Abstract
Facioscapulohumeral muscular dystrophy (FSHD) is a neuromuscular disorder characterized by an insidious onset and progressive course. The disease has a frequency of about 1 in 20,000 and is transmitted in an autosomal dominant fashion with almost complete penetrance. Deletion of an integral number of tandemly arrayed 3.3-kb repeat units (D4Z4) on chromosome 4q35 is associated with FSHD but otherwise the molecular basis of the disease and its pathophysiology remain obscure. Comparison of mRNA populations between appropriate cell types can facilitate identification of genes relevant to a particular biological or pathological process. In this report, we have compared mRNA populations of FSHD and normal muscle. Unexpectedly, the dystrophic muscle displayed profound alterations in gene expression characterized by severe underexpression or overexpression of specific mRNAs. Intriguingly, many of the deregulated mRNAs are muscle specific. Our results suggest that a global misregulation of gene expression is the underlying basis for FSHD, distinguishing it from other forms of muscular dystrophy. The experimental approach used here is applicable to any genetic disorder whose pathogenic mechanism is incompletely understood.
Facioscapulohumeral muscular dystrophy (FSHD) is an autosomal dominant neuromuscular disorder characterized by progressive involvement of facial and limb girdle muscles and significant clinical variability (1). Its incidence is 1 in 20,000 with almost complete penetrance (2). The FSHD genetic locus has been mapped by linkage analysis to the very distal part of the chromosome 4 long arm (3–6). FSHD is associated with the presence of EcoRI fragments shorter than 35 kb belonging to a polymorphic region and containing a 3.3-kb KpnI tandemly arrayed repeat (D4Z4), whose size varies in normal individuals between 35 and 300 kb (7). It thus has been hypothesized that deletions of the tandemly repeated units generating short alleles might cause FSHD through a rearrangement of chromatin structure that silence genes in the vicinity (8). In Drosophila, there is an analogous phenomenon, termed position effect variegation (PEV) (9). As a result of PEV, the expression of an euchromatic gene is altered when it is transposed adjacent to heterochromatic regions (10). However, no transcribed sequences have been identified within the tandemly repeated array (11–13) and the pathogenic mechanism of FSHD has not been elucidated.
The molecular basis of biological or pathological mechanisms might be understood through analysis of the mRNA expression pattern in a specific cell or tissue. In particular, the isolation of differentially expressed genes may allow the characterization of pathways involved in a specific biological function (14) or pathological process (15, 16).
To define the molecular mechanism responsible for FSHD, we compared mRNA expression patterns of FSHD and normal muscle. To isolate genes that are differentially expressed in the dystrophic muscle we performed PCR-based subtractive hybridization (17). We generated two subtractive libraries: one containing genes overexpressed in FSHD muscle (FSHD muscle cDNA subtractive library); the other containing genes underexpressed in FSHD muscle (normal muscle cDNA subtractive library). These two libraries were highly enriched in muscle-specific transcripts. Moreover, we isolated genes whose altered expression was consistent with the established pathophysiological mechanism of the disease. Here we show that a characteristic feature of FSHD muscle is gross misregulation of gene expression. Our results provide information useful for ultimately identifying the genetic defect responsible for FSHD.
Materials and Methods
RNA Extraction and Subtractive Hybridization.
Muscle biopsies were obtained with informed consent from the deltoid muscle of four FSHD patients, one Becker muscular dystrophy (BMD) patient, and one amyotrophic lateral sclerosis (ALS) patient and three sex- and age-matched normal individuals. All of the selected FSHD patients displayed the same degree of severity of the disease. The selection of the site for sampling was based on the clinical examination of the patient’s muscle. Typically, the upper portion of the deltoid muscle is affected and the lower portion is spared (1). All samples underwent histological examination. In particular, the results of the histological analysis of the muscle sample used in the subtractive hybridization experiment have been described (18). Total RNA was extracted from frozen samples by using the TRI Reagent (Sigma). cDNAs were synthesized from 1 μg of DNase-treated total RNA isolated by using the Smart PCR cDNA synthesis kit (CLONTECH). In brief, total RNA was primed with 1 μM of cDNA synthesis primer (5′-AAGCAGTGGTATCAACGCAGAGTACT(30)N−1N-3′, N = A, C, G, or T, N−1 = A, G, or C) and reverse-transcribed. When reverse transcriptase reaches the end of the mRNA, its endogenous terminal transferase activity adds a few additional deoxycytidine residues. This dC stretch pairs with the oligo dG at the 3′ end of a second oligonucleotide (5′-AAGCAGTGGTATCAACGCAGAGTACGCGGG-3′). Reverse transcriptase then switches templates and extends to the end of the second oligonucleotide. First-strand cDNA corresponding to 20 ng of the starting RNA was PCR-amplified with 1 μM amplification primer with sequence corresponding to the 5′ end of both oligonucleotides (5′-AAGCAGTGGTATCAACGCAGAGT-3′). Long-distance PCR was performed at 95°C for 5 sec, 65°C for 5 sec, and 68°C for 6 min for the optimal cycle number that yielded exponential amplification. This method overcomes the scarcity of RNA obtained from the bioptic material and allows synthesis of full-length cDNA. The cDNAs from normal and FSHD-affected muscle were used as tester and driver, alternatively, in two separate subtractive-hybridization experiments to isolate, respectively, genes overexpressed (the FSHD muscle cDNA was the tester) or underexpressed (the normal muscle cDNA was the tester) in the FSHD dystrophic muscle. The experiments were performed by using a PCR-select cDNA subtraction kit (CLONTECH) as described (17). This method overcomes the problem of variable mRNA abundance by introducing a hybridization step that normalizes sequence abundance. The cDNAs from the FSHD-affected muscle and one normal muscle were digested with RsaI before cDNA subtraction and the tester was ligated to adapter DNA. After two hybridizations with the tester and the driver (8 hr and 14 hr) the resulting mixture was diluted 1:1,000 and PCR-amplified by using flanking and nested primers to produce a subtracted and normalized PCR fragment library.
cDNA Library Construction and Analysis.
After hybridization, differentially expressed transcripts selectively amplified by suppression PCR (17) were digested with SmaI/EagI and ligated into pBSSKII vector (Stratagene). Blue-white screening was carried out in Escherichia coli DH5α cells. A total of 132 partial sequences of recombinant clones were determined with dye primers by using the dideoxy chain termination method (19) and compared with entries in the GenBank database by using the blast homology search program (20). Designation of a gene identity for an expressed sequence tag (EST) indicates that the misassignment probability was less than 10−35 and the muscle sequences aligned with 98–100% identity.
Virtual Northern Blots.
Five micrograms of PCR-amplified cDNAs from each sample were fractionated by electrophoresis on a 1.2% agarose gel at 50 V for 4 hr in 1× TBE (90 mM Tris/90 mM boric acid/2.0 mM EDTA, pH 8.0), transferred onto a Zetabind membrane (Bio-Rad) by alkaline blotting and hybridized with the indicated 32P-labeled cDNA probes at 65°C in a 0.5 M NaHPO4, 7% SDS solution. Use of PCR-amplified cDNAs circumvents the scarcity of total RNA obtained from the bioptic materials and provides information similar to those obtained from bona fide Northern blots (21).
Expression Analysis.
Northern blots carrying poly(A)+ mRNA from human multiple tissues (CLONTECH) were hybridized with the indicated radiolabeled cDNA probes at 68°C for 1 hr in ExpressHyb (CLONTECH) hybridization solution to define their expression pattern.
Protein Profile Analysis.
The composition in myosin heavy chain (MHC) isoforms was determined in single muscle fibers by SDS/PAGE. Three bands in the MHC region, corresponding to three isoforms (MHC-1, MHC-2A, and MHC-2B) were separated and detected by silver staining (Amersham Pharmacia). The isoform composition of tropomyosin (TM) was determined in small fragments of bundles by gel electrophoresis on 15% polyacrylamide gels according to the procedure described by Giulian et al. (22). Three TM isoforms were identified by immunoblotting using a primary mouse mAb against sarcomeric TM (Sigma). TM bands were visualized by an enhanced chemiluminescent method (Bio-Rad).
Results
Construction and Analysis of Subtractive Libraries.
To isolate genes that are abnormally expressed in FSHD muscle, we performed PCR-based subtractive hybridization (17) by using poly(A)+ mRNA isolated from deltoid muscle biopsies of one normal individual and one FSHD patient. We generated two subtractive libraries: one containing genes overexpressed in FSHD muscle (FSHD muscle cDNA subtractive library); the other containing genes underexpressed in FSHD muscle (normal muscle cDNA subtractive library).
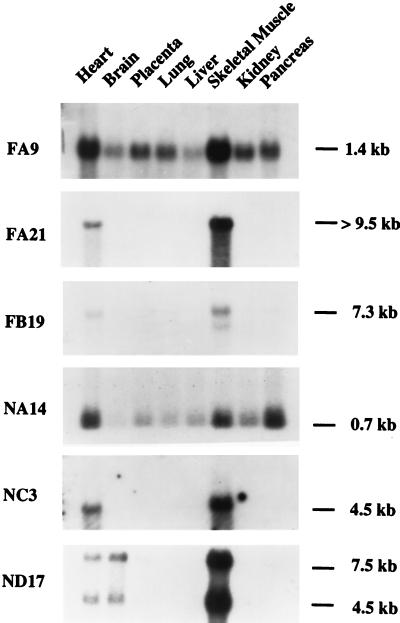
Partial sequences of 79 clones derived from the FSHD muscle cDNA subtractive library and 53 derived from the normal muscle cDNA subtractive library were analyzed for homology in GenBank and European Molecular Biology Laboratory databases. As summarized in Table 1, 24% of the isolated cDNAs did not correspond to any known gene or sequence (ESTs, sequence-tagged sites, or coding sequences). Of the known genes, a significant number (41%) were muscle specific (Table 2), and 29% of the identified ESTs were isolated from heart and skeletal muscle cDNA libraries. This finding suggests that the two subtractive libraries are highly enriched in muscle-specific transcripts. To confirm this idea, Northern blot analysis was performed with several cDNAs (FA9, FA21, FB19, NA14, NC3, and ND17) by using a panel of poly(A)+ mRNAs isolated from various human adult tissues. Fig. 1 shows that all six cDNA probes detected transcripts expressed primarily or exclusively in heart and skeletal muscle.
Table 1.
Sequence analysis of cDNA clones
| Clones | Category | Number |
|---|---|---|
| FSHD muscle subtractive library, 79 (100%) | ||
| Known function | Human | 16 (20%) |
| Other species | 3 (4%) | |
| Total | 19 (24%) | |
| Unknown function | CDS | 3 (4%) |
| STS | 4 (5%) | |
| EST | 15 (19%) | |
| Genomic DNA | 1 (1%) | |
| No homologies | 20 (25%) | |
| Total | 43 (54%) | |
| Repetitive elements | 17 (22%) | |
| Normal muscle subtractive library, 53 (100%) | ||
| Known function | Human | 12 (22.5%) |
| Other species | 3 (5.5%) | |
| Total | 15 (28%) | |
| Unknown function | 31 (59%) | |
| CDS | 0 | |
| STS | 0 | |
| EST | 19 (36%) | |
| Genomic DNA | 1 (2%) | |
| No homologies | 11 (21%) | |
| Total | 31 (59%) | |
| Repetitive elements | 7 (13%) | |
A total of 132 partial sequences of recombinant clones derived from the two muscle cDNA subtractive libraries were determined and compared with entries in GenBank database by using the blast homology search program (20). Designation of a gene identity for an EST indicates that the misassignment probability was less than 10−35 and the muscle sequences aligned with 98–100% identity. Complete sequence information for all cDNA clones is available on request rosella.tupler@umassmed.edu. STS, sequence-tagged sites; CDS, coding sequence. Clones were grouped on the basis of their homology or similarity to published sequences. Clones with homology greater than 95% were considered identical. Clones at least 60% similar to a gene from a heterologous species were considered to have a similar function. Remaining clones, which did not fit into any of the above categories, were considered novel genes.
Table 2.
Summary of cDNA clones corresponding to genes with known function
| Category | FSHD muscle subtractive library | Normal muscle subtractive library |
|---|---|---|
| Muscle structure proteins | TM-α chain (slow skeletal muscle) (M) (FC 30) | β-MHC (fast skeletal muscle) (M) (NC 24) |
| Myoglobin (M) (FA 24) | ||
| Titin (M) (FB 34) | ||
| Nebulin (M) (FC 40) | ||
| Regulatory factors | Human homologue to yeast RAD 6 (FG 16) | XP-C repair complementing protein (NC 2) |
| Histone 4 acetyl transferase (M) (FA 9) | Core binding factor beta transcriptional coactivator (NB 13) | |
| Nonhistone chromosomal protein HMG17 (FF 31) | Myocyte enhancer factor 2C (M) (ND 17) | |
| Early growth response-1 factor (NH 23) | ||
| RING finger protein (NA 27) | ||
| Human apobec-1 binding protein (FC 1) | Eukaryotic initiation factor 4B (NA 34) | |
| Human ribosomal protein L7 (NB 8) | ||
| Serine-threonine phosphtase 2C α (FA 44) | Tyrosine phosphatase (CAAX) (NA 21) | |
| Platelet glycoprotein IV (NF 23) | ||
| Thrombomodulin (NG 19) | ||
| Mitochondrial proteins | Adenine nucleotide translocator 1 (M) (FE 45) | |
| Manganese superoxide dismutase 2 (FG 28) | ||
| Carbonic anydrase II (M) (FG 35) | ||
| Other | Vacuolar H(+) ATPase (FA 13) | H-capping protein (M) (NC 6) |
| Dihydro lipoamide dehydrogenase (FA 10) | ||
| Adenylosuccinate synthetase (M) (FC 31) |
Known human genes from both libraries were grouped according to biological function. Four categories were identified. Three of them (muscle structure, mitochondria, and other) relate to basic activities of the muscle cell. The fourth, regulatory factors, includes components affecting mainly transcription, posttranscriptional processes, translation, and signal transduction. M, muscle-specific gene.
Figure 1.
Transcription profiles of isolated cDNAs. Autoradiograms of multiple tissue Northern blots hybridized with clones FA9, FA21, FB19, NA14, NC3, and ND17. The exposure time ranged from 12 hr to 1 week, indicative of different abundance mRNAs. Clones FA9 and FB19 detected mRNAs of 1.4 and 7.3 kb, respectively, highly expressed in skeletal muscle and heart. Clones FA21 and NC3 detected mRNA transcripts of 4.5 and >9.5 kb, respectively, uniquely expressed in skeletal muscle and heart. Clone NA14 detected a 0.7-kb mRNA abundantly expressed in skeletal muscle, heart, and pancreas. Clone ND17 detected two mRNAs, 7.5 and 4.5 kb, primarily expressed in skeletal muscle, heart, and brain.
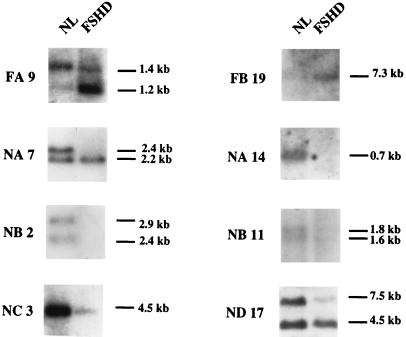
To verify that the identified genes also were differentially expressed, eight cDNAs were used to probe a “virtual” Northern blot containing immobilized, PCR-amplified cDNA derived from normal and FSHD mRNA (see Materials and Methods). Fig. 2 shows that all eight genes were, as expected, differentially expressed: clones FA9 and FB19, isolated from the FSHD muscle cDNA subtractive library, were overexpressed in FSHD muscle; clones NA7, NA14, NB2, NB11, NC3, and ND17, isolated from the normal muscle cDNA subtractive library, were underexpressed in FSHD muscle. These results confirm that the majority of the genes present in the two libraries were indeed differentially expressed.
Figure 2.
Expression of cDNA clones in FSHD and normal muscle (NL). Blots containing an equal amount of PCR-amplified cDNA from normal and FSHD-affected muscle mRNAs were hybridized with 32P-labeled cDNA clones. FA9 and FB19 were derived from the FSHD muscle cDNA subtractive library. Hybridization using radiolabeled FA9 revealed two bands of 1.4 and 1.2 kb. In the FSHD muscle the 1.4-kb species was of lower intensity, whereas in the FSHD muscle the 1.2-kb species was predominant. Hybridization with FB19 detected a single 7.3-kb band only in the FSHD muscle cDNA. Clones NA7, NA14, NB2, NB11, NC3, and ND17, derived from the normal muscle cDNA subtractive library, correspond to reverse-transcribed mRNAs underexpressed in the FSHD muscle. NA7 detected two bands of 2.4 and 2.2 kb in the normal muscle, whereas the 2.4-kb band was absent from the FSHD muscle. NA14 detected one 0.7-kb band in normal muscle cDNA, which was absent from FSHD muscle. NB2 and NB11 detected, in each case, two bands of 2.9 and 2.4, and 1.8 and 1.6 kb, respectively, that were absent from FSHD muscle. NB11 and NC3 detected one band of 2.2 and 4.5 kb, respectively, in the normal muscle, which was less intense in FSHD muscle. ND17 cDNA revealed two bands, 7.5 and 4.5 kb. The 7.5-kb signal was less intense in FSHD muscle.
Analysis of Muscle-Specific Structural Proteins.
The deregulation of muscle-specific transcripts implied that genes encoding muscle structural proteins also would be affected. Consistent with this possibility, Table 2 shows that TM, β-MHC, myoglobin, nebulin, and titin mRNAs also were altered in the FHSD muscle. To confirm that there was also deregulation at the protein level, we analyzed the protein profile of normal and FSHD muscle by silver staining or immunoblotting. Fig. 3 shows that the fast isoform MHC-2B (silver staining) and the fast form of the TM-α (immunoblotting) were absent from the FSHD muscle.
Figure 3.
Analysis of muscle-specific structural proteins in FSHD muscle fibers. MHC isoforms in bioptic samples from a control subject (NL) and a FSHD patient were detected by silver staining. The fast isoform MHC-2B is absent from the FSHD patient muscle (Left). TM isoforms from the same samples were detected by immunoblotting, using a specific anti-TM antibody (Sigma). The TM-α fast is not present in the FSHD patient muscle (Right).
Comparison to Other Neuromuscular Disorders.
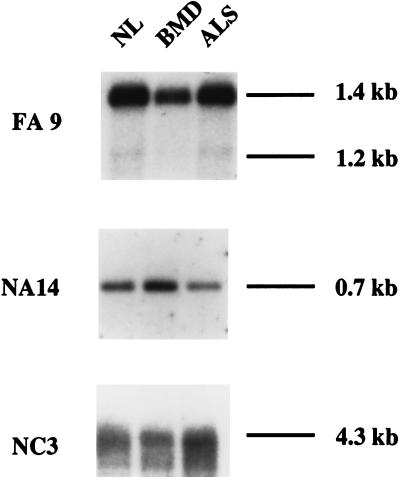
To verify that deregulated expression of muscle-specific genes distinguishes FSHD from other neuromuscular diseases, we analyzed the expression of FA9, NA14, and NC3 in BMD and ALS. These diseases arise from two different types of molecular pathophysiology. In BMD, the dystrophic process results from mutations in the dystrophin gene that alters the interaction between the cytoskeleton and sarcolemma, causing muscle degeneration associated with muscle cell death and regeneration (23). In ALS, an atrophic process occurs in muscle caused by lack of electrical stimuli and resultant α-motor neuron death (24). The results of Fig. 4 show that expression of the three genes in these other neuromuscular diseases was normal.
Figure 4.
Transcription profile in other neuromuscular disorders. Representative differentially expressed transcripts in FSHD muscles (FA9, NA14, and NC3) were used to probe blots containing PCR-amplified cDNAs from muscles affected by BMD and ALS. NL, normal control.
Analysis of Other FSHD Patients.
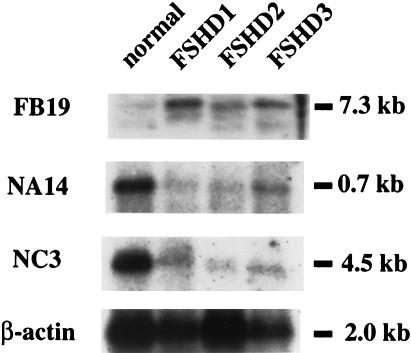
To verify the generality of the deregulated expression of muscle-specific genes observed in FSHD muscle, we extended the expression analysis of FB19, NA14, and NC3 to three other FSHD patients and a normal control. The results of Fig. 5 show that FB19 is overexpressed and NA14 and NC3 are underexpressed in the dystrophic muscles of all three FSHD patients. We conclude that expression of muscle-specific genes is misregulated in FSHD.
Figure 5.
A comparable transcription profile in muscle from multiple FSHD patients. Representative differentially expressed transcripts (FB19, NA14, and NC3) were used to probe blots containing PCR-amplified cDNA isolated from FSHD-affected muscle.
Discussion
FSHD is a complex disease with a peculiar involvement of muscle groups, highly variable severity, and unpredictable progression. In light of this complexity, our experiments sought to identify genes relevant to the abnormal muscle phenotype. Differential screening of mRNA populations between normal and affected tissues is a potentially powerful strategy for identifying genes involved in a particular pathological process. In this report we show that profound changes in gene regulation occur in the dystrophic muscle of FSHD patients. The differentially expressed transcripts could represent genes whose expression depends on the mutated gene or genes that are linked to functional changes secondary to the dystrophic process. At this stage we do not know the primary cause of the global alteration of gene expression in FSHD. It is nevertheless attractive to speculate that a genetic defect in a transcription factor might be the underlying basis of the FSHD dystrophic process.
Interestingly, we found that a significant number of the abnormally expressed genes encode transcription regulators: for example, HMG 17, a nonhistone chromosomal protein (25); HHR6, the human homologue to yeast RAD6 (26); and a new histone 4 acetyl transferase (27) (FA9) are overexpressed in FSHD muscle. Five genes encoding transcription factors are underexpressed in FSHD muscle, including ND17, which corresponds to human myocyte enhancer factor 2C (MEF2C), known to play an important role in myogenesis (28). The deregulated expression of these factors could explain the wide spectrum of genes that are abnormally transcribed in FSHD muscle. Consistent with this idea is our analysis of muscle contractile proteins (Fig. 3) and the prevalence of hypertrophic slow muscle fibers type 1 in histological sections (data not shown). Thus, the ultimate consequence of the deregulated gene expression is loss of normal muscle molecular architecture, explaining the established pathophysiology of the disease (1).
The finding that genes whose expression is altered in FSHD are expressed normally in BMD and ALS indicates that global misregulation of muscle-specific gene expression is not a common feature of neuromuscular disease but rather is specific to FSHD. The discovery of several additional muscle-specific genes suggests that this experimental approach can help elucidate the molecular basis of muscle cell biology and uncover new mechanisms leading to muscular dystrophy. This experimental strategy may make it possible to achieve a general view about complex mechanisms occurring in an affected tissue and is a straightforward approach to analyzing a complex disease phenotype.
Acknowledgments
We thank D. De Grandis and C. Angelini (Telethon Neuromuscular Tissue Bank C07) for providing the muscle tissue samples, and C. Reggiani, P. Maraschio, and L. Tiepolo for helpful suggestions and for critically reading the manuscript. This work was supported by Telethon Grant 1041 (to R.T.). R.T. is a FSH Society-Delta Railroad Construction Fellow, and M.R.G. is an Investigator of the Howard Hughes Medical Institute.
Abbreviations
- FSHD
facioscapulohumeral muscular dystrophy
- BMD
Becker muscular dystrophy
- ALS
amyotrophic lateral sclerosis
- EST
expressed sequence tag
- MHC
myosin heavy chain
- TM
tropomyosin
Footnotes
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AW0055750-69 and AW055377-87).
References
- 1.Munsat T L. In: Myology. Engel A G, Banker B Q, editors. New York: McGraw–Hill; 1994. pp. 1220–1232. [Google Scholar]
- 2.Lunt P W, Harper P S. J Med Genet. 1992;26:755–760. doi: 10.1136/jmg.26.12.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gilbert J R, Stajich J M, Speer M C, Vance J M, Stewart C S, Yamaoka L H, Samson F M, Fardeau M, Potter T G, Roses A D, Pericak-Vance M A. Am J Hum Genet. 1992;51:424–427. [PMC free article] [PubMed] [Google Scholar]
- 4.Mathews K D, Mills K A, Bosch E P, Ionasescu V V, Wiles K R, Buetow K H, Murray J C. Am J Hum Genet. 1992;51:428–431. [PMC free article] [PubMed] [Google Scholar]
- 5.Weiffenbach B, Bagley R, Falls K, Hyser C, Storvick D, Schultz P, Mendell J, van Dijk K W, Milner E C B, Griggs R. Am J Hum Genet. 1992;51:416–423. [PMC free article] [PubMed] [Google Scholar]
- 6.Sarfarazi M, Wijmenga C, Upadhyaya M, Weiffenbach B, Hyser C, Mathews K D, Murray J C, Gilbert J R, Pericak-Vance M A, Lunt P, et al. Am J Hum Genet. 1992;51:396–403. [PMC free article] [PubMed] [Google Scholar]
- 7.Wijmenga C, Hewitt J E, Sandkuijl L A, Clark L N, Wright T J, Dauwerse G H, Gruter A, Hofker M H, Moerer P, Williamson R, et al. Nat Genet. 1992;2:26–30. doi: 10.1038/ng0992-26. [DOI] [PubMed] [Google Scholar]
- 8.Winokur S T, Bengtsson U, Feddersen J, Mathews K D, Weiffenbach B, Bailey H, Markovich R P, Murray J C, Wasmuth J J, Altherr M R, et al. Chromosome Res. 1994;2:225–234. doi: 10.1007/BF01553323. [DOI] [PubMed] [Google Scholar]
- 9.Moehrle A, Paro R. Dev Genet. 1994;15:478–484. doi: 10.1002/dvg.1020150606. [DOI] [PubMed] [Google Scholar]
- 10.Hendrich B D, Willard H F. Hum Mol Genet. 1995;4:1765–1777. doi: 10.1093/hmg/4.suppl_1.1765. [DOI] [PubMed] [Google Scholar]
- 11.Hewitt J E, Lyle R, Clark L N, Valleley E M, Wright T J, Wijmenga C, van Deutekom J C, Francis F, Sharpe P T, Hofker M, et al. Hum Mol Genet. 1994;3:1287–1295. doi: 10.1093/hmg/3.8.1287. [DOI] [PubMed] [Google Scholar]
- 12.Lee J H, Goto K, Matsuda C, Arahata K. Muscle Nerve. 1995;2:S6–S13. [PubMed] [Google Scholar]
- 13.Lyle R, Wright T J, Clark N L, Hewitt J E. Genomics. 1995;28:389–397. doi: 10.1006/geno.1995.1166. [DOI] [PubMed] [Google Scholar]
- 14.Morris M E, Viswanathan N, Kuhlman S, Davis F, Weitz C J. Science. 1998;279:1544–1547. doi: 10.1126/science.279.5356.1544. [DOI] [PubMed] [Google Scholar]
- 15.Lisitsyn N, Lisitsyn N, Wigler M. Science. 1993;259:946–951. doi: 10.1126/science.8438152. [DOI] [PubMed] [Google Scholar]
- 16.Kuang W W, Thompson D A, Hoch R V, Weigel R J. Nucleic Acids Res. 1998;26:1116–1123. doi: 10.1093/nar/26.4.1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Diatchenko L, Lau Y-F C, Campbell A P, Chenchik A, Moqadam F, Huang B, Lukyanov S, Lukyanov K, Gurskaya N, Sverdlov E D, Siebert P D. Proc Natl Acad Sci USA. 1996;93:6025–6030. doi: 10.1073/pnas.93.12.6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tupler R, Barbierato L, Memmi M, Sewry C A, De Grandis D, Maraschio P, Tiepolo L, Ferlini A. J Med Genet. 1998;35:778–783. doi: 10.1136/jmg.35.9.778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sanger F, Nicklen S, Coulson A R. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Altschul S F, Gish W, Miller W, Myers W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 21.Hung H L, Song F, Gewirtz A. Leukemia. 1999;13:295–297. doi: 10.1038/sj.leu.2401274. [DOI] [PubMed] [Google Scholar]
- 22.Giulian G G, Moss R L, Greaser M. Anal Biochem. 1983;129:277–287. doi: 10.1016/0003-2697(83)90551-1. [DOI] [PubMed] [Google Scholar]
- 23.Engel A G, Yamamoto M, Fischbeck K H. In: Myology. Engel A G, Banker B Q, editors. New York: McGraw–Hill; 1994. pp. 1133–1187. [Google Scholar]
- 24.Winderbank A J, Mulder D W. In: Myology. Engel A G, Banker B Q, editors. New York: McGraw–Hill; 1994. pp. 1854–1869. [Google Scholar]
- 25.Paranjape S M, Krumm A, Kadonaga J T. Genes Dev. 1995;9:1978–1991. doi: 10.1101/gad.9.16.1978. [DOI] [PubMed] [Google Scholar]
- 26.Roest H P, van Klaveren J, de Wit J, van Gurp C G, Koken M H M, Vermey M, van Roijen J H, Hoogerbrugge J W, Vreeburg J T M, Baarends W M, et al. Cell. 1996;86:799–810. doi: 10.1016/s0092-8674(00)80154-3. [DOI] [PubMed] [Google Scholar]
- 27.Verrault A, Kaufman P D, Kobayashi R, Stillman B. Curr Biol. 1997;8:96–108. doi: 10.1016/s0960-9822(98)70040-5. [DOI] [PubMed] [Google Scholar]
- 28.Lin Q, Schwarz J, Bucana C, Olson E N. Science. 1997;276:1404–1407. doi: 10.1126/science.276.5317.1404. [DOI] [PMC free article] [PubMed] [Google Scholar]