response to Myc's ability to suppress Bcl-XL expression. These results may explain why MYC-overexpressing tumors are at least initially responsive to chemotherapy, and they suggest that targeting Bcl-XL function or expression is an attractive means to compromise the survival of tumors bearing mutations in p53.
MATERIALS AND METHODS
Cell culture.
MEFs from embryonic day 12.5 (E12.5) embryos (wild type, p53−/−, bcl-x+/−, or bcl-x−/− on a C57BL/6 background and bcl-2+/+, bcl-2+/−, or bcl-2−/− on an Sv129 background) were explanted and maintained on a 3T9 protocol (9 × 105 cells transferred at 3-day intervals) (65) and propagated in Dulbecco's modified Eagle's medium plus 10% fetal bovine serum (FBS), 2 μM glutamine, 0.1 μM nonessential amino acids, 55 mM 2-mercaptoethanol, and 10 mg of gentamicin (GIBCO)/ml. p53-deficient MEFs were derived from mice provided by Gerard Zambetti (St. Jude Children's Research Hospital), and bcl-X- and bcl-2-deficient MEFs were derived from mice provided by Craig Thompson (Abramson Cancer Center, University of Pennsylvania) and David Woo (UCLA), respectively. All results were confirmed using multiple MEF isolates.
Viral infections.
The Myc-ER cDNA, provided by Dean Felsher and J. Michael Bishop (University of California, San Francisco), was subcloned into the EcoRI site of the MSCV-IRES-GFP retroviral vector (38). The MSCV-hBcl-XL-IRES-YFP virus expression construct was kindly provided by J.-C. Marine. MSCV-Myc-ER-IRES-GFP, control MSCV-IRES-GFP, and MSCV-hBcl-XL-IRES-YFP viruses were produced by cotransfection of 293T cells with vector and helper virus plasmids. Viruses were harvested at intervals, pooled, filtered, and used to infect GPE86 cells (American Type Culture Collection) in the presence of 8 mg of Polybrene/ml, generating stable retroviral producer cell lines. After subsequent cell sorting of green fluorescent protein (GFP)- and yellow fluorescent protein (YFP)-positive cells, the three producer lines (GFP-only, Myc-ER-GFP, and hBcl-XL-YFP) were expanded and their supernatants were used to infect early-passage MEF cultures (wild type or p53−/−) in the presence of 8 mg of Polybrene/ml. Infected GFP/YFP-positive MEFs were sorted by fluorescence-activated cell sorter (FACS) analysis and expanded in culture. Expression of the Myc-ER fusion protein and/or human Bcl-XL protein was confirmed by immunoblotting and was comparable in wild-type and p53−/− MEFs.
Clonogenic assays.
Effects of Myc activation and γ-IR on long-term growth of MEFs were assessed by clonogenic assays. Briefly, cells were plated in 10-cm plates (5,000 cells/plate) and allowed to adhere overnight. Cells were then treated with 1 μM 4-hydroxytamoxifen (4-HT; Sigma, St. Louis, Mo.) for 24 h to activate Myc-ER and were then exposed to 5 Gy of irradiation from a 137Cs irradiator. Twenty-four hours following irradiation, cells were washed and refed with fresh medium, and the colony number was scored 10 days later by staining with 1% Coomassie dissolved in 50% methanol-50% phosphate-buffered saline (PBS).
Viability and apoptosis assays.
To assess the effects of Myc activation with and without γ-IR on cell survival, MEFs were seeded at 5 × 105 cells/ml in a 100-mm plate in the absence or presence of 1 μM 4-HT for 24 h at 37°C prior to exposure of 5 Gy of irradiation. Samples were then harvested at specific intervals, and cell viability was determined by trypan blue dye exclusion. Apoptosis was determined by propidium iodide staining, with quantification of the percentage of subdiploid nuclei and verification morphologically on May-Grunwald-Giemsa-stained cytospin smears. Annexin V-fluorescein isothiocyanate (FITC) staining was carried out as described by the manufacturer (BioWhittaker Inc., Walkersville, Md.). Briefly, 2 × 105 to 5 × 105 MEFs were washed in PBS and resuspended in 190 μl of binding buffer (10 mM HEPES [pH 7.4], 140 mM NaCl, 2.5 mM CaCl). Ten microliters of annexin V-FITC was added and incubated for 10 min, and the cells were washed once and resuspended in 190 μl of binding buffer. To this was added 10 μl of 20-μg/ml propidium iodide stock solution. Cells were then analyzed for the presence of apoptotic cells by FACS analysis. To detect cell death in B cells derived from untreated or treated (2-Gy γ-IR) wild-type or Eμ-myc mice, we stained cytospins (5 × 104 cells) with the terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling (TUNEL) method using the ApopTag fluorescent detection kit (Serological Corporation, Norcross, Ga.) according to the instructions of the manufacturer. Slides were then washed 20 times and visualized with FITC. Finally, slides were mounted using Fluoromount-G antifade reagent (Southern Biotechnology Associates, Birmingham, Ala.) and analyzed by confocal microscopy.
Irradiation of Eμ-myc transgenic mice.
The inbred C57BL/6 Eμ-myc transgenic mouse strain was kindly provided by Alan Harris (Walter and Eliza Hall Institute, Melbourne, Australia) and Charles Sidman (University of Cincinnati). At the start of the experiment, four groups (wild type mock irradiated, Eμ-myc mock irradiated, wild type γ-irradiated, and Eμ-myc γ-irradiated) containing eight mice each were analyzed for their differential blood counts. One week after blood sampling, one group each of wild-type and Eμ-myc mice was subjected to 2 Gy of whole-body irradiation, whereas the other two groups were left untreated. Mice were analyzed 4 h following treatment. B cells were purified from the bone marrow and spleens of untreated and γ-IR-treated mice by FACS for B220 and immunoglobulin M (IgM).
Cell cycle analyses.
B220-positive B cells were collected from the bone marrow of wild-type and Eμ-myc mice by FACS, centrifuged, and resuspended in 1 ml of propidium iodide staining solution (0.05 mg of propidium iodide per ml, 0.1% sodium citrate, 0.1% Triton X-100). Immediately prior to flow cytometric analysis, each sample was treated at room temperature with 5 μg of DNase-free RNase (Calbiochem, San Diego, Calif.)/ml for 30 min. Samples were then filtered through a 40-μm-pore-size nylon mesh. Fluorescence (λ = 563 to 607 nm) emitted from propidium iodide-DNA complexes was measured from approximately 2 × 105 cells with a FACScan flow cytometer (Becton Dickinson Immunocytometry, San Jose, Calif.). The ModFit computer program (Verity Software House, Topsham, Maine) was used to determine the percentage of cells within the G1, S, and G2-M phases of the cell cycle.
Immunoblotting.
Whole-cell protein extracts from MEFs and B cells were isolated as previously described (20, 65). Protein (50 μg per lane for MEF cultures and 10 μg per lane for primary B cells) was electrophoretically separated in 10% polyacrylamide gels containing sodium dodecyl sulfate. Proteins were transferred to nitrocellulose membranes (Protran; Schleicher & Schuell, Dassel, Germany) and blotted with antibodies specific for murine p53 (Ab-7; Calbiochem), Ser15 p53 (Cell Signaling, Beverly, Mass.), c-Myc (06-340; Upstate Biotechnology, Lake Placid, N.Y.), Bcl-2 (15021A; PharMingen, San Diego, Calif.), Bcl-XL (B22260; Transduction Labs, San Diego, Calif.), Bax (13686E; PharMingen), Puma (ab9643; Abcam, Cambridge, United Kingdom), caspase-9 (Cell Signaling), cytochrome c (Santa Cruz), poly(ADP)ribose polymerase (PARP; Oncogene Research, Boston, Mass.), and β-actin (Sigma Chemical). To detect human Bcl-XL expression, another antibody (B22630; Transduction Labs) was used. Following incubation with primary antibodies, the blots were then incubated with appropriate anti-mouse or anti-rabbit Ig secondary antibodies (Amersham Pharmacia, Piscataway, N.J.). Bound immunocomplexes were detected by enhanced chemiluminescence (Amersham Pharmacia) or ECL Supersignal (Pierce, Rockford, Ill.).
Northern blotting, RT-PCR, and real-time PCR analyses.
Total RNA was isolated from MEFs by using TRIzol reagent as per the manufacturer's directions (Life Technologies, Grand Island, N.Y.) at the indicated intervals following the addition of 1 μM 4-HT and/or treatment with 5 Gy of γ-IR. For reverse transcription-PCR (RT-PCR), the RNA was first DNase treated and purified using phenol-chloroform extraction. The first-strand reaction was then performed using a first-strand synthesis kit (Roche) according to the manufacturer's instructions. Primers for PCR were bcl-x sense (5′-AGCAACCGGGAGCTGGTGGTCGAC-3′) and bcl-x antisense (5′-GACTGAAGAGTGAGCCCAGCAGA-3′). The PCR product was purified and cloned into pGEM-Teasy (Promega, Inc.) and verified by sequencing. To obtain probes for Northern blot analyses, the clones were cut with NotI and the insert was gel purified. For Northern blot analysis, 20 μg of total RNA per lane was separated on a formaldehyde-containing 1.6% agarose gel and transferred to a positively charged nylon membrane. The membrane was probed with cDNA probes that had been labeled using [32P]dCTP and ReadyPrime (Amersham Pharmacia Biotech). For real-time PCR analysis, p53−/− MEFs infected with MSCV-IRES-GFP or MSCV-Myc-ER-IRES-GFP viruses were treated for 24 h with 4-HT followed by 5 Gy of γ-IR. RNA was extracted using the RNeasy mini kit (Qiagen) with an additional DNase treatment according to the manufacturer's recommendation. Real-time PCR was performed using the TaqMan EZ RT-PCR kit (Applied Biosystems) and primers and probes directed against bcl-x and acidic ribosomal protein P0 (internal positive control), in an ABI Prism 7700 sequence detector. Relative levels were determined using the ΔΔCT method described by ABI. Primer and probe sequences are available upon request.
Comet assays.
Comet assays were performed essentially as described by the manufacturer (Trevigen, Gaithersburg, Md.). Briefly, cells (105) were suspended in 1% low-melting-point agarose in PBS (pH 7.4) and pipetted onto the provided glass microscope slides. The agarose was allowed to set at 4°C for 30 min, and the slides were then immersed in lysis solution at 4°C for 1 h to dissolve cellular proteins and lipids. Slides were then placed in alkali solution for 1 h at room temperature in the dark. Slides were immersed in 1× Tris-borate-EDTA (TBE) buffer for 5 min twice and transferred to a horizontal electrophoretic apparatus containing 1× TBE buffer for 10 min. Slides were visualized using fluorescence microscopy (fluorescein filter), and 100 cells were counted per sample.
Confocal immunofluorescence analysis of Ser139-H2AX.
For analysis of phosphorylation of Ser139 of histone 2AX (H2AX), cells were grown on coverslips and irradiated (5 Gy), and samples were collected at 6 and 12 h postirradiation. Cells were rinsed briefly with PBS, fixed in 1:1 methanol-acetone for 30 min at −20°C, and air dried for 20 min. Fixed cells were blocked with 10% FBS-PBS, incubated at room temperature for 30 min, and stained for 1 h with a polyclonal antibody specific for Ser139-phosphorylated histone H2AX (1:100 dilution in 1% FBS-PBS [Trevigen]). Following 10 washes in PBS, primary antibody binding was visualized using an anti-rabbit Cy3 conjugate incubated for 30 min at room temperature. Coverslips were washed 10 times prior to being mounted onto slides and analyzed by confocal microscopy.
RESULTS
c-Myc sensitizes MEFs and B cells to γ-IR-induced apoptosis.
Recent studies in immortal and tumor-derived cell lines have suggested that Myc can bypass p53-mediated cell cycle arrest following DNA damage (56, 57) and may instead promote an apoptotic phenotype. To investigate Myc's effects on the DNA damage response in a more physiologically relevant setting, we evaluated primary MEFs engineered to express a conditionally activatable form of c-Myc, Myc-ER (which can be activated by the estrogen receptor [ER] agonist 4-HT [34]), as well as precancerous B cells derived from Eμ-myc transgenic mice, which overexpress c-myc in the B-cell compartment by virtue of the Ig heavy chain enhancer (Eμ) (1).
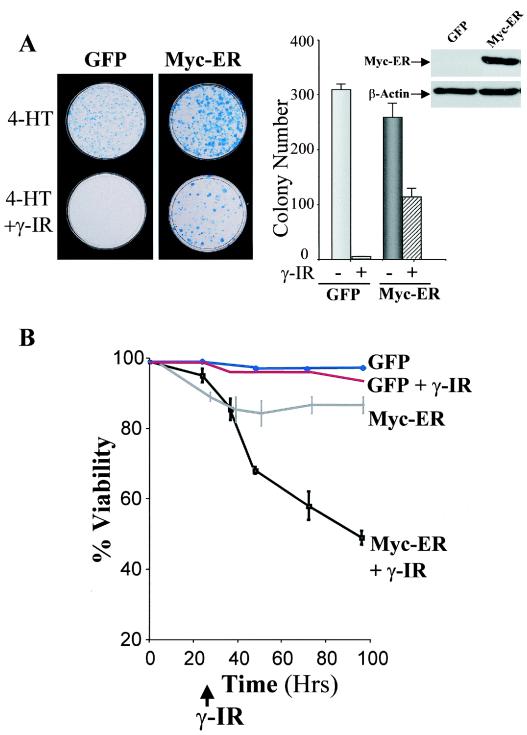
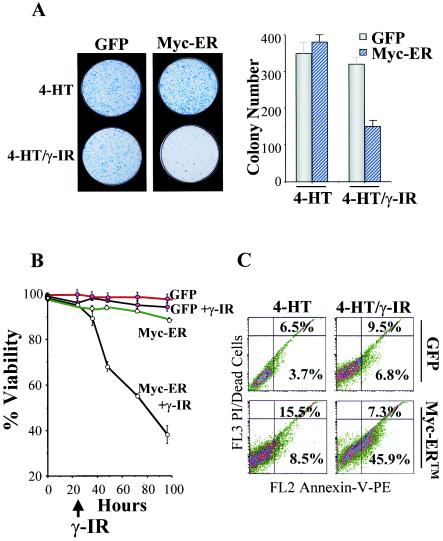
Early-passage MEFs were isolated from E12.5 wild-type embryos and infected with the MSCV-Myc-ER-IRES-GFP-expressing retrovirus or with GFP-only virus, and pools of infected cells expressing GFP were isolated by FACS and expanded in culture. To assess effects of γ-IR on long-term growth, clonogenic assays were performed in GFP-only- and Myc-ER-expressing MEFs either treated with 4-HT alone or also exposed to 5 Gy of γ-IR. As expected for cells having wild-type p53, treatment of GFP-only-expressing cells with γ-IR compromised their growth (Fig. 1A). Colony numbers were also reduced (approximately threefold) in Myc-expressing MEFs following exposure to 5 Gy of γ-IR, and the sizes of these colonies were often smaller (Fig. 1A). Therefore, γ-IR impairs the proliferative potential of MEFs, even when they are engineered to overexpress Myc.
FIG. 1.
The survival of Myc-expressing MEFs is compromised. (A) GFP-only- or Myc-ER-expressing MEFs (right panel inset) were treated with 4-HT (1 μM) for 24 h and irradiated with 5 Gy, and after 24 h 5 × 103 cells were plated in fresh growth medium with 4-HT. Colonies were stained with Wright-Giemsa after 10 days. The right panel shows the mean colony numbers from three separate experiments. (B) γ-IR triggers cell death in Myc-expressing MEFs. GFP-only- and Myc-ER-expressing MEFs were treated with 4-HT (1 μM) for 24 h and then left untreated or were irradiated with 5 Gy. At the indicated intervals, viability was determined by trypan blue dye exclusion.
To determine whether reductions in growth potential of Myc-expressing cells exposed to γ-IR were associated with increases in cell death, their viability was assessed. Although GFP-only-expressing MEFs failed to proliferate, these cells remained viable following γ-IR treatment. As expected (21, 24), Myc-ER-expressing MEFs cultured in serum were generally resistant to apoptosis. In contrast, when these cells were exposed to γ-IR they underwent rampant cell death (Fig. 1B).
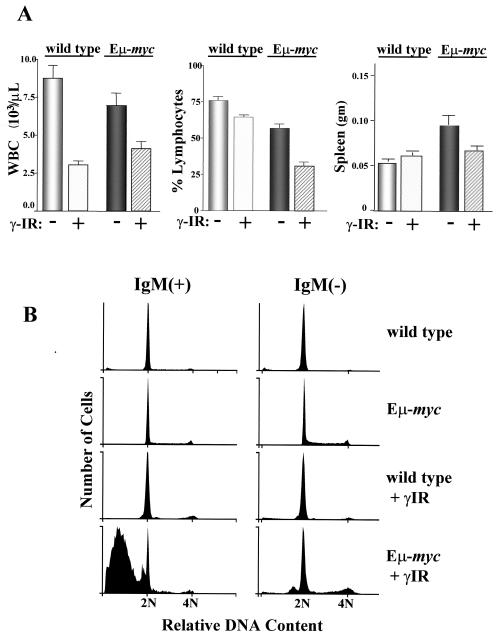
The effects of γ-IR in compromising the survival of Myc-expressing MEFs could also be due to effects of ex vivo culture (58). We therefore addressed whether treatment with low doses of γ-IR would preferentially affect the survival of precancerous (6-week-old) B cells of Eμ-myc transgenic mice. These mice undergo a characteristic hyperproliferation of the B-cell compartment before developing a clonal lymphoma at 4 to 6 months of age (1), and this is usually associated with a pronounced lympho-leukemia. Age-matched wild-type and Eμ-myc transgenics were either mock treated or treated with 2 Gy of whole-body irradiation. Within 4 h of treatment there were significant reductions in total white blood cell counts (WBC) in both wild-type and Eμ-myc transgenic mice (Fig. 2A, left panel). However, in Eμ-myc transgenics there was a selective and marked decrease in the number of peripheral blood lymphocytes (Fig. 2A, middle panel) and in the weight of the spleen (Fig. 2A, right panel). Furthermore, cell cycle analyses of IgM-positive pre-B/B cells and IgM-negative pro/pre-B cells in the bone marrow revealed a dramatic increase in sub-G1 populations of bone marrow cells isolated from irradiated Eμ-myc mice, whereas this was not evident in irradiated B cells of wild-type littermates (Fig. 2B). Therefore, c-Myc-overexpressing cells also exhibit increased sensitivity to γ-IR in vivo.
FIG. 2.
γ-IR selectively compromises B-lymphocyte survival in Eμ-myc transgenic mice. Wild-type and Eμ-myc transgenic littermates were left untreated or were treated with 2 Gy of irradiation (n = 8 for each group). (A) After 4 h the WBC number (left panel) and the percent lymphocytes (middle panel) were evaluated in the peripheral blood of these mice. In addition, the weights of the spleens of these mice were determined (right panel). Although WBC numbers were reduced upon γ-IR treatment in both wild-type and Eμ-myc mice, there were marked reductions in lymphocyte numbers and spleen weights evident in Eμ-myc mice. (B) Bone marrow was harvested from these same animals, and B220+ cells were stained with propidium iodide and analyzed by FACS. Note the marked increase in the numbers of sub-G1 IgM+ and IgM− Eμ-myc cells when exposed to γ-IR.
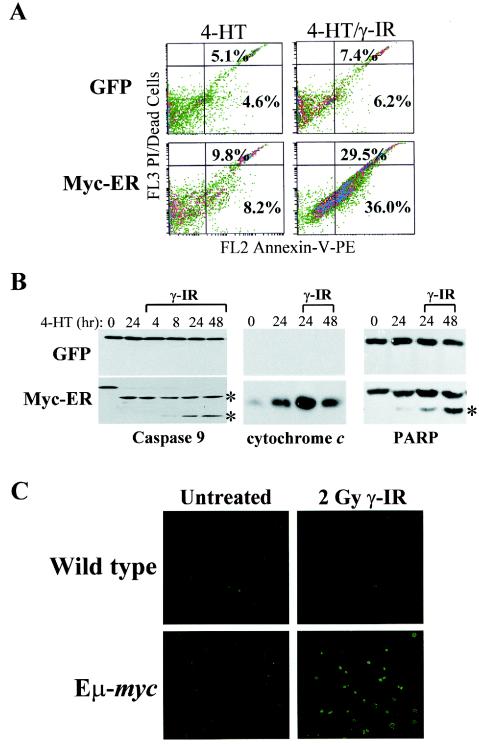
Myc sensitizes cells to many apoptotic insults (2, 21), and we therefore addressed whether the loss in the viability of Myc-expressing cells following γ-IR was indeed due to apoptosis. FACS analysis of annexin V, an early marker of apoptosis, demonstrated modest increases in annexin V in Myc-expressing MEFs treated with 4-HT versus GFP-only-expressing MEFs (Fig. 3A). In contrast, there were marked increases in annexin V positivity in Myc-expressing MEFs also exposed to γ-IR (Fig. 3A). Furthermore, the appearance of other hallmarks of apoptosis were also augmented in Myc-ER-expressing cells exposed to γ-IR, including increases in the release of cytochrome c from mitochondria and the cleavage of caspase-9 and the caspase-3 substrate PARP (Fig. 3B). Finally, TUNEL analysis established that the rampant cell death observed in B cells from the bone marrow of Eμ-myc transgenics exposed to γ-IR was also due to apoptosis (Fig. 3C). Therefore, Myc activation sensitizes cells to γ-IR-induced apoptosis.
FIG. 3.
γ-IR triggers apoptosis in Myc-expressing MEFs and in Eμ-myc transgenic B cells. (A) FACS histograms of PE-annexin V binding. GFP-only- or Myc-ER-expressing MEFs were treated with 4-HT (1 μM) for 24 h and irradiated with 5 Gy, and after 24 h cells were stained with annexin-V-PE and propidium iodide (PI). The percentages of annexin V- and PI-negative and -positive cells are given in each quadrant. Results shown are representative of three independent experiments performed with two different MEF cell cultures. (B) γ-IR triggers caspase-9 and PARP cleavage, and the release of cytochrome c into the cytosol, in Myc-expressing MEFs. The indicated cells were treated for 24 h with 4-HT (1 μM) and treated with γ-IR, and lysates were analyzed at the indicated intervals by Western blotting for caspase-9 (left panel), the cytosolic fraction of cytochrome c (29) (middle panel), and PARP (right panel). The asterisk indicates the cleaved forms of caspase-9 and PARP. (C) γ-IR triggers apoptosis in B cells of 6-week-old (precancerous) Eμ-myc transgenic mice. Wild-type and Eμ-myc transgenic littermates were treated with 2 Gy of γ-IR. After 4 h bone marrow was harvested and sorted for B220+ cells by FACS, and cytospins were assessed for apoptotic cells by immunofluorescence using TUNEL assays. TUNEL-positive apoptotic cells appear bright green. Representative fields are shown.
c-Myc acts in synergy with γ-IR to trigger apoptosis.
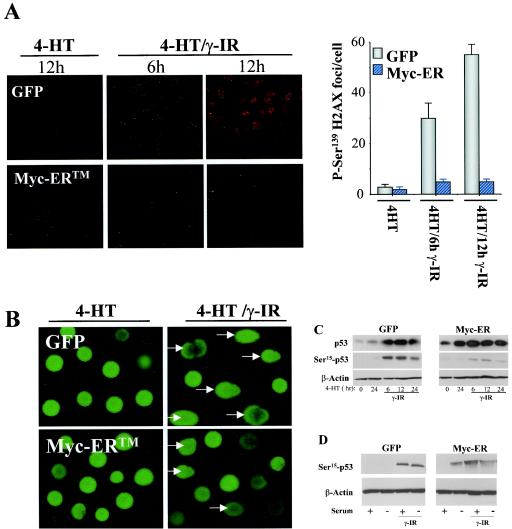
Myc has been suggested to directly induce DNA damage, in part from the detection of Ser139-phosphorylated H2AX, which occurs following double-stranded breaks (61). However, these studies were performed under serum-starved conditions, a condition where Myc is known to trigger apoptosis (21). We therefore evaluated whether Myc activation alone was capable of provoking the DNA damage response in MEFs cultured in serum, a scenario in which Myc rather sensitizes cells to γ-IR-induced apoptosis. Confocal immunofluorescence analyses with anti-Ser139-H2AX antibody demonstrated rapid and sustained induction of Ser139-H2AX-positive foci in GFP-only-expressing MEFs following γ-IR (Fig. 4A). By contrast, Myc activation following 4-HT treatment failed to induce phosphorylation of H2AX on Ser139 (Fig. 4A). On the contrary, rather than enhance the DNA damage response, Myc activation dampened the response, as levels of Ser139-H2AX-positive foci were reduced in Myc-expressing MEFs exposed to γ-IR (Fig. 4A).
FIG. 4.
Myc alone does not induce the DNA damage response in MEFs. (A) Detection of Ser139-H2AX foci in GFP-only- or Myc-ER-expressing MEFs treated with 4-HT with or without γ-IR. MEFs were treated with 4-HT (1 μM) for 24 h and irradiated with 5 Gy, and cells were harvested at the indicated intervals. Following fixation and blocking procedures, cells were incubated with anti-Ser139-H2AX antibody and the respective fluorophore-conjugated secondary antibody (Cy3 anti-rabbit). A representative figure from three separate experiments is shown (upper panels), and at right the data are plotted as the number of fluorescent Ser139-H2AX foci per cell. A minimum of 100 counts for three independent fields is shown. (B) Comet assays of DNA damage in GFP-only- or Myc-ER-expressing MEFs. Cells were treated with 4-HT (1 μM) for 24 h, irradiated with 5 Gy, and exposed to an electric field (migration of the DNA in the electric field is from left to right). Arrows indicate GFP-positive cells with obvious comets. (C) p53 phosphorylation on Ser15 does not occur following Myc activation in MEFs. Cells were treated with 4-HT (1 μM) for 24 h and irradiated with 5 Gy, and whole-cell lysates were evaluated for levels of Ser15-phosphorylated p53 and compared to steady-state levels of total p53. Note the robust induction of Ser15-phosphorylated p53 in GFP-only-expressing MEFs treated with γ-IR; this was not detectable in Myc-ER-expressing MEFs treated with 4-HT, despite the robust induction of total p53. The blots shown were exposed for the same length of time. (D) Serum suppresses Myc's ability to provoke Ser15 phosphorylation of p53. The indicated MEFs were treated with 4-HT and cultured in replete growth medium (lanes 1, 3, 5, and 7) or in 0.1% FCS medium (lanes 2, 4, 6, and 8). Cells were then left untreated (lanes 1, 2, 5, and 6) or were treated with 5 Gy of γ-IR (lanes 3, 4, 7, and 8). After 24 h cells were harvested and levels of phospho-p53Ser15 were determined by immunoblot analysis.
A standard method to detect DNA damage is the comet assay, which directly measures many forms of DNA damage using a single-cell gel electrophoresis assay (32). As expected, exposure of GFP-only-expressing MEFs to 5 Gy of γ-IR induced obvious comets in most cells (Fig. 4B). Comets were also obvious, but again to a lesser extent, in Myc-expressing MEFs following exposure to γ-IR (Fig. 4B). Again, however, activation of Myc alone failed to induce comet formation in any Myc-expressing cells (Fig. 4B).
As a final measure to test whether Myc could directly induce the DNA damage response, we also assessed the phosphorylation status of Ser18 of p53 (the murine equivalent of human Ser15, but for simplicity hereafter referred to as Ser15), which is rapidly phosphorylated by the Atm kinase following exposure to γ-IR (10). As expected, treatment of GFP-only-expressing MEFs with γ-IR induced steady-state levels of p53, and under these conditions p53 was indeed phosphorylated on Ser15 (Fig. 4C). In contrast, activation of c-Myc alone did not induce phosphorylation of p53 on Ser15, despite the fact that Myc induced steady-state levels of total p53 (Fig. 4C). Rather, levels of Ser15 p53 phosphorylation were reduced in Myc-expressing MEFs treated with γ-IR (Fig. 4C), again suggesting that Myc can impair at least some aspects of the DNA damage response. Therefore, by three independent measures, Myc activation in MEFs cultured in growth medium, with its full complement of survival factors, does not lead to direct DNA damage, but rather acts in synergy with γ-IR to trigger apoptosis.
These findings raised questions as to whether Myc's ability to provoke a DNA damage response is suppressed by serum factors. We therefore evaluated GFP-only- and Myc-expressing MEFs for activation of the DNA damage response in cells cultured in replete medium (10% fetal calf serum [FCS]) or in 0.1% FCS. Strikingly, phosphorylation of p53 on Ser15 was evident following Myc activation in the absence, but not presence, of serum (Fig. 4D, compare lanes 5 and 6). Therefore, Myc's ability to provoke a DNA damage response is suppressed by serum factors which also suppress Myc-induced apoptosis (21, 24).
Myc targets Bcl-XL, Bcl-2, and Puma to sensitize MEFs to γ-IR.
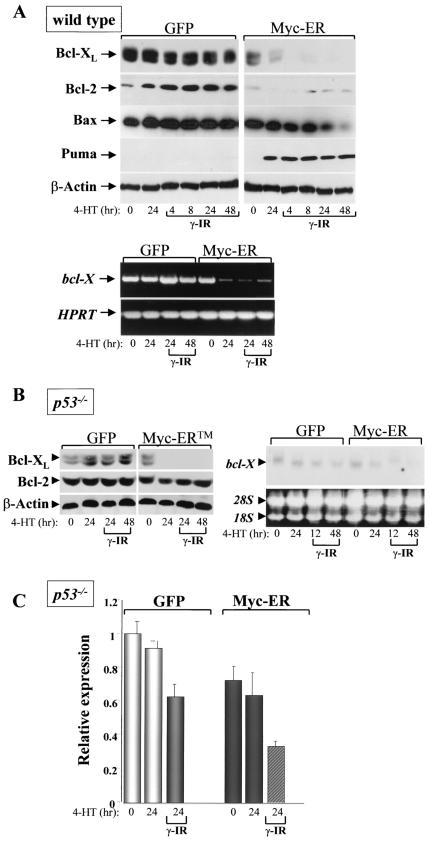
In addition to activating the ARF-p53 apoptotic checkpoint, in primary hematopoietic cells c-Myc also triggers apoptosis by suppressing the expression of the antiapoptotic regulators Bcl-2 and Bcl-XL (20). We therefore assessed the effects of c-Myc on Bcl-2 and Bcl-XL expression in MEFs and in B cells of wild-type and Eμ-myc mice before and following γ-IR. Treatment of GFP-only-expressing MEFs with 5 Gy of γ-IR did not lead to significant changes in the levels of Bcl-2 or Bcl-XL, and there were also no alterations in levels of the proapoptotic Bax protein (Fig. 5A, upper panels). Strikingly, activation of Myc-ER by 4-HT resulted in a profound suppression of both Bcl-XL and Bcl-2 protein levels, and this response was augmented in Myc-expressing cells following exposure to γ-IR (Fig. 5A, upper panels). RT-PCR analyses confirmed that bcl-X suppression in irradiated and nonirradiated Myc-expressing cells occurred at the RNA level (Fig. 5A, lower panels).
FIG.5.
Myc selectively targets the expression of Bcl-2, Bcl-XL, and Puma in MEFs. (A) GFP-only- or Myc-ER-expressing wild-type MEFs were treated for 24 h with 4-HT, irradiated with 5 Gy, and at the indicated intervals harvested for immunoblot analyses (top panels) or RT-PCR analysis of bcl-X and HPRT RNA (lower panels). (B) Only Bcl-XL is suppressed by Myc in p53-defcient MEFs. GFP-only- or Myc-ER-expressing p53-deficent MEFs were treated for 24 h with 4-HT, irradiated with 5 Gy, and at the indicated intervals harvested for immunoblot analyses (left panels) or Northern blot analysis of bcl-X RNA (right panels). Puma expression was not detected in Myc-expressing p53-deficient MEFs (negative data not shown). To control for loading, the levels of 28S and 18S RNA are shown at right. (C) Real-time PCR analysis of bcl-X levels in p53-deficient MEFs engineered to express Myc-ER. The indicated cells were treated for 24 h with 4-HT and irradiated with 5 Gy, and at the indicated intervals RNA was harvested for real-time PCR analysis. The internal control in the real-time reactions is arpp0.
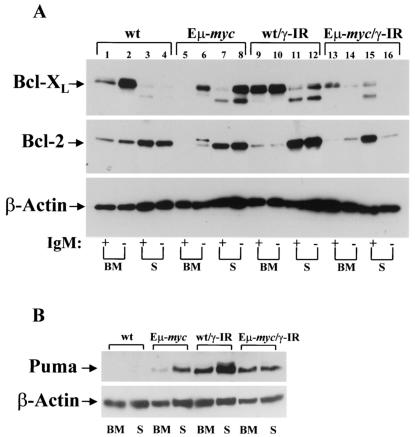
To address whether Myc also influenced Bcl-2 and Bcl-XL expression following γ-IR treatment in vivo, we harvested bone marrow and spleens from 6-week-old wild-type and Eμ-myc transgenic littermates and from these mice treated with 2 Gy of whole-body γ-IR. After 4 h, B cells were isolated using the cell surface markers B220 and IgM by FACS. As expected, in pro/pre- (IgM-negative) B cells from the bone marrow of wild-type mice, Bcl-XL was expressed at high levels, whereas Bcl-2 expression was more prominent in splenic B cells (Fig. 6A, lanes 1 to 4). Interestingly, treatment with γ-IR increased Bcl-XL levels in IgM+ B cells present in bone marrow and in splenic B cells, and it also increased Bcl-2 levels in splenic B cells (Fig. 6A, lanes 9 to 12). In contrast, Bcl-XL and Bcl-2 expression levels were markedly different in Eμ-myc transgenic B cells. First, as expected (20), Bcl-XL and Bcl-2 levels were reduced in bone marrow-derived Eμ-myc B cells (Fig. 6A, lanes 5 and 6). However, these reductions were not evident in splenic B cells, where Bcl-2 levels were comparable to those of wild-type splenic B cells, and Bcl-XL expression was rather elevated in IgM-negative B cells (Fig. 6A, lanes 7 and 8); therefore, the suppression of Bcl-XL and Bcl-2 by Myc is restricted to bone marrow-derived B cells. Second, following treatment with γ-IR, there were marked reductions in both Bcl-XL and Bcl-2 levels in IgM-negative Eμ-myc B cells (Fig. 6A, compare lanes 6 and 8 to lanes 14 and 16). Third, in IgM+ bone marrow-derived transgenic B cells, where the highest levels of apoptosis following γ-IR were observed (Fig. 2B), there were markedly lower levels of Bcl-XL and Bcl-2 compared to those expressed in wild-type cells (Fig. 6A, compare lanes 9 and 13). Therefore, Myc selectively suppresses Bcl-XL and Bcl-2 expression in B cells present in bone marrow, and this response is augmented by γ-IR, which selectively kills these cell populations in vivo (Fig. 2B).
FIG. 6.
Myc targets the expression of Bcl-2, Bcl-XL, and Puma in bone marrow (BM) and splenic (S) B cells of Eμ-myc transgenic mice. Wild-type (lanes 1 to 4 and 9 to 12) and precancerous Eμ-myc transgenic littermates (lanes 5 to 8 and 13 to 16) (n = 8 for each group) were left untreated (lanes 1 to 8) or were treated with 2 Gy of γ-IR (lanes 9 to 16). Four hours later, bone marrow and spleens from each of these four groups of mice were pooled and FACS sorted for B220 and IgM, and 20 μg of extracts was assessed by immunoblotting with antibodies specific for Bcl-2, Bcl-XL, and β-actin (A), or 60 μg of extract from IgM+ B cells was analyzed for Puma levels (B).
p53-induced apoptosis has been variously associated with its ability to induce Bax or Bak (55), which together are required for intrinsic and extrinsic signals that trigger apoptosis (7, 40), and/or to induce the BH3-only factors Puma or Noxa (43, 44, 65). In MEFs, activation of Myc-ER or treatment with γ-IR had little effect on the expression of Bax or Bak (Bax levels actually diminished in Myc-expressing cells treated with γ-IR) or upon the expression of Noxa or Bad, another proapoptotic Bcl-2 family member (Fig. 5A and data not shown). By contrast, the expression of Puma was markedly induced following Myc activation in MEFs (Fig. 5A) and was also elevated in bone marrow and splenic B cells of Eμ-myc transgenic mice (Fig. 6B), but it was not detectable in normal MEFs or B cells (Fig. 5A and 6B). Puma expression was also highly elevated by γ-IR in normal B cells (Fig. 6B) but was not induced by γ-IR in wild-type MEFs (Fig. 5A), indicating there are context-specific effects on Puma induction. In Myc-expressing cells the elevated levels of Puma were sustained following γ-IR treatment (Fig. 5A and 6B). Therefore, Myc's ability to sensitize MEFs and B cells to γ-IR-induced apoptosis is associated with the repression of Bcl-XL and Bcl-2 and with increases in the expression of Puma.
Myc also sensitizes p53-deficient MEFs to γ-IR-induced apoptosis.
Many cancers that overexpress MYC, including Burkitt's lymphoma (BL), respond very well to chemotherapy, despite the fact that one-third of primary BLs harbor mutated p53 alleles (25). We therefore addressed whether Myc might also sensitize p53-deficient MEFs to γ-IR. p53-deficient MEFs were infected with the MSCV-IRES-GFP or MSCV-Myc-ER-IRES-GFP viruses, and infected cells were GFP sorted by FACS. These cells were then expanded in culture, treated with 4-HT for 24 h in growth medium, and challenged with γ-IR. As expected, both GFP-only- and Myc-expressing p53−/− MEFs continued to proceed through the cell cycle following γ-IR (data not shown). Surprisingly, however, γ-IR severely compromised the long-term growth of Myc-expressing p53−/− MEFs (Fig. 7A). Myc-expressing p53−/− MEFs cultured in serum were, as predicted, resistant to apoptosis following the addition of 4-HT (Fig. 7B). However, when further treated with γ-IR, Myc-ER-expressing p53-deficient MEFs underwent rapid apoptosis, as determined by typical changes in cell morphology (data not shown), loss of viability (Fig. 7B), the appearance of annexin V-positive cells (Fig. 7C), and the cleavage of PARP (data not shown). Therefore, Myc also sensitizes p53-deficient MEFs to γ-IR-induced apoptosis.
FIG. 7.
Myc sensitizes p53−/− MEFs to γ-IR-induced apoptosis. (A) GFP-only- or Myc-ER-expressing p53−/− MEFs were treated with 4-HT (1 μM) for 24 h and left untreated or were irradiated with 5 Gy. Twenty-four hours later, 5 × 103 cells were plated in growth medium containing 4-HT. Left panels, colonies were stained with Wright-Giemsa after 10 days. Right panel, the mean colony numbers from three separate experiments is shown. (B) Cells treated as for panel A were examined for apoptosis by trypan blue dye exclusion. (C) Cells treated as for panel A were examined for apoptosis by FACS analyses of propidium iodide (PI)-annexin V-stained cells. The percentages of annexin V- and PI-negative and -positive cells are given in each quadrant. Results shown are representative of three independent experiments performed with two different MEF cell cultures.
In primary hematopoietic cells, Myc suppresses the expression of bcl-X even in cells derived from ARF- and/or p53-deficient mice (20). We therefore evaluated whether this pathway was also operational in p53−/− MEFs and if it was affected by γ-IR. Indeed, Myc activation in p53−/− MEFs suppressed the expression of Bcl-XL, both at the protein and RNA level, and this response was augmented following exposure to γ-IR (Fig. 5B and C). Interestingly, unlike the response in wild-type MEFs (Fig. 5A), Myc failed to suppress Bcl-2 protein expression in p53-deficient MEFs (Fig. 5B); therefore, this response is strictly p53 dependent. In addition, Myc activation failed to induce Puma expression in p53−/− MEFs (data not shown). Therefore, Myc's ability to sensitize MEFs to γ-IR-induced apoptosis is associated with the selective suppression of bcl-X expression.
Targeting of Bcl-XL by Myc is necessary and sufficient to provoke γ-IR-induced apoptosis.
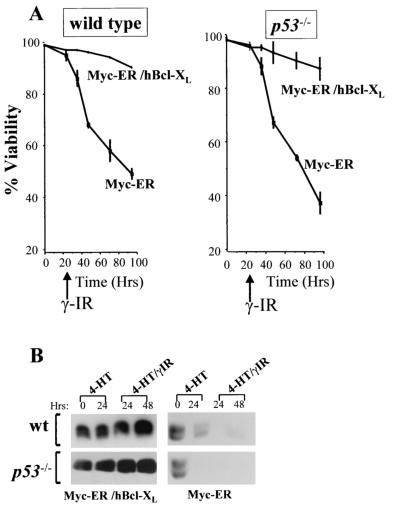
If Myc's ability to suppress Bcl-XL expression was relevant to sensitizing MEFs to γ-IR, then restoration of Bcl-XL should protect Myc-expressing cells from γ-IR-induced apoptosis. We therefore coinfected wild-type and p53−/− MEFs with MSCV-Myc-ER-IRES-GFP plus MSCV-Bcl-XL-IRES-YFP viruses and FACS sorted for doubly fluorescent cells. These cells were expanded in culture, and overexpression of human Bcl-XL was confirmed by immunoblotting using a human-specific Bcl-XL-specific antibody (Fig. 8B). Cells in growth medium were treated with 4-HT for 24 h to activate Myc-ER and then exposed to γ-IR. Bcl-XL overexpression dramatically impaired the apoptotic response of both Myc-expressing wild-type and p53−/− MEFs following exposure to γ-IR (Fig. 8A). Therefore, Myc-mediated suppression of bcl-X is necessary for its ability to sensitize MEFs to γ-IR-induced apoptosis.
FIG. 8.
bcl-X suppression is necessary for Myc to trigger apoptosis following γ-IR. Wild-type and p53−/− MEFs were transduced with MSCV-Myc-ER-IRES-GFP- and MSCV-hBcl-XL-IRES-YFP-expressing viruses, and doubly transduced MEFs were compared to those expressing Myc-ER alone. (A) GFP-only- or Myc-ER-expressing MEFs were treated with 4-HT (1 μM) for 24 h and irradiated with 5 Gy, and viability was determined by trypan blue dye exclusion at the indicated intervals. Viability experiments shown are representative of three independent experiments performed in triplicate. (B) Immunoblot analysis with an antibody specific for human Bcl-XL (left panels) demonstrated that overexpression of human Bcl-XL protein was sustained in Myc-expressing cells exposed to γ-IR, contrary to results with endogenous mouse Bcl-XL (right panels).
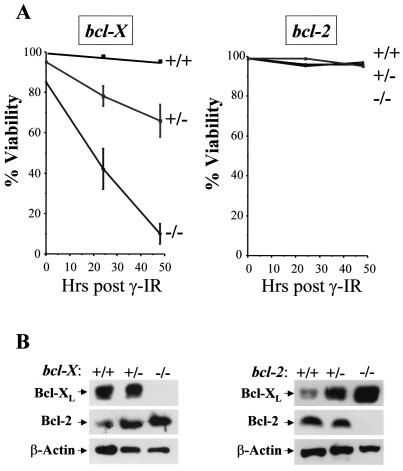
The profound reductions in Bcl-XL levels following Myc activation in either wild-type or p53−/− MEFs suggested that loss of bcl-X alone might render MEFs susceptible to γ-IR-induced apoptosis. Loss of bcl-X in mice results in an embryonic lethality at E13.5 with associated apoptosis of both hematopoietic cells and neurons (42). We therefore derived several wild-type, bcl-X+/−, and bcl-X−/− MEF lines from E12.5 embryos, and early-passage, exponentially growing cells were assessed for their sensitivity to γ-IR. Although wild-type MEFs were totally resistant to γ-IR-induced apoptosis, bcl-X−/− MEFs were exquisitely sensitive to γ-IR and died within 2 days of exposure (Fig. 9A), despite being cultured in growth medium containing a full complement of survival factors. Furthermore, haploinsufficiency effects were also evident, as bcl-X+/− MEFs were intermediate in their sensitivity to γ-IR-induced death. Importantly, the effects of bcl-X loss on rendering MEFs sensitive to γ-IR were specific, as treatment of bcl-2+/− and bcl-2−/− MEFs with 5 Gy of γ-IR did not induce apoptosis (Fig. 9A) and restoration of Bcl-XL expression in bcl-X−/− MEFs rendered these cells resistant to γ-IR-induced death (data not shown). In part this could reflect a compensatory up-regulation of Bcl-XL protein in bcl-2−/− MEFs (Fig. 9B, right panels); however, a similar compensatory up-regulation of Bcl-2 protein levels in bcl-X−/− MEFs (Fig. 9B, left panels) did not protect these cells from γ-IR-induced apoptosis. Therefore, bcl-X loss is also sufficient to selectively sensitize MEFs to γ-IR-induced apoptosis.
FIG. 9.
bcl-X loss, but not loss of bcl-2, radiosensitizes MEFs. (A) MEFs prepared from the indicated embryos were cultured in growth medium and exposed to 5 Gy of γ-IR, and their viability was assessed by trypan blue dye exclusion. Viability experiments shown are representative of three independent experiments with three independent MEF cell cultures, performed in triplicate. (B) Immunoblot analysis of Bcl-XL and Bcl-2 protein levels in bcl-X+/+, bcl-X+/−, and bcl-X−/− MEFs (left panels) and in bcl-2+/+, bcl-2+/−, and bcl-2−/− MEFs (right panels).
DISCUSSION
c-Myc functions as a central oncogenic switch, yet in normal cells c-Myc overexpression also triggers apoptosis (2, 21) and regulators of Myc-induced apoptosis are disabled in most tumor types. This suggests that blocking apoptosis is likely essential for Myc to promote tumorigenesis (19, 47, 54). However, it has also been proposed that Myc provokes genomic instability (37) and that this contributes to Myc's ability to transform a cell. For example, Myc overexpression in immortal and tumor-derived cell lines can lead to gross chromosomal aberrations and amplifications, and BLs bearing the MYC/Ig translocations do have recurrent chromosomal abnormalities (9).
Recently, Myc's capacity to trigger genomic instability has been linked to its ability to directly induce DNA damage (61). This scenario is fundamentally different from one in which Myc overexpression selects for mutations in p53 (66) which in turn result in genomic instability. Here we have shown that Myc does not directly induce the DNA damage response in MEFs but rather acts in synergy with γ-IR to induce apoptosis. Surprisingly, this response occurs even in MEFs lacking p53, demonstrating that (i) p53 is dispensable for the apoptotic response of Myc-expressing cells to γ-IR, and (ii) Myc activates other pathways that cooperate with γ-IR to induce apoptosis. In wild-type MEFs and in B cells derived from Eμ-myc transgenic mice, the response to Myc and γ-IR is a double-edged sword which likely involves the proapoptotic Puma protein and the suppression of the antiapoptotic regulators Bcl-XL and Bcl-2 (Fig. 5 and 6). However, only one of these targets, Bcl-XL, appears to be essential for the response, as Bcl-XL expression is selectively targeted in p53−/− MEFs and bcl-X loss alone renders cells sensitive to γ-IR.
Clonogenic and apoptosis assays of wild-type and p53−/− MEFs revealed that Myc sensitized both cell types to radiation. The sensitivity of p53−/− MEFs expressing Myc to γ-IR was surprising, as p53 loss alone renders cells resistant to DNA damage (35) or to Myc-induced apoptosis when MEFs are deprived of serum (66). Therefore, the apoptotic response is dramatically altered in the context of both Myc overexpression and γ-IR, which alone do not induce death in MEFs cultured in serum but when combined provoke cell suicide. These results contrast with those of Vafa et al. (61), which rather showed that Myc alone was sufficient to directly induce DNA damage. This could reflect differences in cell type and/or the fact that their studies were carried out in the absence of survival factors, a condition known to give rise to oxidative stress (36) and to relieve repression of Myc-mediated apoptosis (24). Indeed, as shown here, serum matters a great deal to the response, as Myc activation in low serum can provoke Ser15 phosphorylation of p53, a hallmark of the response to γ-IR (Fig. 4D). At present it is unclear whether the DNA damage response would be triggered by Myc alone in vivo, as the apoptotic index of B cells from Eμ-myc transgenic mice is rather low but is dramatically enhanced following treatment with γ-IR (Fig. 2B). Furthermore, it is difficult to reconcile how triggering DNA damage could be a selective advantage to a tumor cell. On the other hand, it is also unlikely that in vivo cells are bathed in an excess of survival factors, so one needs to keep an open mind on the issue.
The response of cells to γ-IR is known to be cell context specific, as some cells, for example thymocytes, undergo rapid apoptosis, whereas others (e.g., fibroblasts and epithelial cells) undergo cell cycle arrest (22). Generally, p53 is considered the key regulator of the response to γ-IR, yet DNA damage induced by γ-IR plus cisplatin treatment in mice induces a transient collapse of the hematopoietic compartment in p53-null mice (49). Our analyses of Myc-expressing cells also revealed p53-dependent and -independent components to the γ-IR apoptotic response. One regulator of the p53-dependent pathway appears to be Puma, as marked increases in Puma expression are evident in wild-type MEFs (but not in p53−/− MEFs) following Myc activation, and Puma expression is also elevated in the B cells of Eμ-myc mice and in normal B cells exposed to γ-IR. A second p53-dependent target appears to be Bcl-2, as Bcl-2 expression is suppressed by Myc in MEFs, yet this fails to occur in p53−/− cells. This is in stark contrast to the p53-independent response, where Myc still suppresses bcl-X in p53−/− MEFs and where Myc plus γ-IR still induces rapid cell death. Notably, Myc-mediated suppression of bcl-X also occurs in vivo in the B cells of Eμ-myc transgenic mice and, thus, this pathway also likely contributes to the demise of these B cells when Eμ-myc transgenics are exposed to γ-IR. Second, enforced expression of Bcl-XL in Myc-expressing MEFs abolished γ-IR-induced apoptosis. Finally, the loss of bcl-X, but not bcl-2, was sufficient to render MEFs sensitive to γ-IR-induced apoptosis. Thus, the suppression of bcl-X by Myc is necessary for it to cooperate with γ-IR to trigger apoptosis, and bcl-X loss alone is sufficient to radiosensitize cells.
There are obvious effects of haploinsufficiency in bcl-X+/− MEFs exposed to γ-IR; therefore, a certain threshold of Bcl-XL protein appears necessary to protect MEFs from this apoptotic pathway. Further, there is an unexpected and striking selectivity to this response, as one would predict that bcl-2 loss should have similar effects but it clearly does not. In part, the radioresistance of bcl-2−/− MEFs could reflect a compensatory up-regulation of endogenous Bcl-XL protein in these cells. However, a similar compensatory increase in Bcl-2 protein in bcl-X−/− MEFs is not protective, indicating that physiological levels of Bcl-XL are needed to guard MEFs from γ-IR-induced apoptosis. These findings agree with the recent work of Weintraub and colleagues who have also demonstrated that loss of Bcl-XL function in human tumor-derived cell lines, and in MEFs, compromises their survival when they are treated with chemotherapeutic agents that provoke the DNA damage response (15). Interestingly, at least two levels of control appear to cooperate to compromise the function of Bcl-XL. Firstly, as shown here, Myc activation leads to reductions in the steady-state levels of Bcl-XL protein that follow reductions in bcl-X transcripts, and Myc's ability to suppress bcl-X promoter activity (J. A. Nilsson and J. L. Cleveland, unpublished results) suggests that this is at least in part a transcriptional response. However, Myc's effects on Bcl-XL protein levels appear more profound, and thus the response could also include effects on bcl-X translation and/or upon turnover of the protein. Secondly, the ability of Bcl-XL to block DNA damage-induced apoptosis is compromised by the deamidation of critical asparagine residues located in the unstructured loop of the protein, and this modification occurs in a p53-independent fashion following treatment with some chemotherapeutic agents (15). However, this modification does not appear to occur in wild-type or p53−/− MEFs expressing Myc with or without exposure to γ-IR, as deamidation is accompanied by marked changes in mobility in sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and using the Bcl-X antibody that detects this modified form of Bcl-XL (15) we have failed to see any changes in the mobility of the Bcl-XL protein (data not shown). Finally, the selective effects of Bcl-XL on the DNA damage response do not necessarily preclude a role for other antiapoptotic Bcl-2 family proteins. In particular, overexpression of Bcl-2 can also selectively block c-Myc's ability to sensitize cells to irradiation (52).
Bcl-XL appears to play a unique function as a guardian against both intrinsic and extrinsic signals that provoke apoptosis. For example, bcl-X is required to protect hematopoietic progenitors during development (42), and this is linked to the intrinsic cell death pathway regulated by cytokines, as the Jak2 tyrosine kinase is specifically required to sustain Bcl-XL expression in myeloid and erythroid progenitors (45; T. A. Baudino, U. B. Keller, and J. L. Cleveland, unpublished results). However, Bcl-XL also plays a critical role in protecting cells from DNA damage, regardless of whether or not they have mutations in the p53 pathway (this study and references 15 and 31), and Bcl-XL expression in tumors is an accurate predictor of response to therapy and prognosis (59, 62). Thus, targeting bcl-X expression and/or function should be a selective and attractive approach to sensitize tumors to chemotherapy and/or radiation.
Acknowledgments
We are grateful for the outstanding technical assistance of Elsie L. White, Chunying Yang, Sara Norton, Rob Jeffers, and Kristen Rothhammer. We also thank the staff of our Animal Resources Center for care and monitoring of animals and members of the Flow Cytometry facility. We thank Martine Roussel and Frederique Zindy for discussion. Finally, we also thank Craig Thompson and David Woo for providing bcl-X+/− and bcl-2−/− mice.
This work was supported in part by grants DK44158 and CA76379 (J.L.C.), Cancer Center CORE grant CA21765, and by the American Lebanese Syrian Associated Charities. U.B.K. is a fellow of the Deutsche Forschungsgemeinschaff (grant KE 222/5-1).
REFERENCES
- 1.Adams, J. M., A. W. Harris, C. A. Pinkert, L. M. Corcoran, W. S. Alexander, S. Cory, R. D. Palmiter, and R. L. Brinster. 1985. The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature 318:533-538. [DOI] [PubMed] [Google Scholar]
- 2.Askew, D. S., R. A. Ashmun, B. C. Simmons, and J. L. Cleveland. 1991. Constitutive c-myc expression in an IL-3-dependent myeloid cell line suppresses cell cycle arrest and accelerates apoptosis. Oncogene 6:1915-1922. [PubMed] [Google Scholar]
- 3.Banin, S., L. Moyal, S. Shieh, Y. Taya, C. W. Anderson, L. Chessa, N. I. Smorodinsky, C. Prives, Y. Reiss, Y. Shiloh, and Y. Ziv. 1998. Enhanced phosphorylation of p53 by ATM in response to DNA damage. Science 281:1674-1677. [DOI] [PubMed] [Google Scholar]
- 4.Baudino, T. A., and J. L. Cleveland. 2001. The Max network gone mad. Mol. Cell. Biol. 21:691-702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baudino, T. A., C. McKay, H. Pendeville-Samain, J. A. Nilsson, K. H. Maclean, E. L. White, A. C. Davis, J. N. Ihle, and J. L. Cleveland. 2002. c-Myc is essential for vasculogenesis and angiogenesis during development and tumor progression. Genes Dev. 16:2530-2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bello-Fernandez, C., G. Packham, and J. L. Cleveland. 1993. The ornithine decarboxylase gene is a transcriptional target of c-Myc. Proc. Natl. Acad. Sci. USA 90:7804-7808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bouillet, P., and A. Strasser. 2002. Bax and Bak: back-bone of T cell death. Nat. Immunol. 3:893-894. [DOI] [PubMed] [Google Scholar]
- 8.Bouillet, P., and A. Strasser. 2002. BH3-only proteins—evolutionarily conserved proapoptotic Bcl-2 family members essential for initiating programmed cell death. J. Cell Sci. 115:1567-1574. [DOI] [PubMed] [Google Scholar]
- 9.Boxer, L. M., and C. V. Dang. 2001. Translocations involving c-myc and c-myc function. Oncogene 20:5595-5610. [DOI] [PubMed] [Google Scholar]
- 10.Canman, C. E., D. S. Lim, K. A. Cimprich, Y. Taya, K. Tamai, K. Sakaguchi, E. Appella, M. B. Kastan, and J. D. Siliciano. 1998. Activation of the ATM kinase by ionizing radiation and phosphorylation of p53. Science 281:1677-1679. [DOI] [PubMed] [Google Scholar]
- 11.Cavalieri, F., and M. Goldfarb. 1987. Growth factor-deprived BALB/c 3T3 murine fibroblasts can enter the S phase after induction of c-myc gene expression. Mol. Cell. Biol. 7:3554-3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen, C. R., Y. Kang, P. M. Siegel, and J. Massague. 2002. E2F4/5 and p107 as Smad cofactors linking the TGFβ receptor to c-myc repression. Cell 110:19-32. [DOI] [PubMed] [Google Scholar]
- 13.Coller, H. A., C. Grandori, P. Tamayo, T. Colbert, E. S. Lander, R. N. Eisenman, and T. R. Golub. 2000. Expression analysis with oligonucleotide microarrays reveals that MYC regulates genes involved in growth, cell cycle, signaling, and adhesion. Proc. Natl. Acad. Sci. USA 97:3260-3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Alboran, I. M., R. C. O'Hagan, F. Gartner, B. Malynn, L. Davidson, R. Rickert, K. Rajewsky, R. A. DePinho, and F. W. Alt. 2001. Analysis of c-MYC function in normal cells via conditional gene-targeted mutation. Immunity 14:45-55. [DOI] [PubMed] [Google Scholar]
- 15.Deverman, B. E., B. L. Cook, S. R. Manson, R. A. Niederhoff, E. M. Langer, I. Rosova, L. A. Kulans, X. Fu, J. S. Weinberg, J. W. Heinecke, K. A. Roth, and S. J. Weintraub. 2002. Bcl-xL deamidation is a critical switch in the regulation of the response to DNA damage. Cell 111:51-62. [DOI] [PubMed] [Google Scholar]
- 16.Eilers, M., S. Schirm, and J. M. Bishop. 1991. The MYC protein activates transcription of the alpha-prothymosin gene. EMBO J. 10:133-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eischen, C. M., G. Packham, J. Nip, B. E. Fee, S. W. Hiebert, G. P. Zambetti, and J. L. Cleveland. 2001. Bcl-2 is an apoptotic target suppressed by both c-Myc and E2F-1. Oncogene 20:6983-6993. [DOI] [PubMed] [Google Scholar]
- 18.Eischen, C. M., M. F. Roussel, S. J. Korsmeyer, and J. L. Cleveland. 2001. Bax loss impairs Myc-induced apoptosis and circumvents the selection of p53 mutations during Myc-mediated lymphomagenesis. Mol. Cell. Biol. 21:7653-7662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eischen, C. M., J. D. Weber, M. F. Roussel, C. J. Sherr, and J. L. Cleveland. 1999. Disruption of the ARF-Mdm2-p53 tumor suppressor pathway in Myc-induced lymphomagenesis. Genes Dev. 13:2658-2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eischen, C. M., D. Woo, M. F. Roussel, and J. L. Cleveland. 2001. Apoptosis triggered by Myc-induced suppression of Bcl-XL or Bcl-2 is bypassed during lymphomagenesis. Mol. Cell. Biol. 21:5063-5070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Evan, G. I., A. H. Wyllie, C. S. Gilbert, T. D. Littlewood, H. Land, M. Brooks, C. M. Waters, L. Z. Penn, and D. C. Hancock. 1992. Induction of apoptosis in fibroblasts by c-myc protein. Cell 69:119-128. [DOI] [PubMed] [Google Scholar]
- 22.Fei, P., E. J. Bernhard, and W. S. El-Deiry. 2002. Tissue-specific induction of p53 targets in vivo. Cancer Res. 62:7316-7327. [PubMed] [Google Scholar]
- 23.Fuchs, S. Y., V. Adler, T. Buschmann, Z. Yin, X. Wu, S. N. Jones, and Z. Ronai. 1998. JNK targets p53 ubiquitination and degradation in nonstressed cells. Genes Dev. 12:2658-2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harrington, E. A., M. R. Bennett, A. Fanidi, and G. I. Evan. 1994. c-Myc-induced apoptosis in fibroblasts is inhibited by specific cytokines. EMBO J. 13:3286-3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hecht, J. L., and J. C. Aster. 2000. Molecular biology of Burkitt's lymphoma. J. Clin. Oncol. 18:3707-3721. [DOI] [PubMed] [Google Scholar]
- 26.Heikkila, R., G. Schwab, E. Wickstrom, S. L. Loke, D. H. Pluznik, R. Watt, and L. M. Neckers. 1987. A c-myc antisense oligodeoxynucleotide inhibits entry into S phase but not progress from G0 to G1. Nature 328:445-449. [DOI] [PubMed] [Google Scholar]
- 27.Iritani, B. M., and R. N. Eisenman. 1999. c-Myc enhances protein synthesis and cell size during B lymphocyte development. Proc. Natl. Acad. Sci. USA 96:13180-13185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnstone, R. W., A. A. Ruefli, and S. W. Lowe. 2002. Apoptosis: a link between cancer genetics and chemotherapy. Cell 108:153-164. [DOI] [PubMed] [Google Scholar]
- 29.Juin, P., A. O. Hueber, T. Littlewood, and G. Evan. 1999. c-Myc-induced sensitization to apoptosis is mediated through cytochrome c release. Genes Dev. 13:1367-1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Juin, P., A. Hunt, T. Littlewood, B. Griffiths, L. B. Swigart, S. Korsmeyer, and G. Evan. 2002. c-Myc functionally cooperates with Bax to induce apoptosis. Mol. Cell. Biol. 22:6158-6169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klocke, B. J., C. B. Latham, C. D'Sa, and K. A. Roth. 2002. p53 deficiency fails to prevent increased programmed cell death in the Bcl-XL-deficient nervous system. Cell Death Differ. 9:1063-1068. [DOI] [PubMed] [Google Scholar]
- 32.Lemay, M., and K. A. Wood. 1999. Detection of DNA damage and identification of UV-induced photoproducts using the CometAssay kit. BioTechniques 27:846-851. [DOI] [PubMed] [Google Scholar]
- 33.Li, L. H., C. Nerlov, G. Prendergast, D. MacGregor, and E. B. Ziff. 1994. c-Myc represses transcription in vivo by a novel mechanism dependent on the initiator element and Myc box II. EMBO J. 13:4070-4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Littlewood, T. D., D. C. Hancock, P. S. Danielian, M. G. Parker, and G. I. Evan. 1995. A modified oestrogen receptor ligand-binding domain as an improved switch for the regulation of heterologous proteins. Nucleic Acids Res. 23:1686-1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lowe, S. W., E. M. Schmitt, S. W. Smith, B. A. Osborne, and T. Jacks. 1993. p53 is required for radiation-induced apoptosis in mouse thymocytes. Nature 362:847-849. [DOI] [PubMed] [Google Scholar]
- 36.Maclean, K., H. Yang, and J. L. Cleveland. 2001. Serum suppresses myeloid progenitor apoptosis by regulating iron homeostasis. J. Cell. Biochem. 82:171-186. [DOI] [PubMed] [Google Scholar]
- 37.Mai, S., M. Fluri, D. Siwarski, and K. Huppi. 1996. Genomic instability in MycER-activated Rat1A-MycER cells. Chromosome Res. 4:365-371. [DOI] [PubMed] [Google Scholar]
- 38.Marx, J. C., J. A. Allay, D. A. Persons, S. A. Nooner, P. W. Hargrove, P. F. Kelly, E. F. Vanin, and E. M. Horwitz. 1999. High-efficiency transduction and long-term gene expression with a murine stem cell retroviral vector encoding the green fluorescent protein in human marrow stromal cells. Hum. Gene Ther. 10:1163-1173. [DOI] [PubMed] [Google Scholar]
- 39.Maya, R., M. Balass, S. T. Kim, D. Shkedy, J. F. Leal, O. Shifman, M. Moas, T. Buschmann, Z. Ronai, Y. Shiloh, M. B. Kastan, E. Katzir, and M. Oren. 2001. ATM-dependent phosphorylation of Mdm2 on serine 395: role in p53 activation by DNA damage. Genes Dev. 15:1067-1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mikhailov, V., M. Mikhailova, K. Degenhardt, M. A. Venkatachalam, E. White, and P. Saikumar. 2002. Association of Bax and Bak homo-oligomers in mitochondria. Bax requirement for Bak reorganization and cytochrome c release. J. Biol. Chem. 278:5367-5376. [DOI] [PubMed] [Google Scholar]
- 41.Momand, J., G. P. Zambetti, D. C. Olson, D. George, and A. J. Levine. 1992. The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell 69:1237-1245. [DOI] [PubMed] [Google Scholar]
- 42.Motoyama, N., F. Wang, K. A. Roth, H. Sawa, K. Nakayama, I. Negishi, S. Senju, Q. Zhang, S. Fujii, et al. 1995. Massive cell death of immature hematopoietic cells and neurons in Bcl-x-deficient mice. Science 267:1506-1510. [DOI] [PubMed] [Google Scholar]
- 43.Nakano, K., and K. H. Vousden. 2001. PUMA, a novel proapoptotic gene, is induced by p53. Mol. Cell 7:683-694. [DOI] [PubMed] [Google Scholar]
- 44.Oda, E., R. Ohki, H. Murasawa, J. Nemoto, T. Shibue, T. Yamashita, T. Tokino, T. Taniguchi, and N. Tanaka. 2000. Noxa, a BH3-only member of the Bcl-2 family and candidate mediator of p53-induced apoptosis. Science 288:1053-1058. [DOI] [PubMed] [Google Scholar]
- 45.Packham, G., E. L. White, C. M. Eischen, H. Yang, E. Parganas, J. N. Ihle, D. A. Grillot, G. P. Zambetti, G. Nunez, and J. L. Cleveland. 1998. Selective regulation of Bcl-XL by a Jak kinase-dependent pathway is bypassed in murine hematopoietic malignancies. Genes Dev. 12:2475-2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pelengaris, S., M. Khan, and G. Evan. 2002. c-myc: more than just a matter of life and death. Nat. Rev. Cancer 2:764-776. [DOI] [PubMed] [Google Scholar]
- 47.Pelengaris, S., M. Khan, and G. I. Evan. 2002. Suppression of Myc-induced apoptosis in beta cells exposes multiple oncogenic properties of Myc and triggers carcinogenic progression. Cell 109:321-334. [DOI] [PubMed] [Google Scholar]
- 48.Pelengaris, S., T. Littlewood, M. Khan, G. Elia, and G. Evan. 1999. Reversible activation of c-Myc in skin: induction of a complex neoplastic phenotype by a single oncogenic lesion. Mol. Cell 3:565-577. [DOI] [PubMed] [Google Scholar]
- 49.Pestina, T. I., J. L. Cleveland, C. Yang, G. P. Zambetti, and C. W. Jackson. 2001. Mpl ligand prevents lethal myelosuppression by inhibiting p53-dependent apoptosis. Blood 98:2084-2090. [DOI] [PubMed] [Google Scholar]
- 50.Pomerantz, J., N. Schreiber-Agus, N. J. Liegeois, A. Silverman, L. Alland, L. Chin, J. Potes, K. Chen, I. Orlow, H. W. Lee, C. Cordon-Cardo, and R. A. DePinho. 1998. The Ink4a tumor suppressor gene product, p19Arf, interacts with MDM2 and neutralizes MDM2's inhibition of p53. Cell 92:713-723. [DOI] [PubMed] [Google Scholar]
- 51.Roth, J., M. Dobbelstein, D. A. Freedman, T. Shenk, and A. J. Levine. 1998. Nucleo-cytoplasmic shuttling of the hdm2 oncoprotein regulates the levels of the p53 protein via a pathway used by the human immunodeficiency virus rev protein. EMBO J. 17:554-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rupnow, B. A., A. D. Murtha, R. M. Alarcon, A. J. Giaccia, and S. J. Knox. 1998. Direct evidence that apoptosis enhances tumor responses to fractionated radiotherapy. Cancer Res. 58:1779-1784. [PubMed] [Google Scholar]
- 53.Schmitt, C. A., J. S. Fridman, M. Yang, S. Lee, E. Baranov, R. M. Hoffman, and S. W. Lowe. 2002. A senescence program controlled by p53 and p16INK4a contributes to the outcome of cancer therapy. Cell 109:335-346. [DOI] [PubMed] [Google Scholar]
- 54.Schmitt, C. A., M. E. McCurrach, E. de Stanchina, R. R. Wallace-Brodeur, and S. W. Lowe. 1999. INK4a/ARF mutations accelerate lymphomagenesis and promote chemoresistance by disabling p53. Genes Dev. 13:2670-2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schuler, M., and D. R. Green. 2001. Mechanisms of p53-dependent apoptosis. Biochem. Soc. Trans. 29:684-688. [DOI] [PubMed] [Google Scholar]
- 56.Seoane, J., H. V. Le, and J. Massague. 2002. Myc suppression of the p21Cip1 Cdk inhibitor influences the outcome of the p53 response to DNA damage. Nature 419:729-734. [DOI] [PubMed] [Google Scholar]
- 57.Sheen, J. H., and R. B. Dickson. 2002. Overexpression of c-Myc alters G1/S arrest following ionizing radiation. Mol. Cell. Biol. 22:1819-1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sherr, C. J., and R. A. DePinho. 2000. Cellular senescence: mitotic clock or culture shock? Cell 102:407-410. [DOI] [PubMed] [Google Scholar]
- 59.Sjostrom, J., C. Blomqvist, K. von Boguslawski, N. O. Bengtsson, I. Mjaaland, P. Malmstrom, B. Ostenstadt, E. Wist, V. Valvere, S. Takayama, J. C. Reed, and E. Saksela. 2002. The predictive value of bcl-2, bax, bcl-xL, bag-1, fas, and fasL for chemotherapy response in advanced breast cancer. Clin. Cancer Res. 8:811-816. [PubMed] [Google Scholar]
- 60.Tao, W., and A. J. Levine. 1999. p19ARF stabilizes p53 by blocking nucleo-cytoplasmic shuttling of Mdm2. Proc. Natl. Acad. Sci. USA 96:6937-6941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vafa, O., M. Wade, S. Kern, M. Beeche, T. K. Pandita, G. M. Hampton, and G. M. Wahl. 2002. c-Myc can induce DNA damage, increase reactive oxygen species, and mitigate p53 function: a mechanism for oncogene-induced genetic instability. Mol. Cell 9:1031-1044. [DOI] [PubMed] [Google Scholar]
- 62.Vilenchik, M., A. J. Raffo, L. Benimetskaya, D. Shames, and C. A. Stein. 2002. Antisense RNA down-regulation of bcl-xL expression in prostate cancer cells leads to diminished rates of cellular proliferation and resistance to cytotoxic chemotherapeutic agents. Cancer Res. 62:2175-2183. [PubMed] [Google Scholar]
- 63.Vousden, K. H., and X. Lu. 2002. Live or let die: the cell's response to p53. Nat. Rev. Cancer 2:594-604. [DOI] [PubMed] [Google Scholar]
- 64.Weber, J. D., L. J. Taylor, M. F. Roussel, C. J. Sherr, and D. Bar-Sagi. 1999. Nucleolar Arf sequesters Mdm2 and activates p53. Nat. Cell Biol. 1:20-26. [DOI] [PubMed] [Google Scholar]
- 65.Yu, J., L. Zhang, P. M. Hwang, K. W. Kinzler, and B. Vogelstein. 2001. PUMA induces the rapid apoptosis of colorectal cancer cells. Mol. Cell 7:673-682. [DOI] [PubMed] [Google Scholar]
- 66.Zindy, F., C. M. Eischen, D. H. Randle, T. Kamijo, J. L. Cleveland, C. J. Sherr, and M. F. Roussel. 1998. Myc signaling via the ARF tumor suppressor regulates p53-dependent apoptosis and immortalization. Genes Dev. 12:2424-2433. [DOI] [PMC free article] [PubMed] [Google Scholar]