Abstract
The majority of cytosolic proteins in eukaryotes contain a covalently linked acetyl moiety at their very N terminus. The mechanism by which the acetyl moiety is efficiently transferred to a large variety of nascent polypeptides is currently only poorly understood. Yeast Nα-acetyltransferase NatA, consisting of the known subunits Nat1p and the catalytically active Ard1p, recognizes a wide range of sequences and is thought to act cotranslationally. We found that NatA was quantitatively bound to ribosomes via Nat1p and contained a previously unrecognized third subunit, the Nα-acetyltransferase homologue Nat5p. Nat1p not only anchored Ard1p and Nat5p to the ribosome but also was in close proximity to nascent polypeptides, independent of whether they were substrates for Nα-acetylation or not. Besides Nat1p, NAC (nascent polypeptide-associated complex) and the Hsp70 homologue Ssb1/2p interact with a variety of nascent polypeptides on the yeast ribosome. A direct comparison revealed that Nat1p required longer nascent polypeptides for interaction than NAC and Ssb1/2p. Δnat1 or Δard1 deletion strains were temperature sensitive and showed derepression of silent mating type loci while Δnat5 did not display any obvious phenotype. Temperature sensitivity and derepression of silent mating type loci caused by Δnat1 or Δard1 were partially suppressed by overexpression of SSB1. The combination of data suggests that Nat1p presents the N termini of nascent polypeptides for acetylation and might serve additional roles during protein synthesis.
Nα-Terminal acetylation of proteins is a common phenomenon in eukaryotes. About 90% of mammalian proteins and 50% of the soluble proteins in lower eukaryotes are Nα-terminally acetylated (7). It is generally accepted that Nα-terminal acetylation of mammalian proteins occurs cotranslationally when newly synthesized polypeptides have just emerged from the ribosomal exit tunnel (16, 32, 34, 57). Proteins with serine and alanine N termini are the most frequently acetylated substrates, and these residues along with methionine, glycine, and threonine account for more than 95% of the N-terminally acetylated residues (40). Very little is known about the composition, function, and importance of mammalian Nα-acetyltransferases. NatH, a human homologue, was recently found to be overexpressed in papillary thyroid carcinomas (9).
Three Nα-terminal acetyltransferase complexes termed NatA, NatB, and NatC have been identified in Saccharomyces cerevisiae (28, 33, 39, 50). They differ in substrate specificity and subunit composition, but all contain a catalytic subunit homologous to the GNAT family of acetyltransferases (30, 41). NatA is the major Nα-terminal acetyltransferase in the yeast cytosol, responsible for the acetylation of serine, alanine, threonine, and glycine (4, 39, 48). NatA has been shown to contain two subunits, Nat1p, a protein of 98 kDa, and the 27-kDa GNAT homologue Ard1p (28, 33). Both subunits are required for activity, and deletion strains of either Δnat1 or Δard1 fail to Nα-terminally acetylate the same set of substrate proteins (28). In agreement with these observations, Δnat1 and Δard1 strains display the same set of phenotypes, such as reduced sporulation efficiency, failure to enter G0 under specific conditions, defect in silencing of the silent mating-type loci, and decreased survival after heat shock (28, 39, 54, 55). The pleiotropic phenotypes are thought to reflect functional defects of various target proteins lacking proper acetylation at their N termini. However, there are surprisingly few examples demonstrating the biological importance of Nα-terminal protein acetylation (40). For NatA, no clear-cut example has been described. It was discussed that the mating defect of a Δnat1 or Δard1 strain might be due to the lack of Nα-acetylation of the silent information regulator protein Sir3p (47). However, experimental evidence is missing so far. Recent evidence suggests that the multiple phenotypes observed in the absence of NatB might be caused by the lack of acetylation of actin and tropomyosin (38). A well-documented example has been reported for Mak3p, the catalytic subunit of NatC. Nα-Acetylation by Mak3p is essential for the coat protein assembly of the L-A double-stranded RNA virus in Saccharomyces cerevisiae (49, 50).
Newly synthesized polypeptides exit the large ribosomal subunit through a tunnel. In yeast, several proteins are located close to the tunnel exit. Two of them, NAC (nascent polypeptide-associated complex) and the Hsp70 homologue Ssb1/2p (Ssb1p and Ssb2p, two 99% identical proteins with identical function and similar expression levels), have been shown to be in close proximity to a variety of nascent polypeptides (14, 36, 43). Recently, it was found that the ribosome-associated complex (consisting of the Hsp70 homologue Ssz1p and the Hsp40 homologue zuotin) is required for efficient cross-linking of the nascent polypeptide to Ssb1/2p (14). During the course of these experiments an additional high-molecular-mass cross-link product was observed. Here, we identify this high-molecular-mass cross-link to the nascent polypeptide as Nat1p, the noncatalytic subunit of NatA (41).
Purification and biochemical analysis of the cross-link-inducing factor revealed that the NatA complex contains a third subunit. This previously uncharacterized subunit, termed Nat5, like Ard1p, belongs to the GNAT family of acetyltransferases. We find that Nat1p interacts not only with the nascent polypeptide chain but also with the ribosome, thereby anchoring the NatA complex quantitatively to ribosomes.
MATERIALS AND METHODS
Yeast strains and plasmids.
Standard yeast genetic techniques were used (46). MH272-3f a/α (ura3/ura3 leu2/leu2 his3/his3 trp1/trp1 ade2/ade2 HMLa/HMLa) is the parental wild-type strain of the derivatives used in this study (13, 20). The generation of the deletion strains lacking ARD1 was done either by replacing the entire gene with the kanMX module (52) or by replacing the 306-bp AflII/XbaI fragment in the middle of ARD1 with URA3. NAT1 was deleted by replacing the 2,384-bp XhoI/NruI fragment with LEU2. NAT5 was deleted by replacing the 225-bp Bpu10I/BbsI fragment with ADE2. The resulting strains are Δard1 (IDA7, MH272-3fα ard1::kanR; IDA7B, MH272-3fα ard1::URA3), Δnat5 (IDA8, MH272-3fα nat5::ADE2), Δnat1 (IDA9, MH272-3fα nat1::LEU2), and Δnat1Δard1Δnat5 (IDA789, MH272-3fa nat1::LEU2 ard1::kanR nat5::ADE2). Δnat1Δssb1Δssb2 (IDA56C9, MH272-3f nat1::LEU2 ssb1::ADE2 ssb2::HIS3) was generated by mating IDA56Cα (ssb1::ADE2 ssb2::HIS3) with IDA9a (MH272-3fa nat1::LEU2) followed by sporulation and tetrad dissection. PCR with appropriate primer combinations and Western blotting with anti-ard1, anti-nat5, and anti-nat1 sera were used to confirm deletion strains. To test ARD1, NAT5, and NAT1 as multicopy suppressors, two methods were applied. Either the genes were cloned into multicopy vectors YEplac195 (2μm, URA3), YEplac112 (2μm, TRP1) (15), or pRS423 (2μm, HIS3) (6) or the genes were cloned downstream of the GAL1-10 promoter of vectors of the pESC series (Stratagene). As the anti-nat5 serum available did not produce strong signals in immunoblots, NAT5 under control of the GAL promoter was fused to an N-terminal FLAG tag for better detection. Resulting multicopy plasmids are 2μm-ARD1, 2μm-NAT5, and 2μm-NAT1. Galactose-inducible constructs are designated pESC-ARD1, pESC-NAT5, and pESC-NAT1. The plasmid overexpressing SSB1 is based on YEplac195 (2μm, URA3) and was previously described (14). Yeast strain ts-187 harbors the S518F point mutant of prt1 encoding translation initiation factor eIF3. ts-187 is defective in initiation of translation at 37°C (19, 53). To generate monosomes in vivo, ts-187 was incubated at 37°C for 20 min as described previously (19). The mating efficiency was tested and quantified as described previously (8). Strains derived from W303 (MATa ade2-101 his3-11,15 trp1-1 leu2-3,112 ura3-1), JRM5 (W303 Δard1), and AMR1 (W303 Δnat1) were obtained from Rolf Sternglanz and are described elsewhere (3, 28). NAT5 was deleted by replacing the 225-bp Bpu10I/BbsI fragment with ADE2, resulting in W303 nat5::ADE2.
Purification of NatA.
Cytosol was prepared as described previously (10) from a 12-liter culture of YRG16 (MATa ura3 leu2 his3 trp1 ade2 HMLa egd2::ADE2 egd1::URA3) (13) and was centrifuged for 1.5 h at 200,000 × g at 4°C. The ribosomal pellet was resuspended in 20 mM HEPES-KOH (pH 7.4), 700 mM potassium acetate, 5 mM magnesium acetate, and 1 mM phenylmethylsulfonyl fluoride (PMSF) and was centrifuged again for 1.5 h at 200,000 × g at 4°C. The ribosome-free supernatant, termed ribosomal salt wash, was diluted with 6 volumes of 40 mM HEPES-KOH (pH 7.4) and loaded onto a ResourceQ (6-ml) anion-exchange column (Amersham-Pharmacia). Bound proteins were eluted with a 100 to 800 mM 25-ml linear potassium acetate gradient in 40 mM HEPES-KOH (pH 7.4). NatA eluted at a potassium acetate concentration of 450 to 550 mM. NatA-containing fractions were pooled and diluted with 1.5 volumes of 40 mM HEPES-KOH (pH 7.4) and applied to a MonoQ (5/5) anion-exchange column (Amersham-Pharmacia). Bound proteins were eluted with a 200 to 600 mM 22-ml linear potassium acetate gradient in 40 mM HEPES-KOH (pH 7.4). NatA eluted with a 250 to 350 mM concentration of potassium acetate. NatA-containing fractions were pooled and loaded onto a Hi-Load (16/60) Superdex 200 prep grade gel filtration column (Amersham-Pharmacia) preequilibrated with 40 mM HEPES-KOH (pH 7.4) and 150 mM potassium acetate. NatA eluted at a volume of 66 ml, and these fractions were stored at −80°C and used for further experiments.
Identification of the NatA subunits.
Purified NatA complex was transferred to a polyvinylidene difluoride membrane. Ard1p and Nat5p were identified by N-terminal Edman degradation. Nat1p was N-terminally blocked and was identified by mass spectrometry of a tryptic digest. To estimate the relative amounts of Ard1p, Nat5p, and Nat1p, the purified NatA complex was separated by reverse-phase high-performance liquid chromatography (HPLC) (Nucleosil 500-5, C3PPN; Macherey-Nagel) by applying a 30 to 60% 6-ml gradient of 0.09% trifluoroacetic acid-CH3CN in 0.08% trifluoroacetic acid-H2O. The molar ratios of the different subunits in the preparation were calculated by using the peak area of the HPLC profile and the following calculated molar extinction coefficients at 280 nm: ɛ (Nat1p), 105,680 M−1 cm−1; ɛ (Ard1p), 21,050 M−1 cm−1; and ɛ (Nat5p), 13,370 M−1 cm−1.
Generation of antibodies.
Polyclonal antibodies specifically recognizing Ard1p (anti-ard1), Nat5p (anti-nat5), or Nat1p (anti-nat1) were generated against His-tagged versions of the proteins expressed in Escherichia coli (EUROGENTEC, Seraing, Belgium).
In vitro transcription, translation, preparation of RNCs, and cross-link assay.
Yeast translation extract was prepared as described previously (11) from strain JK93dα (20) or YRG16a (13). Translation reactions were performed in the presence of [35S]methionine (Amersham-Pharmacia). Ribosome-bound nascent chains for cross-link experiments were produced as described previously (10). Different lengths of DNA fragments for transcription reactions were generated by PCR with plasmids pSP65-mdh1 (carrying yeast MDH1), pSP65-rpl4 (carrying yeast RPL4A), or pDJ100 (carrying the sequence for prepro α-factor as a template) (14). In the cases of RPL4A and prepro α-factor, the reverse primer changed the penultimate amino acid to methionine for better detection. Point mutants of Rpl4A [Rpl4A-S(2)K] and of prepro α-factor [prepro α-R(2)K], in which the respective amino acids were changed to lysine, were used when indicated. Translation reactions were primed with truncated mRNA encoding proteins of different lengths, leading to the generation of the following ribosome-nascent chain complexes (RNCs): the N-terminal 50, 70, or 87 amino acids of prepro α-factor or prepro α-factor R(2)K; the N-terminal 50, 70, or 86 amino acids of yeast ribosomal protein Rpl4A or Rpl4A-S(2)K; and the N-terminal 40, 45, 50, 55, 60, 80, 85, 104, 150, or 200 amino acids of mitochondrial malate dehydrogenase (Mdh1p). For the detailed determination of the minimal length required for cross-link formation to the different ribosome-bound factors, the precursor of Mdh1p was chosen as a nascent polypeptide, since this protein contains 13 lysines evenly distributed within the first 200 amino acids (see Fig. 4D). In some experiments, prepro α-R(2)K was used to increase cross-link efficiency.
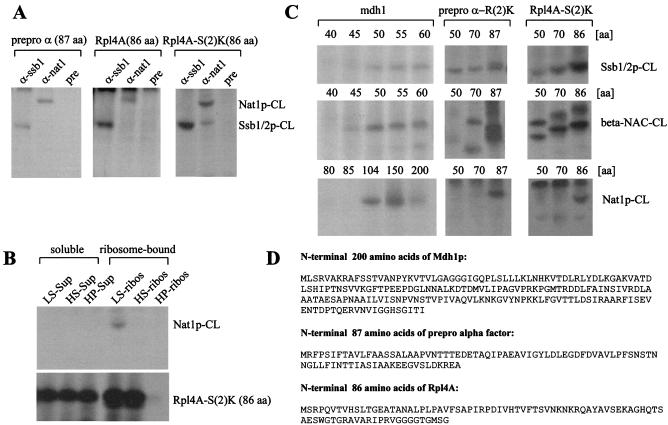
FIG. 4.
NatA contacts a variety of ribosome-bound nascent chains. (A) Cross-link reactions were performed with low-salt-treated RNCs carrying the prepro α-factor, Rpl4A, or Rpl4A-S(2)K as a nascent chain. Aliquots of the cross-link reactions were subjected to immunoprecipitation under denaturing conditions. Anti-ssb1 (α-ssb1), anti-nat1 (α-nat1), or preimmune serum (pre) was used for the immunoprecipitations. The material bound to the respective antibody was run on 10% Tris-Tricine gels and subsequently analyzed by autoradiography. (B) RNCs carrying Rpl4A-S(2)K were generated and subsequently separated from soluble proteins by centrifugation through a sucrose cushion under low-salt conditions (LS-Sup and LS-ribos), under high-salt conditions (HS-Sup and HS-ribos), and after treatment with puromycin (HP-Sup and HP-ribos) as described in Materials and Methods. Each sample was subjected to a cross-link reaction and subsequently to immunoprecipitation under denaturating conditions with anti-nat1. The material bound to anti-nat1 (upper panel) and a total of the cross-linking reaction (lower panel) was run on a 10% Tris-Tricine gel and subsequently analyzed by autoradiography. Note, as shown in lane 1, a fraction of Rpl4A-S(2)K was not associated with the ribosome under low-salt conditions. This fraction does not contain NatA, which is retained in the LS-ribos fraction (compare Fig. 2B). (C) Low-salt RNCs carrying different length yeast malate dehydrogenase (mdh1), prepro α-R(2)K, or Rpl4A-S(2)K as indicated were subjected to cross-linking reactions. The numbers indicate the length of the nascent chains in amino acids (aa). Each sample was split into three aliquots and subjected to immunoprecipitation reactions under denaturating conditions with anti-nat1, anti-ssb1, or anti-beta-nac antibodies. The material bound to the respective antibody was run on 10% Tris-Tricine gels and subsequently analyzed by autoradiography. Nat1p-CL, cross-link of the nascent chain to Nat1p; Ssb1/2p-CL, cross-link of the nascent chain to Ssb1/2p; beta-NAC-CL, cross-link of the nascent chain to beta-NAC. (D) Protein sequences of the nascent chains used for cross-linking experiments.
The initial methionine of Rpl4A is cleaved cotranslationally, and the processed protein contains a single methionine at position 84. Prepro α-factor contains a single methionine at position 1, which is not cleaved. Approximately 40 amino acids of the ribosome-bound nascent chains are covered by the ribosome. The homobifunctional cross-linker bis-(sulfosuccinimidyl)-suberate (BS3) (spacer length, 1.14 nm; Pierce) was used for cross-linking reactions. BS3 covalently cross-links primary amines, like the ɛ-amino group of lysines or the Nα-amino group of a nascent polypeptide. For cross-linking reactions, RNCs were isolated as described under high (presence of 700 mM potassium acetate)- or low (presence of 120 mM potassium acetate)-salt conditions as described previously (14). Cross-linking reactions (typically 40-μl final volume) were started by the addition of BS3 to a final concentration of 400 μM and were incubated for 30 min on ice. The reaction was quenched for 10 min on ice by the addition of glycyl-glycine to a final concentration of 5 mM. Reaction mixtures were analyzed on 10% Tris-Tricine gels (45) followed by autoradiography. Quantification of 125I and 35S on immunoblots or autoradiographies, respectively, was by Aida ImageAnalyzer (Raytest; Isotopenmessgeräte GmbH).
Preparation of ribosomes.
In a typical experiment, 60 μl of yeast cytosol was diluted 1:2 with either low-salt dilution buffer (20 mM HEPES-KOH [pH 7.4], 120 mM potassium acetate, 2 mM dithiothreitol [DTT], 5 mM magnesium acetate, 1 mM PMSF) or high-salt dilution buffer (20 mM HEPES-KOH [pH 7.4], 1.4 M potassium acetate, 2 mM DTT, 5 mM magnesium acetate, 1 mM PMSF). After removal of an aliquot (cytosol), 40 μl of the samples was layered on top of a 100 μl of low-salt sucrose cushion (20 mM HEPES-KOH [pH 7.4], 25% sucrose, 120 mM potassium acetate, 2 mM DTT, 5 mM magnesium acetate, 1 mM PMSF) or a 100 μl of high-salt sucrose cushion (20 mM HEPES-KOH [pH 7.4], 25% sucrose, 700 mM potassium acetate, 2 mM DTT, 5 mM magnesium acetate, 1 mM PMSF). Accordingly, potassium acetate concentrations of between 100 and 700 mM were applied to determine the salt sensitivity of NatA interaction with the ribosome. After centrifugation at 200,000 × g in an RC M120 GX ultracentrifuge (Sorvall) for 3 h at 4°C, samples were separated into supernatant and pellet and corresponding amounts were analyzed on Tris-Tricine gels followed by immunoblotting. To generate nascent chain-free ribosomes, a mock translation reaction was performed in the presence of 1 mM puromycin, without the addition of mRNA. After an incubation of 30 min at room temperature, potassium acetate was added to a final concentration of 800 mM and ribosomes (ribosomal subunits) were separated from cytosolic proteins and released polypeptides by centrifugation on a high-salt sucrose cushion as described above. The nascent chain-free ribosomal pellet was resuspended in resuspension buffer (20 mM HEPES-KOH [pH 7.4], 100 mM potassium acetate, 2 mM DTT, 5 mM magnesium acetate, 0.5 mM PMSF, 2 U of RNase inhibitor/ml) and used for further experiments. In a control experiment, the addition of puromycin after the translation of Rpl4A-S(2)K released all nascent chains quantitatively from the ribosomal pellet (see Fig. 4B; data not shown).
Immunoprecipitation reactions.
Immunoprecipitation reactions were performed with protein A-Sepharose beads (CL-4B; Amersham-Pharmacia) coated with immunoglobulin directed against proteins as indicated in the figure legends. To identify the cross-link partners of the nascent polypeptides, immunoprecipitations were performed under denaturing conditions as described previously (14). Immunoprecipitations were performed under native conditions to characterize the subunit composition of the NatA complex as described previously (13, 44). Starting material for native immunoprecipitations was the ribosomal salt wash (compare to “Purification of NatA” above).
Miscellaneous.
Protein concentrations were determined by the Bradford assay (Bio-Rad), with bovine serum albumin as a standard. [125I]-labeled protein A was used to develop the immunoblots (17). Zymolyase was from Seikagaku, Falmouth, Mass.; all other reagents were from Sigma or Roche Molecular Biochemicals. Sequence alignments were performed with T-COFFEE, version 1.37 (31).
RESULTS
Purification of a protein complex that forms a novel, high-molecular-mass cross-link to nascent polypeptides in yeast.
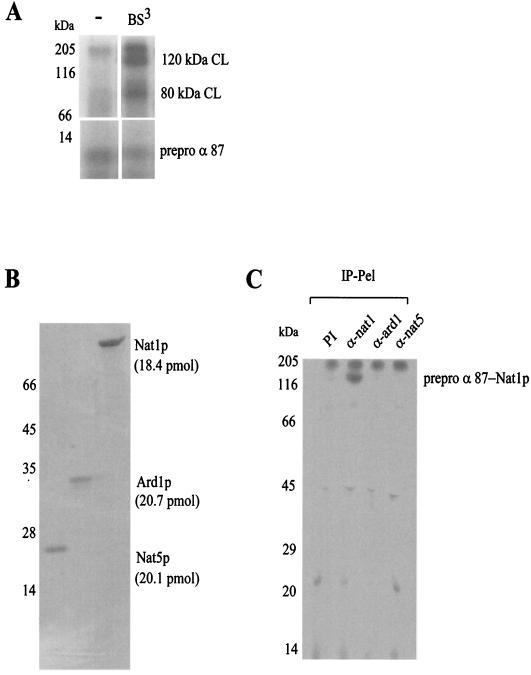
Cross-linking to ribosome-bound nascent chains, as e.g., to the N-terminal 87 amino acids of prepro α-factor, generated two high-molecular-mass cross-link products with an efficiency of 15 to 20% (Fig. 1A) (4, 36). The cross-link product of about 80 kDa was to stably ribosome-bound Ssb1/2p (14), whereas the protein forming the 120-kDa cross-link has not been characterized previously. High-salt treatment of RNCs eliminated both the 80- and 120-kDa cross-links (data not shown) (14). The ribosome-associated material that was removed by high-salt treatment (ribosomal salt wash) served as the starting material for the purification of the protein forming the 120-kDa cross-link to the nascent polypeptide. Fractionated ribosomal salt wash was subjected to cross-linking experiments in the presence of high-salt RNCs and analyzed for the appearance of the 120-kDa cross-link (see Materials and Methods). This assay led to the identification of three copurifying proteins: Nat1p (98 kDa), Ard1p (27 kDa), and Nat5p (20 kDa). Approximation of the molar concentrations in the purified complex suggested that the three polypeptides were present in a 1:1:1 ratio (Fig. 1B) (for quantification refer to Materials and Methods). The finding that Nat1p, Ard1p, and Nat5p were present in equimolar amounts, combined with published data on the size of the NatA complex (24, 33), suggests that the complex is trimeric.
FIG. 1.
Nat1p, a subunit of yeast NatA interacts with RNCs. (A) RNCs carrying 87 N-terminal amino acids of 35S-labeled prepro α-factor (prepro α 87) were incubated in the absence (−) or presence of the homobifunctional cross-linker BS3 as indicated. Aliquots were run on 10% Tris-Tricine gels and were subsequently analyzed by autoradiography. The amounts of prepro α 87 and the cross-link products (CL) were quantified by densitometry to approximate the cross-linking efficiency. The upper panel (120 kDa CL, 80 kDa CL) shows a four-times-longer exposure than the lower panel (prepro α 87); the signal was linear in relation to the exposure time. The amount of prepro α 87 was set to 100%. (B) Subunits of the cross-link-inducing complex were separated by reverse-phase HPLC and analyzed on a Coomassie-stained gel. The amount of protein contained in the different fractions was determined by using the peak area of the HPLC profile and the calculated molar extinction coefficients (see Materials and Methods). (C) Cross-link reactions were performed with low-salt treated RNCs carrying 35S-labeled prepro α 87 as a nascent chain. Aliquots of the cross-link reactions were subjected to immunoprecipitation under denaturing conditions with preimmune serum (PI), anti-nat1 (α-nat1), anti-ard1 (α-ard1), and anti-nat5 (α-nat5) antibodies. Shown is the material bound to the respective antibody (IP-Pel). Immunoprecipitation of the 120-kDa cross-link with anti-nat1 was quantitative (data not shown).
Immunoprecipitation reactions were performed to confirm the identity of the 120-kDa cross-link to Nat1p. The 120-kDa cross-link product was quantitatively immunoprecipitated with anti-nat1 under denaturing conditions. No cross-link products between the nascent polypeptide and Ard1p or Nat5p were observed under the conditions tested (Fig. 1C and data not shown).
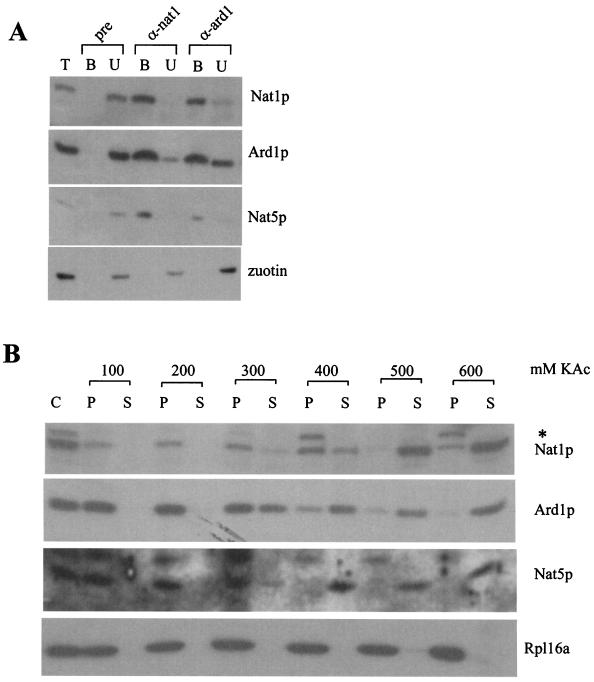
Nat1p and Ard1p are known to form a complex termed NatA (33). Nat5p, like Ard1p, belongs to the GNAT family of Nα-acetyltransferases (30, 41). The fact that Nat1p, Ard1p, and Nat5p copurified on a variety of chromatography columns combined with the sequence homology between Ard1p and Nat5p suggested that NatA might contain an additional subunit. From a ribosomal salt wash, anti-nat1 antibodies immunoprecipitated Nat1p and coimmunoprecipitated Ard1p and Nat5p, with efficiencies of 90 to 100% under native conditions (Fig. 2A). The immunoprecipitation efficiency of Ard1p with anti-ard1 was approximately 70%, and Nat1p and Nat5p were coimmunoprecipitated with similar efficiency (Fig. 2A). As a control, zuotin, a protein also contained in the ribosomal salt wash, was coimmunoprecipitated neither with anti-nat1 nor with anti-ard1 (Fig. 2A). The combined data suggest that the bulk of Nat1p, Ard1p, and Nat5p forms a complex.
FIG. 2.
Nat5p, a member of the GNAT family of acetyltransferases, is a subunit of NatA. (A) Immunoprecipitation reactions with ribosomal salt-wash as the starting material were performed under native conditions. Aliquots of the ribosomal salt-wash (T), of the material bound to the respective antibody (B), and of the unbound material (U) were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and analyzed by immunodecoration with antibodies specifically recognizing Nat1p, Ard1p, Nat5p, and zuotin. Protein A-Sepharose beads were coated with preimmune serum (pre), antibody recognizing Nat1p (α-nat1), or antibody recognizing Ard1p (α-ard1). (B) Separation of yeast cytosol (C) into a ribosomal pellet (P) and a postribosomal supernatant (S) at increasing concentrations of potassium acetate (KAc) as indicated. Aliquots were separated on 10% Tris-Tricine gels, transferred to nitrocellulose, and decorated with antibodies directed against Nat1p, Ard1p, Nat5p, and Rpl16a (ribosomal marker). The band labeled with an asterisk is a polypeptide cross-reacting with anti-nat1 on Western blots.
NatA is quantitatively bound to the ribosome via Nat1p.
We had purified NatA from the ribosomal salt wash, suggesting that a fraction of NatA was bound to ribosomes. To determine the distribution of NatA, the yeast cytosol was separated into a ribosomal pellet and a postribosomal supernatant at increasing concentrations of salt. At concentrations of potassium acetate up to 200 mM, NatA was quantitatively recovered in the ribosomal pellet. At higher potassium acetate concentrations, all three subunits were simultaneously released from the ribosomal fraction (Fig. 2B). The result suggests that the bulk of NatA is associated with ribosomes at physiological salt concentrations.
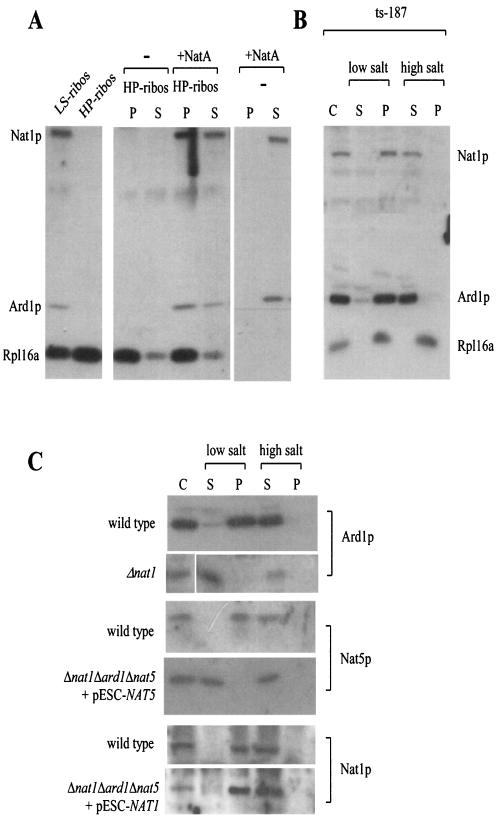
Efficient cross-linking of Nat1p to the nascent polypeptide (Fig. 1A and C) raised the question of whether binding of NatA to ribosomes was mediated exclusively via the interaction of Nat1p to the nascent polypeptide or whether NatA was also interacting directly with the ribosome. To distinguish between these possibilities, we applied two different approaches. First, we tested whether NatA would bind to nascent chain-free ribosomes in vitro. To this end, nascent chains were released by puromycin treatment and ribosomal subunits were subsequently incubated in the presence of NatA. Indeed, NatA was recovered in the ribosomal pellet (Fig. 3A). This was not due to aggregation, since NatA remained in the supernatant fraction when ribosomes were absent (Fig. 3A). Second, we made use of a yeast strain (ts-187) expressing a temperature-sensitive version of the β-subunit of eIF3, which at 37°C quickly leads to the generation of nontranslating ribosomes in vivo (19, 53). When the cytosol of heat-shocked ts-187 cells was separated into a ribosomal pellet and a postribosomal supernatant, NatA was recovered exclusively in the ribosomal pellet, from which it was released upon treatment with high salt (Fig. 3B and data not shown). The combination of data indicates that NatA binds directly to ribosomes, even in the absence of nascent polypeptides.
FIG. 3.
Nat1p anchors Ard1p and Nat5p to the ribosome. (A) Nascent chain-free ribosomes, generated by high-salt-puromycin treatment, were isolated, resuspended, incubated in the absence (−) or presence (+NatA) of partially purified NatA, and subsequently reisolated by centrifugation. As a control, NatA was subjected to centrifugation in the absence of ribosomes. LS-ribos, ribosomes isolated at low-salt conditions; HP-ribos, high-salt-puromycin treated ribosomes; P, ribosomal pellet; S, postribosomal supernatant. (B) The yeast strain ts-187, defective in translationinitiation at elevated temperatures, was incubated at 37°C for 20 min prior to harvest. Subsequently, cytosol (C) was prepared as described in Materials and Methods and separated into the ribosomal pellet (P) and a postribosomal supernatant (S) in the presence of low salt (120 mM potassium acetate) or high salt (700 mM potassium acetate) concentrations. (C) Cytosol (C) from a wild-type yeast strain, a strain lacking Nat1p (Δnat1), a strain lacking Ard1p and Nat1p but expressing NAT5 under control of a GAL promoter (Δnat1Δard1Δnat5 + pESC-NAT5), and a strain lacking Nat5p and Ard1p but expressing NAT1 under control of a GAL promoter (Δnat1Δard1Δnat5 + pESC-NAT1) was separated into a postribosomal supernatant (S) and a ribosomal pellet (P) in the presence of low or high salt concentrations as described above. (A to C) Samples were separated on 10% Tris-Tricine gels, transferred onto nitrocellulose and decorated with antibodies directed against Nat1p, Ard1p, Nat5p, and Rpl16a (ribosomal marker) as indicated.
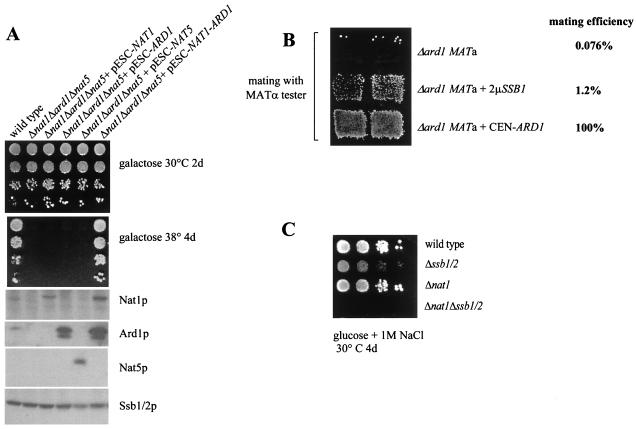
To determine which of the NatA subunits were responsible for stable association with the ribosome, we tested the ribosome association of Ard1p in Δnat1. A direct comparison between the amount of ribosomes and Ard1p in wild-type and Δnat1 strains revealed that Ard1p was significantly reduced in Δnat1 (data not shown). The remainder, however, did not bind to ribosomes and was recovered in the postribosomal supernatant even under low-salt conditions (Fig. 3C, Ard1p). Destabilization of Nat1p and Nat5p in Δard1 was even more severe. In the absence of Ard1p, neither Nat1p nor Nat5p could be detected by immunoblotting (data not shown). To test the localization of Nat1p and Nat5p in the absence of Ard1p, we placed NAT1 and NAT5 under the control of the strong inducible GAL1-10 promoter (22). When strains Δnat1Δard1Δnat5 plus pESC-NAT1 and Δnat1Δard1Δnat5 plus pESC-NAT5 were grown on galactose-containing medium, Nat1p and Nat5p were readily detectable by immunoblotting (see Fig. 6A, lower panel). Analysis of the localization of Nat5p revealed that in the absence of Nat1p-Ard1p, the protein was not associated with ribosomes (Fig. 3C, Nat5p). Ribosome association of Nat1p, however, remained unaltered in the absence Ard1p-Nat5p. As in the wild-type strain, Nat1p was detected in the ribosomal fraction under low-salt conditions and was released from the ribosome by high-salt treatment (Fig. 3C, Nat1p). We conclude that stable association of Ard1p and Nat5p with the ribosome depends on the presence of Nat1p and that Nat1p is able to bind to the ribosome by itself.
FIG. 6.
(A) None of the NatA subunits by itself supports growth at elevated temperature. Δnat1Δard1Δnat5 strains overexpressing NAT1, ARD1, or NAT5 as indicated from a GAL-inducible promoter were grown on galactose as a carbon source. Serial 10-fold dilutions were analyzed on galactose at 30 or 38°C. Aliquots of each strain were also analyzed on immunoblots with antibodies directed against Nat1p, Ard1p, Nat5p, and Ssb1/2p as indicated. (B) Mating efficiency in Δard1 is increased by high levels of Ssb1/2p. The mating efficiencies of Δard1 MATa, Δard1 MATa complemented with ARD1 on a centromeric plasmid (Δard1+ CEN-ARD1), and Δard1 MATa expressing SSB1 on a multicopy plasmid (Δard1+ 2μm-SSB1) are shown. The mating efficiency with a MATα tester strain was quantified as described in reference 8. The mating efficiency of Δard1 MATa ± CEN-ARD1 was set at 100%. (C) Simultaneous lack of nat1 and ssb1/2 results in synthetic phenotype enhancement. The wild type, Δssb1/2, Δnat1, and Δnat1Δssb1/2 were grown to early log phase at 30°C on minimal glucose medium. Serial 10-fold dilutions containing the same number of cells were spotted from left to right and incubated as indicated. Note that the weak growth defect of Δnat1 on 1 M NaCl-containing medium is only visible upon shorter incubation times.
NatA is in close proximity to the N termini of a variety of nascent polypeptides.
Methionine, the N-terminal amino acid of prepro α-factor, the nascent chain used for the initial purification of NatA, is not a substrate for Nα-acetylation by NatA (40). Nevertheless, Nat1p efficiently cross-links to prepro α-factor, indicating that it contacts a nonsubstrate nascent chain (Fig. 1C and 4A). To test whether NatA interaction was affected by its ability to recognize the nascent chain as a substrate, RNCs carrying the first 86 amino acids of the ribosomal protein Rpl4A, an in vivo substrate of NatA (4, 48), were generated and subjected to cross-linking. During in vitro translation, the initial methionine was efficiently removed from Rpl4A (data not shown) and Nat1p formed a cross-link to Rpl4A (Fig. 4A). Replacement of serine with lysine at position 2 changes Rpl4A to a nonsubstrate protein and, at the same time, generates an additional free amino group for the cross-link reaction. Rpl4A-S(2)K efficiently formed a cross-link to Nat1p (Fig. 4A). Nat1p also formed cross-link products to the precursor and the mature form of mitochondrial malate dehydrogenase (14). We conclude that Nat1p interacts with a variety of ribosome-bound nascent polypeptides, independently of their amino acid sequence or their Nα-terminal residue. In this respect, NatA resembles proteins with chaperone-like properties, like NAC and Ssb1/2p. NAC and Ssb1/2p only interact efficiently with nascent polypeptides in the context of the ribosome (36, 37, 56). To test whether NatA would also tightly interact with a polypeptide in the absence of ribosomes, we performed cross-link reactions with released nascent polypeptides. Nat1p formed a cross-link only when both the nascent polypeptide and Nat1p were bound to the ribosome (Fig. 4B). A less stable interaction between Nat1p and a polypeptide, however, may not be detected with this experimental setup.
NatA forms cross-link products to nascent polypeptides downstream of NAC and Ssb1/2p.
Yeast NAC interacts with nascent chains of 72 and 86 amino acids (42). Ssb1/2p was shown to bind to nascent chains of 54 amino acids; however, it failed to interact with a 34-amino-acid nascent chain (21). We have now, in parallel experiments, determined the minimal length requirement for the interaction of yeast malate dehydrogenase (Mdh1p) to NAC, Ssb1/2p, and NatA (Fig. 4C and D). NAC and Ssb1/2p showed interaction with an Mdh1p of at least 45 amino acids. However, Nat1p only formed a cross-link to an Mdh1p longer than 100 amino acids (Fig. 4C). The cross-link of Nat1p to Mdh1p required somewhat longer nascent chains than the cross-link to Rpl4A or the prepro α-factor (Fig. 4A and data not shown). This variability might be due to positioning of the primary amino groups or might be caused by conformational differences in the nascent polypeptides. Nascent chains of 50 and 70 amino acids, derived from prepro α-factor or Rpl4A, were tested for cross-link formation to confirm that Nat1p, in general, required longer nascent chains than NAC and Ssb1/2p. In agreement with the data obtained for Mdh1p, both prepro α-factor and Rpl4A of 50 and 70 amino acids formed cross-link products with NAC and Ssb1/2p; however, Nat1p did not (Fig. 4C). We conclude that cross-link formation of different nascent chains with Nat1p requires significantly longer nascent chains than cross-link formation with either NAC or Ssb1/2p.
Deletion of either NAT1 or ARD1 results in temperature sensitivity.
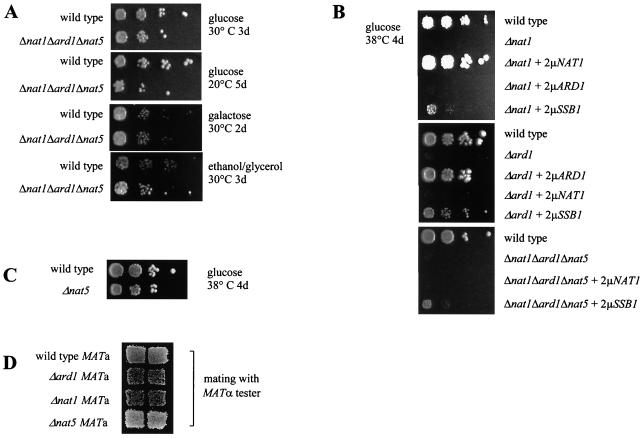
No general growth defect was associated with the deletion of all three NatA subunits (Δnat1Δard1Δnat5) on a variety of media at 20 and 30°C (Fig. 5A). Growth rates of the wild type and Δnat1Δard1Δnat5 were determined in parallel experiments. Both strains displayed doubling times of 1.7 h on yeast extract-peptone-dextrose and of 2.6 h on yeast extract-peptone-galactose at 30°C. At 20°C on yeast extract-peptone-dextrose, the wild type and Δnat1Δard1Δnat5 displayed doubling times of 4.3 and 4.2 h, respectively. At 38°C, however, deletion of either NAT1 or ARD1 results in temperature sensitivity (Fig. 5B). The triple deletion strain Δnat1Δard1Δnat5 also displayed temperature sensitivity (Fig. 5B and Fig. 6A). Growth at 38°C was tested in the triple deletion strain Δnat1Δard1Δnat5 overexpressing NAT1, ARD1, or NAT5. None of the subunits by itself was able to support growth at 38°C, whereas cooverexpression of Nat1p and Ard1p restored growth at 38°C (Fig. 5B and Fig. 6A). These data are compatible with a model in which the biological function of NatA strictly depends on the Nat1p-Ard1p complex, as was suggested earlier (28). In light of our findings, the simplest explanation might be that Nat1p in the absence of the catalytic subunit Ard1p is not an active Nα-acetyltransferase, whereas Ard1p cannot act as an Nα-acetyltransferase if not anchored to the ribosome via Nat1p (Fig. 3C and Fig. 7).
FIG. 5.
Absence of either Nat1p or Ard1p causes temperature sensitivity. (A) A haploid wild-type strain and the corresponding mutant strain lacking nat1, ard1, and nat5 (Δnat1Δard1Δnat5) were grown to early log phase at 30°C on minimal glucose medium. Serial 10-fold dilutions containing the same number of cells were spotted from left to right onto plates containing different carbon sources and additives and were incubated as indicated. (B) The wild type, Δard1, Δnat1, and Δnat1Δard1Δnat5 expressing ARD1, NAT1, or SSB1 on multicopy plasmids (all based on pYEPlac195; see Materials and Methods) were grown as described above. (C) Serial dilutions of the wild type and Δnat5 were grown at 38°C. (D) Patch mating assay. A W303 (MATa HMLα) wild-type strain and its Δnat1, Δard1, and Δnat5 derivatives were assayed with a MATα tester strain for mating efficiency in a patch mating assay.
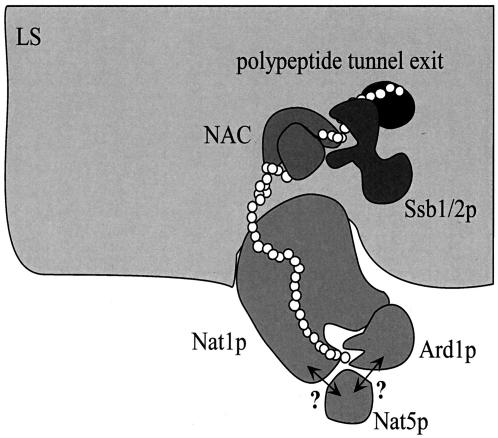
FIG. 7.
A model for cotranslational Nα-acetylation of polypeptides by NatA. Nat1p, the noncatalytic NatA subunit, mediates the stable interaction of the complex with the large ribosomal subunit (LS). In addition, Nat1p contacts the nascent polypeptide when approximately 40 amino acids have emerged from the exit tunnel (i.e., the nascent chain has a total length of approximately 80 amino acids). This interaction, possibly mediated via TPRs contained in Nat1p, may guide the growing polypeptide to the catalytic subunit Ard1p and the putatively catalytic subunit Nat5p, which transfer an acetyl moiety from acetyl-coenzyme A to specific N-terminal amino acids (see the introduction). Ard1p requires the presence of Nat1p for stable association with the ribosome. It is unknown whether Nat5p interacts with the ribosome via Nat1p, Ard1p, or both (indicated by question marks). The catalytic subunits may have little or no affinity for the nascent polypeptide in the absence of Nat1p; hence, a stable complex with Nat1p is essential for Nα-acetyltransferase activity. Complex formation between Nat1p and Ard1p might occur via predicted coiled coil domains in their C-terminal regions. Two other ribosome-bound factors, NAC and the Hsp70 homolog Ssb1/2p bind to nascent chains between 40 and 50 amino acids in length. Binding of NAC and Ssb1/2p precedes the binding of Nat1p. Whether or not NAC, Ssb1/2p, and NatA simultaneously bind to one and the same ribosome (as suggested here) or independently to different ribosome molecules is currently unknown. For details, see Results and Discussion. Please note that other factors bound to the ribosome or the nascent chain have been omitted for simplicity.
Deletion of NAT5 does not cause temperature sensitivity or HML derepression.
In contrast to the Δnat1 and Δard1 strains, Δnat5 did not display temperature sensitivity (Fig. 5C). As Δnat1 (MATa HMLα) and Δard1 (MATa HMLα) were previously shown to cause derepression of the HML silent mating-type locus (54), we tested whether Δnat5 would display the same phenotype. In a patch mating assay, Δnat5 (MATa HMLα) showed wild-type mating efficiency (Fig. 5D). To examine whether Δnat5 caused more subtle HML silencing defects, we also determined the response to α-factor mating pheromone in an α-factor confrontation assay (27). Like the wild type, Δnat5 showed 100% shmoo formation, whereas only approximately 10% of Δnat1 and Δard1 strains formed shmoos (data not shown). The combined data indicate that HML in the Δnat5 strain was repressed as efficiently as in the wild-type control. It has been suggested that HML derepression might be caused by the lack of Nα-acetylation of a so far unidentified substrate (40). According to this hypothesis, Ard1p, not Nat5p, would be responsible for that activity.
Sequence-independent interaction with nascent polypeptides, as observed for Nat1p, are properties reminiscent of chaperone-like proteins (18). To test the possibility that chaperones might be able to suppress the temperature-sensitive phenotype of strains lacking functional NatA, we have tested overexpression of the Hsp70 homologs SSA1, SSB1, and SSZ1; the Hsp40 homologs YDJ1 and ZUO1; HSP26; and HSP104 and co-overexpression of both subunits of NAC (EGD1 and EGD2). Only a high level of Ssb1/2p resulted in partial suppression of temperature sensitivity in Δnat, Δard1, and Δnat1Δard1Δnat5 (Fig. 5B and data not shown). Overexpression of Ssb1/2p was also tested for its effect on derepression of the HML silent mating-type locus caused by lack of NatA. When compared to the corresponding wild-type strain, the mating efficiency of Δard1 (MATa) with a MATα tester strain was reduced to 0.076%. Δard1 overexpressing Ssb1/2p (Δard1 MATa plus 2μm-SSB1) displayed a more-than-10-fold increase in mating efficiency (1.2% of the wild type) (Fig. 6B). We conclude that both temperature sensitivity and derepression of the HML locus are partly suppressed by overexpression of Ssb1/2p.
Our data indicated that Ssb1/2p and Nat1p are associated with the ribosome and localize in close proximity to the nascent polypeptide chain and that SSB1 acts, albeit weakly, as a multicopy suppressor in Δnat1Δard1Δnat5. However, the phenotype of Δnat1Δard1Δnat5 differs significantly from the phenotype of Δssb1/2. The absence of Ssb1/2p results in a complex set of phenotypes including slow growth, cold sensitivity, and sensitivity to high concentrations of salt (29). We have generated Δnat1Δssb1/2 to test for synthetic effects between Δssb1/2 and Δnat1. Δnat1Δssb1/2 displayed the combined set of phenotypes. Like Δssb1/2, the strain displayed slow growth and cold sensitivity, and like Δnat1, the strain was unable to grow at 38°C (data not shown). Δnat1 and Δssb1/2 strains share a moderate salt-sensitive phenotype. Δnat1Δssb1/2, however, was entirely unable to grow in the presence of salt (Fig. 6C). We conclude that simultaneous lack of nat1 and ssb1/2 results in a synthetic enhancement, compatible with parallel but functionally related roles of these two ribosome-bound factors.
DISCUSSION
A model of NatA interaction with the ribosome and the nascent polypeptide.
Nat1p is predicted to contain a minimum of 5 and up to 8 tetratricopeptide repeat (TPR) motifs (2). Clusters of TPR motifs have been found in a variety of proteins, including chaperones and proteins involved in protein transport (5). The partial crystal structure of the peroxisomal import receptor Pex5p in complex with a peroxisomal targeting peptide indicates that multiple TPR motifs form the peptide binding site (12). By analogy, interaction of Nat1p with the nascent polypeptide might be mediated by at least part of its TPR motifs. Nat1p also contains a highly charged region between amino acids 620 and 670 with the potential to form a coiled coil (25). This region of Nat1p is poised to interact with Ard1p, which also contains a potential coiled coil in its C-terminal domain (amino acids 194 to 224). This idea would be in agreement with earlier data indicating that a C-terminally truncated version of Ard1p did not stably interact with Nat1p (33). Interestingly, Nat5p lacks the C-terminal charged region detected in its homologue Ard1p. Based on a combination of structural features and biochemical data, we propose a model of subunit interactions for NatA, its binding to the ribosome, and its interaction with the nascent polypeptide (for details, see Fig. 7).
Nat1p plays a pivotal role in cotranslational Nα-terminal acetylation.
There is long-standing evidence from the mammalian system suggesting that Nα-terminal acetylation occurs cotranslationally (16, 32, 35, 51). Our results indicate that NatA indeed acts cotranslationally and that it is the Nat1p subunit that ensures cotranslational acetylation by dual means. Nat1p not only anchors the catalytic subunits to the ribosome but also presents the nascent chains to Ard1p (and possibly Nat5p), which in turn transfer the acetyl moiety from acetyl coenzyme A to the N terminus of the polypeptide chain (Fig. 7). Ard1p and Nat5p are unable to stably interact with their substrates by themselves, as indicated by their inability to bind to the ribosome. Furthermore, overexpression of Ard1p or Nat5p failed to suppress temperature sensitivity in the absence of Nat1p.
A complex arrangement around the ribosomal tunnel exit.
NatA shares the ability to contact nascent chains independent of their amino acid sequence with at least two other ribosome-associated proteins: NAC and Ssb1/2p. In addition, Srp54p, a subunit of the signal recognition particle, specifically interacts with nascent secretory polypeptides of 50 to 70 amino acids in length (23). The presence of these various factors at the ribosomal tunnel exit implies a highly ordered arrangement and sequence of binding events.
NAC is a heterodimeric complex which can reversibly bind to eukaryotic ribosomes. NAC is located in direct proximity to newly synthesized polypeptide chains as they emerge from the ribosome and is known to interact with nascent chains at a very early stage (43). A direct comparison between NAC and Ssb1/2p in our experimental setup revealed that both components bind to nascent chains as short as 45 to 50 amino acids. NAC and Ssb1/2p might coreside on the ribosome; however, our experiments cannot distinguish between simultaneous binding of both proteins to a single nascent chain or mutually exclusive binding to different nascent chains of equal length. Simultaneous binding of NatA and Ssb1/2p or NAC seems sterically possible, as NatA interacts with more-mature nascent chains (Fig. 4). Our future goal will be to determine the arrangement of NAC, Ssb1/2p, NatA and their dynamic interaction with the nascent polypeptide.
Nat5p, a subunit of NatA without function?
We found that Nat5p, a predicted member of the GNAT family of Nα-acetyltransferases, is bound to the functional NatA complex, consisting of Nat1p and Ard1p, in stoichiometric amounts. In contrast to Nat1p and Ard1p, which are essential for NatA function, the absence of Nat5p causes neither a growth defect at 38°C nor HML derepression. Attempts to detect Nat5p substrates in yeast by two-dimensional gel electrophoresis have failed so far (41). Possibly, Nat5p is responsible for the Nα-acetylation of a small subset of proteins that are not involved in mating type silencing or affected at elevated temperature. Results from a deletion suppressor screen hint at a putative function for Nat5p (1). Temperature sensitivity of two different double null mutant strains involved in ubiquitin-dependent protein degradation (mck1 mds1 and bul1 bul2) was suppressed by the deletion of NAT5 (termed ROG2 in reference 1).
The in vivo function of NatA is unknown.
As previously discussed by others, it is quite surprising that, although Ard1p and Nat1p are responsible for the Nα-acetylation of a large number of proteins, only a few specific phenotypes are observed in Δnat1 and Δard1 mutant strains at normal growth temperatures (33). However, Δnat1 and Δard1 strains displayed a general growth defect at 38°C.
Two different scenarios might account for temperature sensitivity in the absence of functional NatA. In the first hypothesis, temperature sensitivity would be caused by the lack of Nα-terminal acetylation of one or more proteins. De novo folding, thermal stability, or protease resistance might be impaired if proteins carry an unprotected Nα-amino group. However, up to now, experimental evidence in support of such direct effects is rare. Degradation by the proteasome is not prevented by Nα-terminal acetylation (26). Purified protein pairs identical but for Nα-acetylation are not easily obtained, and as a consequence, biochemical and biophysical properties of such protein pairs have not been studied (40). In the second scenario, the lack of Nα-acetylation would not be the sole reason for the defects observed in the absence of NatA. The ability of Nat1p to interact with nascent polypeptides is a property shared with chaperones. In this respect, it is interesting that the ribosome-bound chaperone Ssb1/2p can partly suppress temperature sensitivity and derepression of the HML locus. It seems unlikely that high levels of Ssb1/2p would restore Nα-acetylation of NatA substrates. Suppression of both temperature sensitivity and silencing defect suggests that Ssb1/2p is able to complement for a general function of NatA. Whether Ssb1/2p can partly substitute for Nat1p function on the ribosome or whether it suppresses defects caused by specific proteins that lack Nα-terminal acetylation awaits further investigation.
Acknowledgments
This work was supported by the Fonds der Chemischen Industrie (to S.R.) and by SFB 610 (to S.R.).
We thank Thierry Mini and Paul Jenö for the initial identification of Nat1p, Hans Trachsel for strain ts-187 (originally described in reference 19), and Hauke Lilie and members of the lab for critically reading of the manuscript.
REFERENCES
- 1.Andoh, T., Y. Hirata, and A. Kikuchi. 2000. Yeast glycogen synthase kinase 3 is involved in protein degradation in cooperation with Bul1, Bul2, and Rsp5. Mol. Cell. Biol. 20:6712-6720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andrade, M. A., C. P. Ponting, T. J. Gibson, and P. Bork. 2000. Homology-based method for identification of protein repeats using statistical significance estimates. J. Mol. Biol. 298:521-537. [DOI] [PubMed] [Google Scholar]
- 3.Aparicio, O. M., B. L. Billington, and D. E. Gottschling. 1991. Modifiers of position effect are shared between telomeric and silent mating-type loci in S. cerevisiae. Cell 66:1279-1287. [DOI] [PubMed] [Google Scholar]
- 4.Arnold, R. J., B. Polevoda, J. P. Reilly, and F. Sherman. 1999. The action of N-terminal acetyltransferases on yeast ribosomal proteins. J. Biol. Chem. 274:37035-37040. [DOI] [PubMed] [Google Scholar]
- 5.Blatch, G. L., and M. Lassle. 1999. The tetratricopeptide repeat: a structural motif mediating protein-protein interactions. Bioessays 21:932-939. [DOI] [PubMed] [Google Scholar]
- 6.Christianson, T. W., R. S. Sikorski, M. Dante, J. H. Shero, and P. Hieter. 1992. Multifunctional yeast high-copy-number shuttle vectors. Gene 110:119-122. [DOI] [PubMed] [Google Scholar]
- 7.Driessen, H. P., W. W. de Jong, G. I. Tesser, and H. Bloemendal. 1985. The mechanism of N-terminal acetylation of proteins. Crit. Rev. Biochem. 18:281-325. [DOI] [PubMed] [Google Scholar]
- 8.Ehrenhofer-Murray, A. E., D. H. Rivier, and J. Rine. 1997. The role of Sas2, an acetyltransferase homologue of Saccharomyces cerevisiae, in silencing and ORC function. Genetics 145:923-934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fluge, O., O. Bruland, L. A. Akslen, J. E. Varhaug, and J. R. Lillehaug. 2002. NATH, a novel gene overexpressed in papillary thyroid carcinomas. Oncogene 21:5056-5068. [DOI] [PubMed] [Google Scholar]
- 10.Fünfschilling, U., and S. Rospert. 1999. Nascent polypeptide-associated complex stimulates protein import into yeast mitochondria. Mol. Biol. Cell 10:3289-3299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garcia, P. D., W. Hansen, and P. Walter. 1991. In vitro protein translocation across microsomal membranes of Saccharomyces cerevisiae. Methods Enzymol. 194:675-682. [DOI] [PubMed] [Google Scholar]
- 12.Gatto, G. J., Jr., B. V. Geisbrecht, S. J. Gould, and J. M. Berg. 2000. Peroxisomal targeting signal-1 recognition by the TPR domains of human PEX5. Nat. Struct. Biol. 7:1091-1095. [DOI] [PubMed] [Google Scholar]
- 13.Gautschi, M., H. Lilie, U. Fünfschilling, A. Mun, S. Ross, T. Lithgow, P. Rücknagel, and S. Rospert. 2001. RAC, a stable ribosome-associated complex in yeast formed by the DnaK-DnaJ homologs Ssz1p and zuotin. Proc. Natl. Acad. Sci. USA 98:3762-3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gautschi, M., A. Mun, S. Ross, and S. Rospert. 2002. A functional chaperone triad on the yeast ribosome. Proc. Natl. Acad. Sci. USA 99:4209-4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gietz, R. D., and A. Sugino. 1988. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 74:527-534. [DOI] [PubMed] [Google Scholar]
- 16.Green, R. M., J. S. Elce, and R. Kisilevsky. 1978. Acetylation of peptidyl-tRNA on rat liver polyribosomes. Can. J. Biochem. 56:1075-1081. [DOI] [PubMed] [Google Scholar]
- 17.Haid, A., and M. Suissa. 1983. Immunochemical identification of membrane proteins after sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Methods Enzymol. 96:192-205. [DOI] [PubMed] [Google Scholar]
- 18.Hartl, F. U., and M. Hayer-Hartl. 2002. Molecular chaperones in the cytosol: from nascent chain to folded protein. Science 295:1852-1858. [DOI] [PubMed] [Google Scholar]
- 19.Hartwell, L. H., and C. S. McLaughlin. 1969. A mutant of yeast apparently defective in the initiation of protein synthesis. Proc. Natl. Acad. Sci. USA 62:468-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heitmann, J., N. R. Movva, P. C. Hiestand, and M. N. Hall. 1991. FK 506-binding protein proline rotamase is a target for the immunosuppressive agent FK 506 in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 88:1948-1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hundley, H., H. Eisenman, W. Walter, T. Evans, Y. Hotokezaka, M. Wiedmann, and E. Craig. 2002. The in vivo function of the ribosome-associated Hsp70, Ssz1, does not require its putative peptide-binding domain. Proc. Natl. Acad. Sci. USA 99:4203-4208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnston, M., and R. W. Davis. 1984. Sequences that regulate the divergent GAL1-GAL10 promoter in Saccharomyces cerevisiae. Mol. Cell. Biol. 4:1440-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kurzchalia, T. V., M. Wiedmann, A. S. Girshovich, E. S. Bochkareva, H. Bielka, and T. A. Rapoport. 1986. The signal sequence of nascent preprolactin interacts with the 54K polypeptide of the signal recognition particle. Nature 320:634-636. [DOI] [PubMed] [Google Scholar]
- 24.Lee, F. J., L. W. Lin, and J. A. Smith. 1988. Purification and characterization of an N alpha-acetyltransferase from Saccharomyces cerevisiae. J. Biol. Chem. 263:14948-14955. [PubMed] [Google Scholar]
- 25.Lupas, A., M. Van Dyke, and J. Stock. 1991. Predicting coiled coils from protein sequences. Science 252:1162-1164. [DOI] [PubMed] [Google Scholar]
- 26.Mayer, A., N. R. Siegel, A. L. Schwartz, and A. Ciechanover. 1989. Degradation of proteins with acetylated amino termini by the ubiquitin system. Science 244:1480-1483. [DOI] [PubMed] [Google Scholar]
- 27.Meijsing, S. H., and A. E. Ehrenhofer-Murray. 2001. The silencing complex SAS-I links histone acetylation to the assembly of repressed chromatin by CAF-I and Asf1 in Saccharomyces cerevisiae. Genes Dev. 15:3169-3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mullen, J. R., P. S. Kayne, R. P. Moerschell, S. Tsunasawa, M. Gribskov, M. Colavito-Shepanski, M. Grunstein, F. Sherman, and R. Sternglanz. 1989. Identification and characterization of genes and mutants for an N-terminal acetyltransferase from yeast. EMBO J. 8:2067-2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nelson, R. J., T. Ziegelhoffer, C. Nicolet, M. Werner-Washburne, and E. A. Craig. 1992. The translation machinery and 70 kd heat shock protein cooperate in protein synthesis. Cell 71:97-105. [DOI] [PubMed] [Google Scholar]
- 30.Neuwald, A. F., and D. Landsman. 1997. GCN5-related histone N-acetyltransferases belong to a diverse superfamily that includes the yeast SPT10 protein. Trends Biochem. Sci. 22:154-155. [DOI] [PubMed] [Google Scholar]
- 31.Notredame, C., D. G. Higgins, and J. Heringa. 2000. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 302:205-217. [DOI] [PubMed] [Google Scholar]
- 32.Palmiter, R. D., J. Gagnon, and K. A. Walsh. 1978. Ovalbumin: a secreted protein without a transient hydrophobic leader sequence. Proc. Natl. Acad. Sci. USA 75:94-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Park, E. C., and J. W. Szostak. 1992. ARD1 and NAT1 proteins form a complex that has N-terminal acetyltransferase activity. EMBO J. 11:2087-2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pestana, A., and H. C. Pitot. 1975. Acetylation of nascent polypeptide chains on rat liver polyribosomes in vivo and in vitro. Biochemistry 14:1404-1412. [DOI] [PubMed] [Google Scholar]
- 35.Pestana, A., and H. C. Pitot. 1975. Acetylation of ribosome-associated proteins in vitro by an acetyltransferase bound to rat liver ribosomes. Biochemistry 14:1397-1403. [DOI] [PubMed] [Google Scholar]
- 36.Pfund, C., N. Lopez-Hoyo, T. Ziegelhoffer, B. A. Schilke, P. Lopez-Buesa, W. A. Walter, M. Wiedmann, and E. A. Craig. 1998. The molecular chaperone Ssb from Saccharomyces cerevisiae is a component of the ribosome-nascent chain complex. EMBO J. 17:3981-3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Plath, K., and T. A. Rapoport. 2000. Spontaneous release of cytosolic proteins from posttranslational substrates before their transport into the endoplasmic reticulum. J. Cell Biol. 151:167-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Polevoda, B., T. S. Cardillo, T. C. Doyle, G. S. Bedi, and F. Sherman. 2003. Nat3p and Mdm20p are required for function of yeast NatB Nα-terminal acetyltransferase and of actin and tropomyosin. J. Biol. Chem. 278:30686-30697. [DOI] [PubMed] [Google Scholar]
- 39.Polevoda, B., J. Norbeck, H. Takakura, A. Blomberg, and F. Sherman. 1999. Identification and specificities of N-terminal acetyltransferases from Saccharomyces cerevisiae. EMBO J. 18:6155-6168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Polevoda, B., and F. Sherman. 2000. Nalpha-terminal acetylation of eukaryotic proteins. J. Biol. Chem. 275:36479-36482. [DOI] [PubMed] [Google Scholar]
- 41.Polevoda, B., and F. Sherman. 2003. N-terminal acetyltransferases and sequence requirements for N-terminal acetylation of eukaryotic proteins. J. Mol. Biol. 325:595-622. [DOI] [PubMed] [Google Scholar]
- 42.Reimann, B., J. Bradsher, J. Franke, E. Hartmann, M. Wiedmann, S. Prehn, and B. Wiedmann. 1999. Initial characterization of the nascent polypeptide-associated complex in yeast. Yeast 15:397-407. [DOI] [PubMed] [Google Scholar]
- 43.Rospert, S., Y. Dubaquié, and M. Gautschi. 2002. Nascent-polypeptide-associated complex. Cell. Mol. Life Sci. 59:1632-1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rospert, S., and R. L. Hallberg. 1995. Interaction of HSP 60 with proteins imported into the mitochondrial matrix. Methods Enzymol. 260:287-292. [DOI] [PubMed] [Google Scholar]
- 45.Schägger, H., and G. von Jagow. 1987. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166:368-379. [DOI] [PubMed] [Google Scholar]
- 46.Sherman, F., G. R. Fink, and J. B. Hicks. 1986. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 47.Stone, E. M., M. J. Swanson, A. M. Romeo, J. B. Hicks, and R. Sternglanz. 1991. The SIR1 gene of Saccharomyces cerevisiae and its role as an extragenic suppressor of several mating-defective mutants. Mol. Cell. Biol. 11:2253-2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Takakura, H., S. Tsunasawa, M. Miyagi, and J. R. Warner. 1992. NH2-terminal acetylation of ribosomal proteins of Saccharomyces cerevisiae. J. Biol. Chem. 267:5442-5445. [PubMed] [Google Scholar]
- 49.Tercero, J. C., J. D. Dinman, and R. B. Wickner. 1993. Yeast MAK3 N-acetyltransferase recognizes the N-terminal four amino acids of the major coat protein (gag) of the L-A double-stranded RNA virus. J. Bacteriol. 175:3192-3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tercero, J. C., and R. B. Wickner. 1992. MAK3 encodes an N-acetyltransferase whose modification of the L-A gag NH2 terminus is necessary for virus particle assembly. J. Biol. Chem. 267:20277-20281. [PubMed] [Google Scholar]
- 51.Uy, R., and F. Wold. 1977. Posttranslational covalent modification of proteins. Science 198:890-896. [DOI] [PubMed] [Google Scholar]
- 52.Wach, A., A. Brachat, R. Pohlmann, and P. Philippsen. 1994. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10:1793-1808. [DOI] [PubMed] [Google Scholar]
- 53.Welch, E. M., and A. Jacobson. 1999. An internal open reading frame triggers nonsense-mediated decay of the yeast SPT10 mRNA. EMBO J. 18:6134-6145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Whiteway, M., R. Freedman, S. Van Arsdell, J. W. Szostak, and J. Thorner. 1987. The yeast ARD1 gene product is required for repression of cryptic mating-type information at the HML locus. Mol. Cell. Biol. 7:3713-3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Whiteway, M., and J. W. Szostak. 1985. The ARD1 gene of yeast functions in the switch between the mitotic cell cycle and alternative developmental pathways. Cell 43:483-492. [DOI] [PubMed] [Google Scholar]
- 56.Wiedmann, B., H. Sakai, T. A. Davis, and M. Wiedmann. 1994. A protein complex required for signal-sequence-specific sorting and translocation. Nature 370:434-440. [DOI] [PubMed] [Google Scholar]
- 57.Yamada, R., and R. A. Bradshaw. 1991. Rat liver polysome N alpha-acetyltransferase: isolation and characterization. Biochemistry 30:1010-1016. [DOI] [PubMed] [Google Scholar]