Abstract
Activation of PERK following the accumulation of unfolded proteins in the endoplasmic reticulum (ER) promotes translation inhibition and cell cycle arrest. PERK function is essential for cell survival following exposure of cells to ER stress, but the mechanisms whereby PERK signaling promotes cell survival are not thoroughly understood. We have identified the Nrf2 transcription factor as a novel PERK substrate. In unstressed cells, Nrf2 is maintained in the cytoplasm via association with Keap1. PERK-dependent phosphorylation triggers dissociation of Nrf2/Keap1 complexes and inhibits reassociation of Nrf2/Keap1 complexes in vitro. Activation of PERK via agents that trigger the unfolded protein response is both necessary and sufficient for dissociation of cytoplasmic Nrf2/Keap1 and subsequent Nrf2 nuclear import. Finally, we demonstrate that cells harboring a targeted deletion of Nrf2 exhibit increased cell death relative to wild-type counterparts following exposure to ER stress. Our data demonstrate that Nrf2 is a critical effector of PERK-mediated cell survival.
Mammalian cells possess a signaling network that senses endoplasmic reticulum (ER) stress and determines cell fate following exposure to stress. The ER signaling network consists of three related transmembrane protein kinases, Ire1α, Ire1β, and PERK (31). Ire1α and Ire1β are composed of a luminal domain that senses stress, a single membrane-spanning domain, and a cytosolic tail that contains both a serine/threonine kinase domain and an RNase domain (40). Ire1-dependent signals induce expression of ER chaperones (40) and CHOP (42), a transcription factor that may participate in the apoptotic program. Activation of PERK, which lacks an RNase domain, following ER stress promotes phosphorylation of the translation initiation factor eukaryotic initiation factor 2α (eIF2α), thereby attenuating translation initiation (15, 22). PERK is one of at least four eIF2α protein kinases that include the heme-regulated kinase (HRI), the interferon-inducible, RNA-dependent protein kinase (PKR), and GCN2 (3, 39). Among these, only PERK function is required for the cellular response to ER stress (22).
Activation of PERK via ER stress initiates cell cycle arrest via inhibition of cyclin D1 translation (5). PERK-dependent signaling is also critical for cell survival following the initiation of an ER stress response. Targeted deletion of PERK dramatically reduces survival of embryonic stem cells following exposure to ER stress-inducing agents (21). Mice that contain inactivated PERK exhibit severe defects in the exocrine pancreas due to a high rate of secretory cell apoptosis (20, 44). While the mechanism that underlies PERK-dependent cell cycle arrest is established, the nature of PERK-dependent cell survival is unresolved.
A majority of PERK's biological activities have been attributed to its function as an eIF2α kinase. However, the possibility that PERK targets additional downstream substrates that function as cellular effectors has not been addressed. In order to identify novel PERK substrates, we performed a yeast two-hybrid screen with the PERK catalytic domain. We describe the identification of the Cap 'n' Collar transcription factor Nrf2 as a PERK substrate. In unstressed cells, Nrf2 is maintained in latent cytoplasmic complexes via association with the cytoskeletal anchor, Keap1. Previous work revealed that oxidative stress triggers dissociation of this complex via an uncharacterized mechanism, thereby allowing Nrf2 nuclear import, where it promotes expression of its downstream target genes (30). We demonstrate that PERK-dependent phosphorylation is both necessary and sufficient to trigger dissociation of Nrf2/Keap1 complexes and thereby promote Nrf2 nuclear import. Furthermore, Nrf2 nuclear translocation is independent of eIF2α phosphorylation. Finally, targeted deletion of Nrf2 reduces cell survival following ER stress. Collectively, these data support a model wherein Nrf2 functions as an effector of PERK cell survival signaling.
MATERIALS AND METHODS
Tissue culture conditions.
Cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum (FCS), antibiotics, nonessential amino acids, and glutamine (Mediatech, Inc.). Wild-type and Nrf2−/− mouse embryonic fibroblasts (MEFs) were immortalized via a standard 3T9 protocol (41). Transfections were performed with Lipofectamine Plus reagent (Life Technologies, Inc.).
Plasmids.
ΔNPERK was generated from murine PERK cDNA using PCR (primers 5′-CCCGGGTCTAGAGAGCGCGCCACCCCGGCCCGG-3′ and 5′-GGGCCCGAATTCTCAGGAGAAGGAGCTTGACTT-3′). ΔNPERKK618A was generated through site-directed PCR mutagenesis (primers 5′-CAATTACGCTATCGCGAGGATCCGGCT-3′ and 5′-AGCCGGATCCTCGCGATAGCGTAATTG-3′ [mutated residues are underlined]). ΔNPERK and ΔNPERKK618A were subcloned into pGEX4T1 and pAS2 (Amersham) vectors using EcoRI sites. Murine Nrf1 and MafG were cloned from a mouse cDNA library (Nrf1, 5′-TCTAGAATGGAAATGCAGGCTATG-3′ and 5′-CAGCTGGTATTTGGACAGCAG-3′; MafG, 5′-GTCCCCCGGGTTATGACGAC-3′ and 5′-GAATTCGTGTCCCTATGACCGAGCATC-3′). Cloned cDNAs were ligated into the pcDNA3 vector (Invitrogen) using EcoRI sites. Myc-Keap1 and His-Keap1 were generated from Keap1 cDNA and cloned into pcDNA3 or pET15b vectors. HA-Nrf2 and GST-Nrf2 were generated from Nrf2 cDNA and cloned into the pCI and pGEX5T2 vectors. The antioxidant response element (ARE) reporter plasmid was generated by cloning four ARE motifs (36) upstream of the TATA sequence in pGL2 (Promega). The mutant ARE reporter contains a minimal initiator sequence upstream of the TATA sequence in pGL2. PERKΔC and myc-PERK plasmids and viruses were described previously (5).
Yeast two-hybrid screen.
ΔNPERK(K618A) was cloned into the pAS2(TRP) vector using EcoRI sites, resulting in GAL4-ΔNPERK(K618A). Saccharomyces cerevisiae strain AH109 was transformed with GAL4-ΔNPERK(K618A) and pACT(LEU) plasmids containing cDNAs from MEFs. Double transformants were selected for their ability to grow on medium lacking tryptophan, leucine, and histidine and supplemented with 50 mM 3-amino-1,2,4-triazole. Transformants were further selected for their ability to drive lacZ expression in a GAL4-ΔNPERKK618A-dependent manner. cDNAs from positive transformants were recovered and sequenced.
EMSA.
A consensus ARE oligomer (primers 5′-CTACGATTTCTGCTTAGTCATTGTCTTCC-3′ and 5′-GGAAGACAATGACTAAGCACAATCGTAG-3′) was end labeled with [γ-32P]ATP and incubated with 10 μg of cell extract (lysed in 50 mM Tris, 250 mM NaCl, 5 mM EDTA, 0.1% Triton, and 4 mM dithiothreitol [DTT]) for 30 min at room temperature in a reaction mixture containing 20 mM HEPES (pH 8.0), 1 mM EDTA, 20 mM KCl, 4 mM MgCl2, 4% glycerol, 2 μg of poly(dI-dC), and 5 mM DTT. Where indicated, proteins were first preincubated for 1 h at 4°C in the presence of antibodies. To achieve maximum resolution of in vivo-derived protein-DNA adducts, native gels were run at low voltage for 4 h with the free probe routinely running off the gel. For in vitro reactions, recombinant Nrf2 (0.5 μg) and in vitro-transcribed and -translated MafG (Promega) were combined in electrophoretic mobility shift assay (EMSA) reaction mixtures as described above.
Immunofluorescence.
Cells proliferating on glass coverslips were transfected and treated as indicated. Cells were fixed in 3% paraformaldehyde and permeabilized in 0.1% Triton in phosphate-buffered saline (PBS). The hemagglutinin (HA) epitope was detected with either the 12CA5 or the 3F10 monoclonal antibody. The myc epitope was detected with either the 9E10 or the Jac6 monoclonal antibody. Calreticulin was detected using a polyclonal anticalreticulin antibody (Stressgen). Cells were stained with either fluorescein isothiocyanate (FITC)-conjugated or biotinylated immunoglobulin G and Texas Red-streptavidin (Vector). DNA was detected with Hoechst 33258 dye (Sigma). Cells were visualized using a Nikon microscope fitted with appropriate filters or a Bio-Rad confocal imaging system.
Reporter assays.
NIH 3T3 cells transfected with the indicated plasmids were left untreated or treated with 5 μg of tunicamycin/ml or 100 μM tert-butylhydroquinone (tBHQ) for the indicated intervals. Luciferase assays were carried out according to the manufacturer's instructions (Dual Luciferase Reporter assay system; Promega) with a luminometer (PE Applied Biosystems). Firefly luciferase activity was normalized against Renilla luciferase activity from the same lysates.
Cellular fractionation, immunoblotting, and Northern blot analysis.
Wild-type, PERK−/−, and eIF2α(S51A) MEFs were treated with 5 μg of tunicamycin/ml for the indicated time intervals. Nuclei were collected in buffer A (10 mM HEPES [pH 8.0], 10 mM KCl, 1.5 mM MgCl2, 0.1 mM EDTA, 0.1 mM EGTA, 0.1% IGEPAL) and buffer C (420 mM NaCl, 1.5 mM MgCl2, 1.0 mM EDTA, 1.0 mM EGTA, 20% glycerol) and stored in buffer D (20 mM HEPES [pH 8.0], 50 mM KCl, 0.2 mM EDTA, 0.2 mM EGTA, 20% glycerol). All buffers were supplemented with 0.1 mM phenylmethylsulfonyl fluoride, 1 μg of aprotinin/ml, 1 μg of leupeptin/ml, 1 μg of pepstatin/ml, and 25 mM β-glycerol phosphate. Proteins were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), transferred to nitrocellulose membranes (Osmonics), and blotted with an anti-Nrf2 polyclonal antibody (Santa Cruz). For detection of eIF2α, cells were lysed in EBC buffer, proteins were resolved by SDS-PAGE and transferred to a nitrocellulose membrane (Osmonics), and total and phosphorylated eIF2α was detected by immunoblotting (Cell Signaling). RNA was extracted using TRIzol reagent (Life Technologies, Inc.). RNA immobilized on nylon membranes (Osmonics) was hybridized with 32P-labeled probes specific for either luciferase, γ-glutamylcysteine synthetase ligase (GCLC), or γ-actin (5). Signals were detected by phosphorimaging.
Protein kinase assays, metabolic labeling, and immunoprecipitation.
Affinity-purified recombinant GST-ΔNPERK was used as the kinase, and equivalent amounts of affinity-purified GST-Nrf2, glutathione S-transferase (GST), and His-eIF2α were used as substrates in the in vitro kinase assays. ΔNPERK was preincubated with kinase buffer (50 mM HEPES [pH 8.0], 10 mM MgCl2, 2.5 mM EGTA, 20 μM ATP) at 30°C for 1 h prior to addition of substrates in order to reduce incorporation of radioactive phosphate into PERK itself due to its propensity to autophosphorylate (22). Kinase reactions were initiated following addition of substrates by the addition of 10 μCi of [γ-32P]ATP and placement at 30°C for 30 min. Reactions were terminated by the addition of sample buffer containing SDS and incubation at 100°C for 3 min. Proteins were resolved by SDS-PAGE and visualized by autoradiography. To obtain PERK produced in mammalian cells, NIH 3T3 cells infected with retrovirus encoding PERK were lysed in radioimmunoprecipitation assay buffer (150 mM NaCl, 10 mM sodium phosphate, 2 mM EDTA, 1% NP-40, 0.1% SDS, 1% deoxycholic acid,) and immunoprecipitated with anti-PERK antiserum. Proteins were mixed with the indicated recombinant substrates in kinase reactions as described above. For metabolic labeling, NIH 3T3 cells were cultured for 1 h in phosphate-free DMEM supplemented with 0.1% dialyzed FCS and then labeled with 1 mCi of [32P]orthophosphate (ICN) and in the presence of 5 μg of tunicamycin/ml. Cells were lysed in NP-40 lysis buffer (50 mM Tris [pH 7.5], 150 mM NaCl, 1% NP-40, 0.5% deoxycholic acid). HA-Nrf2 was precipitated from cell lysates with the 12CA5 antibody and visualized by autoradiography.
In vitro binding assays.
Nrf2, in vitro transcribed and translated (Promega) in the presence of [35S]methionine (ICN), was mixed with purified His-Keap1 or His-eIF2α immobilized on Talon beads in binding buffer (20 mM Tris, 150 mM NaCl, 0.5% IGEPAL) for 2 h at 4°C. After washes with buffer, Nrf2-containing complexes were incubated with ΔNPERK or ΔNPERKK618A in the absence or presence of 500 μM ATP at 30°C for the indicated intervals in kinase buffer lacking EGTA.
Annexin V staining and clonogenic survival assays.
Cells proliferating on glass coverslips were left untreated or were treated with 5 μg of tunicamycin/ml, 4 μM staurosporine, or tumor necrosis factor alpha (TNF-α; 5 ng/ml) plus cycloheximide (10 μg/ml) for the indicated intervals. Cells were then washed with PBS and stained with a FITC-conjugated annexin V antibody and propidium iodide (Pharmingen) and examined by fluorescence and light microscopy. Annexin V-positive cells are expressed as the percentage of total cells. For colony outgrowth, cells were plated at 2 × 104/60-mm dish, and 24 h later cells were left untreated or treated with 1 μg of tunicamycin/ml for 8 h. Following treatment, cells were washed with PBS, refed with complete medium, and allowed to grow for 7 days.
RESULTS
Activation of the Nrf2 transcription factor and Nrf2 target genes by the UPR.
To identify novel PERK substrates, we performed a yeast two-hybrid screen with the catalytic domain of murine PERK. Sequence analysis of these clones revealed three overlapping cDNAs predicted to encode the C-terminal 219 amino acids of the Nrf1 transcription factor. Nrf1 belongs to the Cap 'n' Collar family of transcription factors (9), which includes NF-E2 (1) and Nrf2 (34). Nrf1 and -2 are homologous within their C-terminal DNA-binding domains, but they are divergent at their N-terminal transactivation domains (25). We confirmed that PERK could bind directly to both Nrf1 and Nrf2 in an in vitro binding assay (data not shown). To determine the likely PERK target, we evaluated the capacity of the unfolded protein response (UPR)-inducing agent, tunicamycin (Fig. 1A), an inhibitor of N-linked glycosylation, or phorbol myristate acetate (PMA) (100 nM), a phorbol ester, to induce Nrf1 or Nrf2 DNA-binding activity. Extracts prepared from cells left untreated or treated for the indicated intervals were evaluated by EMSA reactions using a 32P-labeled oligonucleotide containing a single ARE (36). Increased DNA-binding activity was detected in extracts from both PMA- and tunicamycin-treated cells by 30 min (Fig. 1A, lanes 3 and 4 and lanes 5 and 6, respectively). Similar results were observed upon thapsigargin treatment (data not shown), which induces ER stress via perturbations in calcium homeostasis. To identify the Nrf family member that bound to DNA, extracts were preincubated with antibodies specific for individual Nrf proteins. An antibody specific for Nrf2, but not for Nrf1, abolished the formation of the shifted complex (compare lanes 8 and 9), indicating that Nrf2, but not Nrf1, is a required component of the tunicamycin-induced DNA-binding complex. Neither Nrf1 nor Nrf2 bound to an oligonucleotide containing a mutant ARE (negative data not shown).
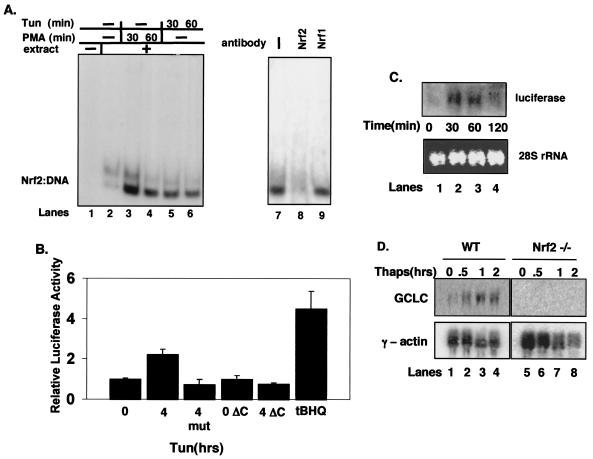
FIG. 1.
Nrf2, and not Nrf1, is activated by ER stress. (A) Whole-cell extracts prepared from NIH 3T3 cells treated with PMA (lanes 3 and 4) or tunicamycin (tun) (1 μg/ml) (lanes 5 and 6) were mixed with a radiolabeled oligonucleotide containing a consensus ARE in an EMSA reaction. In lanes 7 to 9, tunicamycin-treated (60 min) whole-cell extracts were exposed to antibodies against Nrf1 (lane 9) or Nrf2 (lane 8) in the EMSA reactions. (B) NIH 3T3 cells were cotransfected with plasmids encoding the 4xARE reporter or a reporter lacking an ARE (mut) along with the cytomegalovirus-Renilla control (internal normalization control) with or without PERKΔC. Cells were treated with 5 μg of tunicamycin/ml or 100 μM tBHQ for the indicated time intervals (in hours), harvested, and assayed for luciferase and Renilla activities. Luciferase activity is shown as the ratio of reporter activity to Renilla luciferase activity; activity in untreated cells was set as 1. Error bars represent the standard deviation from three independent experiments. (C) NIH 3T3 cells were transfected with the 4xARE reporter and treated with tunicamycin (5 μg/ml) for the indicated intervals. RNA was extracted, and expression of luciferase was assessed by Northern blot analysis. (D) Wild-type (lanes 1 to 4) and Nrf2−/− (lanes 5 to 8) fibroblasts were treated with thapsigargin (1 μM) for the indicated intervals. RNA was extracted, and expression of GCLC and γ-actin was determined by Northern blot analysis.
To evaluate whether tunicamycin elevates Nrf2-dependent gene expression, NIH 3T3 cells were cotransfected with a firefly luciferase plasmid containing four ARE sequences (4xARE) and a cytomegalovirus-directed Renilla luciferase plasmid as an internal control. Treatment with tunicamycin resulted in a reproducible induction of luciferase activity (Fig. 1B, panel 2). Treatment with tBHQ, a known inducer of Nrf2 activity (28), also increased ARE-dependent gene expression (Fig. 1B, panel 6). Neither tunicamycin nor tBHQ elicited expression of a luciferase reporter which lacked the ARE motifs (Fig. 1B, panel 3, and data not shown). To confirm these results, we directly assessed luciferase gene expression by Northern blotting. Increased luciferase mRNA was evident by 30 min of tunicamycin treatment (Fig. 1C). We noted a decrease in luciferase message by 120 min. This decrease likely reflects the relative instability of the luciferase message compounded by the transient nature of Nrf2 activation following ER stress (see Fig. 3 and 4, below).
FIG. 3.
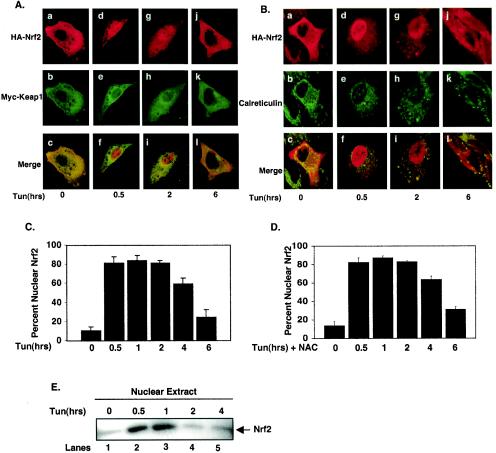
The UPR triggers PERK-dependent Nrf2 nuclear translocation. (A and B) NIH 3T3 cells proliferating on glass coverslips were cotransfected with plasmids encoding HA-Nrf2 and myc-Keap1. Transfected cells were treated with 5 μg of tunicamycin/ml for the indicated periods of time and fixed, and proteins were visualized by confocal microscopy with antibodies specific for the HA epitope, the myc epitope, and calreticulin. (C) Graphical representation of data in panels A and B. Error bars represent the standard deviations from three independent experiments. (D) NIH 3T3 cells cotransfected with plasmids encoding HA-Nrf2 and myc-Keap1 were pretreated with 5 mM NAC and then treated as described for panels A and B. Error bars represent the standard deviations from three independent experiments. (E) Nuclear extracts prepared from NIH 3T3 cells treated with 5 μg of tunicamycin/ml for the indicated periods of time were resolved by SDS-PAGE, transferred to a nitrocellulose membrane, and probed with an antibody specific for Nrf2.
FIG. 4.
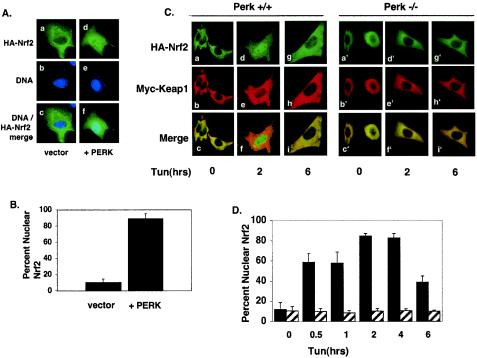
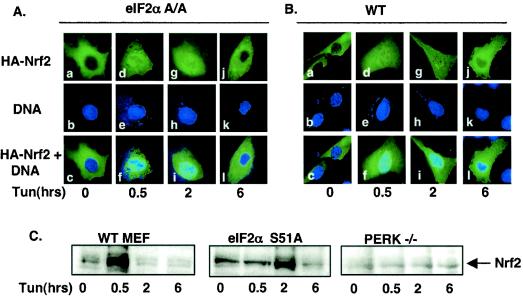
PERK activity is necessary and sufficient for Nrf2 nuclear translocation. (A) NIH 3T3 cells proliferating on glass coverslips were cotransfected with plasmids encoding HA-Nrf2, Myc-Keap1 (panels a to c), or HA-Nrf2, myc-Keap1 and myc-PERK (panels d to f). Twenty-four hours posttransfection, cells were fixed and proteins were detected by indirect immunofluorescence with an antibody specific for the HA epitope. DNA was stained with Hoechst dye. (B) Graphical representation of data in panel A. Error bars represent standard deviations from three independent experiments. (C) Wild-type (PERK+/+) or PERK−/− murine fibroblasts proliferating on glass coverslips were cotransfected with plasmids encoding HA-Nrf2 and myc-Keap1 and treated with 5 μg of tunicamycin/ml for the indicated time intervals. Proteins were detected with antibodies specific for the HA and myc epitopes and visualized by confocal microscopy. (D) Graphical representation of data presented in panel C. Solid bars represent wild-type cells, and hatched bars represent PERK−/− cells. Error bars represent the standard deviations from three independent experiments.
To determine if UPR-dependent induction of the ARE reporter required PERK activity, we measured expression of the ARE reporter in NIH 3T3 cells cotransfected with a dominant-negative PERK (PERKΔC) (5). While the cotransfection of PERKΔC did not influence basal luciferase gene expression (Fig. 1B, bar 4), cotransfection of PERKΔC eliminated tunicamycin-dependent induction of 4xARE-luciferase (bar 5).
We next assessed the capacity of ER stress-inducing agents to induce expression of an endogenous Nrf2 target gene. For this experiment, we utilized fibroblasts prepared from either wild-type or Nrf2-null mouse embryos. Wild-type and Nrf2−/− MEFs were challenged with thapsigargin, total RNA was collected, and expression of the Nrf2 target gene, GCLC (7), was assessed. Thapsigargin treatment resulted in a rapid increase in GCLC mRNA in wild-type (Fig. 1D, lanes 1 to 4) but not in Nrf2−/− (lanes 5 to 8) MEFs. Induction of GCLC was also noted in MEFs containing a homozygous knock-in of a nonphosphorylatable allele of eIF2α, eIF2α(S51A) (data not shown) (37). These data demonstrate that induction of the UPR triggers the activation of Nrf2 and Nrf2-dependent gene expression and suggest that PERK function is essential for Nrf2 activation.
Nrf2 is a PERK substrate.
To evaluate the possibility that Nrf2 is a PERK substrate, we produced an N-terminally truncated PERK kinase, ΔNPERK. ΔNPERK purified from bacteria was mixed with equivalent amounts of GST, His-eIF2α, or GST-Nrf2 (Fig. 2A, lanes 4 to 6) in the presence of [γ-32P]ATP. PERK phosphorylated His-eIF2α and GST-Nrf2 (Fig. 2A, lanes 2 and 3) but not GST (lane 1). We also assessed whether PERK expressed in mammalian cells could phosphorylate Nrf2. PERK was precipitated from cell lysates prepared from NIH 3T3 cells transduced with a retrovirus encoding PERK (38); overexpression of PERK results in its activation via enforced oligomerization in the absence of cell stress (4, 22). PERK precipitates phosphorylated both GST-Nrf2 and His-eIF2α, but not GST (Fig. 2B). As a specificity control, we also evaluated the ability of Ire1β precipitated from infected NIH 3T3 cells to phosphorylate GST-Nrf2 or GST. In contrast to PERK, Ire1β failed to phosphorylate either GST or Nrf2 (Fig. 2B, lanes 1 and 2). Expression of both PERK and Ire1 was confirmed in parallel by immunoblotting (data not shown). This control illustrates two important points. First, Nrf2 is not a general substrate of all the known ER protein kinases, and second, our precipitates do not contain significant quantities of contaminating protein kinases that nonspecifically phosphorylate Nrf2. These data demonstrate that Nrf2 is a direct PERK substrate.
FIG. 2.
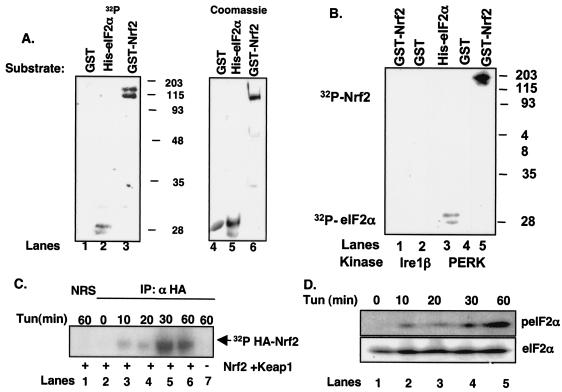
Phosphorylation of Nrf2 by PERK and during the UPR. (A) Recombinant GST (lane 1), GST-Nrf2 (lane 3), or His-eIF2α (lane 2) was mixed with the catalytic domain of PERK along with [γ-32P]ATP and incubated at 30° for 30 min. Phosphorylated proteins were resolved on denaturing polyacrylamide gels and visualized by autoradiography. Lanes 4, 5, and 6 show Coomassie brilliant blue staining of GST, His-eIF2α, and GST-Nrf2, respectively. (B) Lysates prepared from NIH 3T3 cells infected with PERK or Ire1β were precipitated with an antibody against PERK or a C-terminal myc epitope, respectively. The precipitated protein was mixed with recombinant His-eIF2α, GST, or GST-Nrf2 along with [γ-32P]ATP. Phosphorylated proteins were resolved on denaturing gels and visualized by autoradiography. (C) NIH 3T3 cells cotransfected with HA-Nrf2 and myc-Keap1 were treated with 5 μg of tunicamycin/ml and [32P]orthophosphate for the indicated times. Lysates were precipitated with the 12CA5 antibody; complexes were resolved by SDS-PAGE and visualized by autoradiography. (D) Lysates prepared from NIH 3T3 cells were treated with 5 μg of tunicamycin/ml and resolved by SDS-PAGE, and membranes were probed with antibodies specific for total and phosphorylated eIF2α.
We next determined if Nrf2 is phosphorylated in vivo following induction of an ER stress response. NIH 3T3 cells cotransfected with plasmids encoding Nrf2 engineered to express an amino-terminal HA epitope tag (HA-Nrf2) and Keap1 were cultured in medium containing [32P]orthophosphate and subsequently challenged with tunicamycin for the indicated intervals (Fig. 2C). Lysates were prepared and subjected to precipitation with the 12CA5 monoclonal antibody. Nrf2 phosphorylation was evident by 10 min of tunicamycin treatment (lane 3) and reached peak levels by 30 min (lane 5), indicating that Nrf2 is rapidly phosphorylated in response to ER stress conditions. Similar results were observed in cells challenged with tBHQ (data not shown) (28). No phosphorylation was detected in mock-transfected cells (lane 7) or in control precipitates (lane 1). Additionally, Nrf2 phosphorylation was eliminated in cells that had been transfected with a plasmid encoding PERKΔC (negative data not shown), implicating PERK as the Nrf2 kinase in intact cells. The rapid phosphorylation of Nrf2 led us to compare the kinetics of Nrf2 phosphorylation with that of eIF2α, a known in vivo substrate of PERK. NIH 3T3 cells were treated with tunicamycin for the indicated intervals, and eIF2α phosphorylation was assessed by immunoblotting using an eIF2α phospho-specific antibody. Increased eIF2α phosphorylation was detectable by 10 to 20 min (Fig. 2D, lanes 2 and 3) and was maximal at 60 min (lane 4). These data demonstrate that Nrf2 is phosphorylated during an ER stress response in a PERK-dependent fashion, with kinetics that are temporally similar to that of the other known PERK substrate, eIF2α.
ER stress triggers PERK-dependent Nrf2 nuclear import.
Under homeostatic conditions, Nrf2 is maintained in inactive cytoplasmic complexes by Keap1 (30). Dissociation of Nrf2/Keap1 complexes and subsequent Nrf2 nuclear import can be achieved by treatment of cells with electrophiles such as tBHQ (28). While the mechanism underlying activation of Nrf2 remains unclear, models invoking oxidation of redox-sensitive cysteines within Keap1 (13, 14, 24) or phosphorylation of Nrf2 (28, 43, 45) have been proposed. Given the capacity of ER stress to induce both Nrf2 DNA-binding activity and Nrf2-dependent gene expression, we reasoned that ER stress must also induce Nrf2 nuclear translocation. To assess regulated nuclear entry of Nrf2, NIH 3T3 cells were cotransfected with plasmids encoding HA-Nrf2 and Keap1 engineered with an N-terminal myc epitope (myc-Keap1). Cells expressing HA-Nrf2 and myc-Keap1 were left untreated or treated with tunicamycin for various intervals, and Nrf2 localization was visualized using epitope-specific antibodies followed by confocal microscopy. In the absence of stimulus, Nrf2 was cytoplasmic (Fig. 3A, panel a, B, panel a, and C); by 30 min of tunicamycin treatment, HA-Nrf2 was primarily nuclear (Fig. 3A, panel d, B, panel d, and C). In contrast, myc-Keap1 remained cytoplasmic at all intervals (Fig. 3A, panels b, e, h, and k). While nuclear Nrf2 staining remained evident at 2 h (Fig. 3A, panel g, B, panel g, and C), by 6 h Nrf2 increasingly relocalized to the cytoplasm (Fig. 3A, panel j, B, panel j, and C). Similar results were observed following treatment with other known inducers of ER stress, including thapsigargin (data not shown). Transient nuclear accumulation of endogenous Nrf2 was also observed following ER stress as determined by subcellular fractionation (Fig. 3E). Loss of endogenous Nrf2 nuclear localization was evident by 2 h, suggesting that overexpression may result in some attenuation of Nrf2 downregulation.
Because the UPR is known to elicit oxidative stress (18, 31), a known inducer of Nrf2 nuclear translocation, we assessed whether tunicamycin-dependent activation of Nrf2 was directed by a burst of reactive oxygen species (ROS) generated as a consequence of ER stress. NIH 3T3 cells transfected with plasmids encoding both HA-Nrf2 and myc-Keap1 were pretreated with the ROS scavenger N-acetylcysteine (5 mM) prior to treatment with tunicamycin. While this dose was sufficient to reduce levels of tunicamycin-induced oxidative stress by over 80%, as measured by dichlorodihydrofluorescein diacetate fluorescence (data not shown), tunicamycin still induced the nuclear accumulation of Nrf2 (Fig. 3D). These results suggest that Nrf2 nuclear transport is unlikely to result from the generation of ROS following ER stress.
The capacity of ER stress to induce Nrf2 nuclear import is consistent with the notion that a subset of cytoplasmic Nrf2 complexes must be in the proximity of the ER. We thus determined whether HA-Nrf2 colocalized with calreticulin, an ER resident protein, in either untreated or tunicamycin-treated cells. As anticipated, in unstimulated cells HA-Nrf2 colocalized with endogenous, ER-localized calreticulin (Fig. 3B, panel c); however, treatment of cells with tunicamycin resulted in HA-Nrf2 nuclear import and a loss of HA-Nrf2-calreticulin colocalization (Fig. 3B, panels f and i). In addition, myc-Keap1 colocalized with calreticulin in the absence or presence of ER stress (data not shown).
The above data suggest that ER stress triggers Nrf2-Keap1 dissociation and Nrf2 nuclear import, but they do not address whether PERK activity is required for this action. To assess the role of PERK in Nrf2 nuclear import, we cotransfected NIH 3T3 cells with plasmids encoding HA-Nrf2 and Keap1 with or without PERK. Cexpression of wild-type PERK efficiently promoted Nrf2 nuclear localization in the absence of a stress-inducing agent (Fig. 4A, panel d, and B), demonstrating that PERK activity is sufficient for Nrf2 nuclear import. To assess whether PERK is required for Nrf2 nuclear import following ER stress, we examined Nrf2 localization in fibroblasts derived from PERK null embryos (PERK−/−). PERK−/− fibroblasts or fibroblasts derived from wild-type littermates (PERK+/+) were cotransfected with plasmids encoding HA-Nrf2 and myc-Keap1. In PERK−/− cells, Nrf2 remained cytoplasmic in the absence or presence of tunicamycin (Fig. 4C, panels a′, d′, and g′, and D), where it colocalized with Keap1 (Fig. 4C, panels c′, f′, and i′). In contrast, in wild-type cells that express endogenous PERK, tunicamycin triggered Nrf2 nuclear localization (Fig. 4C, compare panels a and d).
To determine whether Nrf2 nuclear translocation was an indirect consequence of PERK-dependent eIF2α phosphorylation, we assessed Nrf2 localization in MEFs containing a homozygous knock-in of a nonphosphorylatable eIF2α allele, eIF2α(S51A) (37). Fibroblasts derived from either wild-type or eIF2α(S51A) embryos were transfected with vectors encoding HA-Nrf2 or myc-Keap1. Treatment of both wild-type and eIF2α(S51A) cells resulted in Nrf2 nuclear accumulation (Fig. 5A and B).
FIG. 5.
Nrf2 nuclear translocation occurs independently of eIF2α phosphorylation. (A and B) Wild-type fibroblasts (B) or eIF2α(S51A) fibroblasts (A) were cotransfected with plasmids encoding HA-Nrf2 and myc-Keap1 and treated with 5 μg of tunicamycin/ml for the indicated time intervals. HA-Nrf2 was detected with the 12CA5 monoclonal antibody and visualized by confocal microscopy. (C) Nuclear extracts were prepared from wild-type (WT), eIF2α(S51A), or PERK−/− MEFs treated with 5 μg of tunicamycin/ml for the indicated intervals, resolved by SDS-PAGE, transferred to a nitrocellulose membrane, and probed with an antibody specific for Nrf2.
We also assessed ER stress-dependent nuclear accumulation of endogenous Nrf2. Nuclear extracts were prepared from wild-type, PERK−/−, and eIF2α(S51A) MEFs treated with tunicamycin for the indicated intervals and assessed for the presence of Nrf2 via Western blotting. Following tunicamycin treatment, Nrf2 translocated to the nucleus in both wild-type and eIF2α(S51A) MEFs; however, no accumulation of Nrf2 was observed in PERK−/− MEFs (Fig. 5C). Efficacy of the fractionation was confirmed by immunoblotting for β-tubulin (cytosolic) and CREB (nuclear) (data not shown). These data strongly support a model wherein nuclear accumulation of Nrf2 following ER stress is a direct consequence of PERK activation. We did note that the rate of Nrf2 nuclear accumulation was more gradual in the eIF2α(S51A) MEFs. The reason for this is unclear, given that we detected similar kinetics of nuclear Nrf2 entry by immunofluorescence. The important point is that Nrf2 nuclear entry occurs in a PERK-dependent and eIF2α-independent fashion.
Phosphorylation of Nrf2 promotes its dissociation from Keap1.
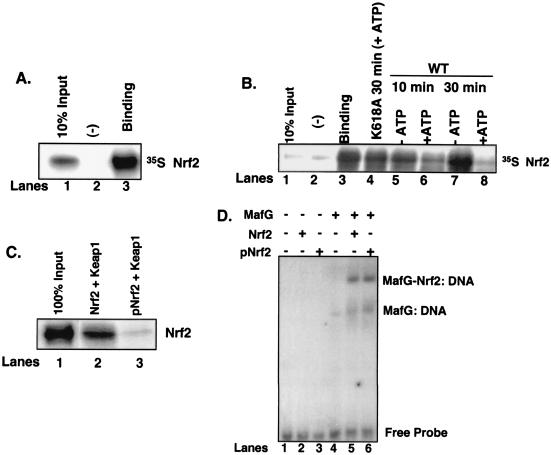
The capacity of tunicamycin to induce PERK-dependent Nrf2 nuclear accumulation, while Keap1 remains cytoplasmic, suggests that PERK phosphorylation triggers Nrf2/Keap1 dissociation. To directly assay complex dissociation, Nrf2 was in vitro transcribed and translated in the presence of [35S]methionine and mixed with recombinant His-Keap1 that was immobilized on nickel affinity resin. Nrf2 efficiently bound Keap1 (Fig. 6A, lane 3) but not the negative control, His-eIF2α (lane 2). After binding and extensive washing, the Nrf2/Keap1 complexes were divided equally into aliquots and incubated in the absence of ΔNPERK (Fig. 6B, lane 3), with ΔNPERK but without ATP (lanes 5 and 7), with ΔNPERK and unlabeled ATP (500 μM) (lanes 6 and 8), or with the catalytically inactive ΔNPERK K618A (22) but with ATP (500 μM) (lane 4) for the indicated times at 30°C. Nrf2/Keap1 complexes were then washed extensively, and the presence of Keap1-associated Nrf2 was determined by SDS-PAGE and visualized by autoradiography. Phosphorylation of Nrf2/Keap1 complexes by PERK led to a time-dependent decrease in Keap1-associated Nrf2 (compare lanes 3 and 6). By 30 min in the presence of the PERK catalytic domain, Nrf2/Keap1 binding was equal to that of the negative control (compare lanes 2 and 8); however, incubation of Nrf2/Keap1 complexes with PERK(K618A) for 30 min had no effect on Nrf2/Keap1 association (compare lanes 3 and 4). The inability of PERK(K618A) to dissociate Nrf2/Keap1 complexes illustrates two important points. First, PERK catalytic activity is essential for Nrf2/Keap1 dissociation rather than simple physical association, as both wild-type PERK and PERK(K618A) can associate with Nrf2 (data not shown). Second, addition of ATP is not sufficient to trigger dissociation in the absence of catalytically active PERK. These data demonstrate that PERK-dependent phosphorylation is sufficient to trigger Nrf2/Keap1 dissociation.
FIG. 6.
Phosphorylation-dependent dissociation of Nrf2 from Keap1. (A) Nrf2 in vitro transcribed and translated in the presence of [35S]methionine was mixed with recombinant Keap1 (lane 3) or eIF2α (lane 2). Complexes were affinity purified by nickel chromatography, resolved by SDS-PAGE, and visualized by autoradiography. Lane 1 is 10% Nrf2 input. (B) Nrf2/Keap1 complexes prepared as described for panel A and mixed with the catalytic domain of PERK in the absence (lanes 5 and 7) or presence (lanes 6 and 8) of ATP (500 μM) or a kinase-dead PERK in the presence of ATP (lane 4) for the indicated intervals. Lane 1 is 10% Nrf2 input, and lane 3 is Nrf2-Keap1 binding. Lane 2 is binding to the negative control, eIF2α. Upon completion of the kinase reactions, the complexes were washed, resolved by SDS-PAGE, and visualized by autoradiography. (C) Nrf2 was mixed with the catalytic domain of PERK in the absence (lane 2) or presence (lane 3) of ATP and then mixed with recombinant Keap1 as described above. After binding, the complexes were washed extensively and proteins were resolved by SDS-PAGE, transferred to a nitrocellulose membrane, and probed with an antibody specific for Nrf2. Lane 1 shows 100% Nrf2 input. (D) Nrf2 (lanes 2 and 5) or PERK-phosphorylated Nrf2 (lanes 3 and 6) was mixed with in vitro-transcribed and -translated MafG (lanes 5 and 6) along with a radiolabeled oligonucleotide containing a consensus ARE in an EMSA reaction. Also shown are EMSA reactions with MafG alone (lane 4) or no protein (lane 1) mixed with the radiolabeled oligonucleotide.
Having shown that PERK-dependent phosphorylation triggers dissociation of Nrf2/Keap1 complexes, we asked whether PERK phosphorylation of Nrf2 would inhibit reassociation of Nrf2/Keap1 complexes. In vitro-transcribed and -translated Nrf2 was incubated with ΔNPERK in the presence (Fig. 6C, lane 3) or absence (lane 2) of ATP for 30 min at 30°C. Following phosphorylation, Nrf2 was mixed with recombinant Keap1. Nrf2 phosphorylation was monitored in parallel reactions by using [γ-32P]ATP (data not shown). After extensive washing, Western analysis revealed that the level of Nrf2 associated with Keap1 was markedly decreased when Nrf2 was prephosphorylated by ΔNPERK (compare lanes 2 and 3). These results demonstrate that not only does PERK-dependent phosphorylation promote Nrf2/Keap1 dissociation, but also that once phosphorylated by PERK, Nrf2 cannot stably associate with Keap1. These data demonstrate that PERK-dependent phosphorylation triggers Nrf2/Keap1 dissociation and prevents reassociation.
To determine whether phosphorylation affects Nrf2 DNA-binding activity, purified recombinant Nrf2 that had been phosphorylated with purified recombinant ΔNPERK, or left unphosphorylated, was mixed with in vitro-transcribed and -translated MafG. The DNA-binding activity of these complexes was then assessed in an EMSA. Monomeric Nrf2, whether phosphorylated or unphosphorylated, exhibited no detectable DNA-binding activity (Fig. 6D, lanes 2 and 3). However, when mixed with a heterodimeric DNA-binding partner, MafG, both forms of Nrf2 exhibited indistinguishable DNA-binding activity (lanes 5 and 6).
Nrf2 expression increases cell survival following ER stress.
Chronic ER stress triggers apoptosis (6, 8, 31). Nrf2 activation plays a role in cell survival following oxidative stress (7, 26). To directly assess the role of Nrf2 in survival following activation of the UPR, wild-type and Nrf2−/− MEFs plated at a low density were challenged with tunicamycin for 8 h and subsequently fed with complete medium lacking tunicamycin. The cells were allowed to recover for 7 days, at which point surviving cells were visualized with Giemsa stain. The expression of Nrf2 greatly enhanced the ability of cells to survive following ER stress (Fig. 7A). Similarly, Nrf2 overexpression increased cell survival in tunicamycin-treated NIH 3T3 cells (data not shown).
FIG. 7.
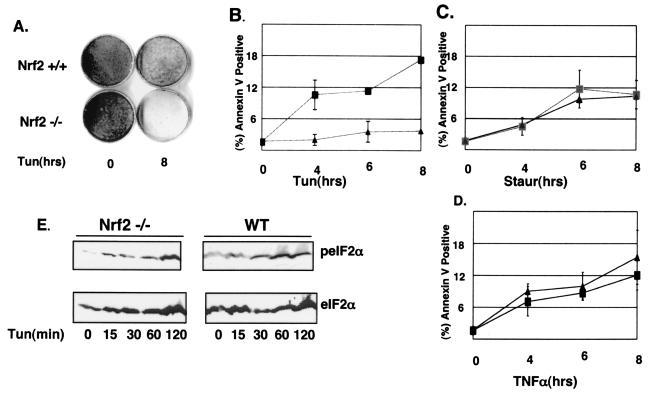
Nrf2 promotes increased cell survival after chronic ER stress. (A) Wild-type and Nrf2−/− MEFs were left untreated or treated with 1 μg of tunicamycin/ml for 8 h and then allowed to recover for 7 days in normal medium. Colonies were visualized by using Giemsa stain. (B to D) Wild-type (▴) and Nrf2−/− (▪) MEFs proliferating on glass coverslips were left untreated or treated with 5 μg of tunicamycin/ml (B), 4 μM staurosporine (C), or 5 ng of TNF-α/ml plus 10 μg of cycloheximide/ml (D) for the indicated intervals (in hours). Apoptotic cells were visualized with a FITC-conjugated annexin V antibody. Error bars represent the standard deviations from three independent experiments. (E) Wild-type and Nrf2−/− MEFs were treated with 5 μg of tunicamycin/ml for the indicated intervals. Proteins were resolved by SDS-PAGE, and membranes were probed with antibodies specific for total and phosphorylated eIF2α.
To further assess the ability of Nrf2 to provide a protective advantage to cells during ER stress, we assayed for early signs of commitment to apoptosis in wild-type and Nrf2−/− MEFs. Cells were treated with tunicamycin for the indicated intervals (Fig. 7B) and then stained with annexin V. A sixfold increase in annexin V staining observed in Nrf2−/− MEFs, relative to that in the wild type, was detectable by 4 h of tunicamycin treatment (Fig. 7B), and levels of annexin V-positive cells increased through 8 h of treatment. To rule out the possibility that the increased rate of death is due to a generalized sensitivity of Nrf2−/− MEFs to all death stimuli, we treated both wild-type and Nrf2−/− MEFs with non-ER stress-inducing death stimuli: staurosporine (Fig. 7C) and TNF-α (Fig. 7D). Nrf2−/− MEFs were no more susceptible to these death stimuli than wild-type MEFs. The fact that eIF2α phosphorylation is induced by tunicamycin treatment in Nrf2−/− MEFs (Fig. 7E) demonstrates that the increased sensitivity of Nrf2−/− MEFs does not reflect compromised eIF2α phosphorylation. These results provide support for a model wherein Nrf2 plays a prosurvival role during the UPR.
DISCUSSION
Identification of Nrf2 as a PERK substrate.
The ER-localized PERK kinase regulates cell fate decisions following cellular insults that perturb ER homeostasis. Upon exposure to ER stress-inducing agents, activated PERK phosphorylates its only documented substrate, eIF2α (22, 38). PERK function is crucial for cell survival following exposure of cells to ER stress (21). While translational regulation is clearly a critical component of PERK-mediated signaling (21), it is also reasonable to consider that PERK signals contribute directly to the expression of prosurvival genes. We performed a yeast two-hybrid screen with the goal of identifying novel PERK substrates that contribute to cell survival during an ER stress response. We have identified the prosurvival transcription factor Nrf2 as a novel PERK substrate. PERK directly phosphorylates Nrf2, thereby identifying Nrf2 as only the second known substrate for PERK. Preliminary data have failed to reveal PERK-dependent phosphorylation of other Nrf family members. Our experiments reveal a role for this phosphorylation in the dissolution of latent, cytoplasmic Nrf2 complexes, ultimately facilitating Nrf2 nuclear import and induction of Nrf2-dependent gene expression.
Under homeostatic conditions, Nrf2 is sequestered in a cytoplasmic complex through its interaction with the actin-binding protein Keap1 (30). Induction of oxidative stress triggers Nrf2 nuclear import and activation, presumably due to its capacity to enforce Nrf2/Keap1 dissociation (28). Our results demonstrate that PERK signaling from the stressed ER also promotes Nrf2 nuclear import. The UPR triggers Nrf2 nuclear import while Keap1 remains cytoplasmic, demonstrating that the UPR promotes the dissolution of Nrf2/Keap1 complexes. Indeed, PERK activity triggers dissociation of Nrf2 from Keap1 in vitro.
Previous models for the regulation of Nrf2/Keap1 dissociation have focused on the possibility that ROS promote complex dissociation (46). While it is possible that under certain conditions ROS could act directly on Nrf2 or Keap1, our data reveal that PERK phosphorylation of Nrf2 is sufficient for Nrf2/Keap1 dissociation and that ROS are not required. While we have not yet identified the residues within Nrf2 targeted by PERK, our preliminary data indicate that the phosphorylation site lies within the amino terminus of Nrf2. This portion of Nrf2 contains the Neh2 domain, which is critical for interaction with Keap1 (30). It seems likely that Nrf2 phosphorylation triggers a conformational change that disrupts Nrf2/Keap1 complexes.
Distinct mechanisms of PERK-dependent regulation of gene expression.
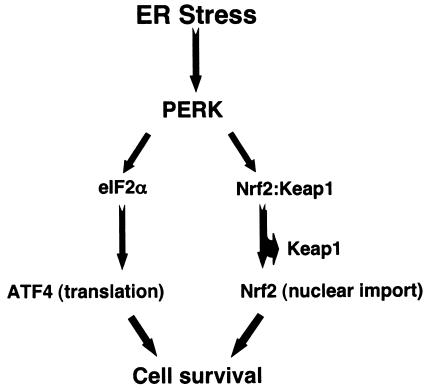
While cellular protein translation is repressed due to eIF2α phosphorylation following ER stress, translation of ATF4 is selectively increased (19). ATF4 translation is not augmented in PERK−/− cells (19), indicating that during the UPR, ATF4 synthesis is PERK dependent. While PERK signaling is necessary for ATF4 translation, an intermediate signaling step, eIF2α phosphorylation, is also required (21, 37). In contrast, our data demonstrate that Nrf2 is an immediate PERK substrate and its activation is independent of eIF2α phosphorylation. Consistent with this idea, we found that stress-dependent Nrf2 nuclear import was lost in PERK−/− cells; furthermore, dominant-negative PERK molecules blocked the nuclear import (data not shown) and transcriptional activation of Nrf2, indicating that PERK activity is required for Nrf2 activation. Unlike ATF4, Nrf2 is synthesized in unstressed cells, but is maintained in latent cytoplasmic complexes, by virtue of its interaction with Keap1 (30). PERK signals Nrf2 directly, resulting in a rapid response to ER stress. As illustrated in Fig. 8, our data demonstrate that following PERK activation signals bifurcate, resulting in the regulation of cell survival via two distinct pathways. The first involves the translation-dependent accumulation of ATF4, and the second is mediated by PERK-dependent phosphorylation of inactive Nrf2.
FIG. 8.
Model depicting the bifurcation of PERK-dependent survival signals (details in text).
PERK, Nrf2, and cell survival signaling.
While most experiments have utilized cell culture systems to evaluate the consequences of increased ER stress, cells in the context of the whole organism are also vulnerable to perturbations in ER homeostasis. Indeed, PERK−/− mice exhibit massive levels of apoptosis of pancreatic beta cells, leading to the development of diabetes (20, 44). Mice homozygous for a mutant, nonphosphorylatable eIF2α (S51A) also display a beta cell deficiency and die soon after birth (37), indicating the importance of translational control during the UPR. These results suggest an important role for PERK signaling in cell survival following ER stress.
Our results demonstrate that Nrf2 activation regulates cell survival following UPR activation. The absence of Nrf2 increased apoptosis following ER stress, but not in response to treatment with other death stimuli. Nrf2-dependent protection from UPR-induced cell death likely results from its function as a regulator of genes encoding phase II detoxifying enzymes (7, 26). Nrf2 is required for inducible expression of phase II detoxifying enzymes following cellular stress (10). As the UPR induces oxidative stress due to accumulation of ROS (23, 33), it is likely that Nrf2-dependent induction of phase II detoxifying enzyme expression will contribute to survival during the UPR response. Consistent with this notion is the recent demonstration that cells containing a targeted deletion of ATF4, a heterodimeric partner for Nrf2, accumulate high levels of ROS resulting in increased cell death (23).
Taken together, the available data suggest a model wherein under conditions of ER stress, PERK senses the accumulation of unfolded proteins in the ER and phosphorylates both Nrf2 and eIF2α (2, 22). While protein translation is globally inhibited (11), translation increases for selected proteins, including ATF4 (19) and ER resident chaperones, which facilitate proper protein folding (16, 32). Nrf2 phosphorylation promotes dissolution of Nrf2/Keap1 complexes and triggers Nrf2 nuclear import. Once in the nucleus, Nrf2 heterodimerizes with small Maf proteins (12, 17, 29, 35) or ATF4 (27) to regulate a program of gene expression. This regulation likely includes the induction of phase II detoxifying enzymes, whose actions maintain cellular redox homeostasis (7, 26), and may also include repression of genes that promote apoptosis.
Acknowledgments
We thank Y.W. Kan and J. Chan for providing the Nrf2−/− MEFs and the Nrf2 cDNA; D. Scheuner for the eIF2α(S51A) MEFs; M. Olson for providing the Jac6 and 3F10 monoclonal antibodies; R. Wek for providing anti-PERK antiserum; D. Ron and H. Harding for providing the PERK cDNA and the PERK−/− fibroblasts; C. B. Thompson for providing TNF-α; and M. C. Simon for providing luciferase cDNA.
This work was supported by AHA grant 0060360Z and the Abramson Family Cancer Research Institute (J.A.D.) and grants GM59213 and ES11721 (M.H.).
REFERENCES
- 1.Andrews, N. C., H. Erdjument-Bromage, M. B. Davidson, P. Tempst, and S. H. Orkin. 1993. Erythroid transcription factor NF-E2 is a haematopoietic-specific basic-leucine zipper protein. Nature 362:722-728. [DOI] [PubMed] [Google Scholar]
- 2.Balachandran, S., C. N. Kim, W.-C. Yeh, T. W. Mak, K. Bhalla, and G. N. Barber. 1998. Activation of the dsRNA-dependent protein kinase, PKR, induces apoptosis through FADD-mediated death signaling. EMBO J. 17:6888-6902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berlanga, J. J., J. Santoyo, and C. Haro. 1999. Characterization of a mammalian homolog of the GCN2 eukaryotic initiation factor 2α kinase. Eur. J. Biochem. 265:754-762. [DOI] [PubMed] [Google Scholar]
- 4.Bertolotti, A., Y. Zhang, L. M. Hendershot, H. P. Harding, and D. Ron. 2000. Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat. Cell Biol. 2:326-332. [DOI] [PubMed] [Google Scholar]
- 5.Brewer, J. W., and J. A. Diehl. 2000. PERK mediates cell-cycle exit during the mammalian unfolded protein response. Proc. Natl. Acad. Sci. USA 97:12625-12630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brostrom, C. O., and M. A. Brostrom. 1998. Regulation of translational initiation during cellular responses to stress. Prog. Nucleic Acid Res. Mol. Biol. 58:79-125. [DOI] [PubMed] [Google Scholar]
- 7.Buetler, T. M., E. P. Gallagher, C. Wang, D. L. Stahl, J. D. Hayes, and D. L. Eaton. 1995. Induction of phase I and phase II drug-metabolizing enzyme mRNA, protein, and activity by BHA, ethoxyquin, and oltipraz. Toxicol. Appl. Pharmacol. 135:45-57. [DOI] [PubMed] [Google Scholar]
- 8.Carlberg, M., and O. Larsson. 1993. Role of N-linked glycosylation in cell-cycle progression and initiation of DNA synthesis in tumor-transformed human fibroblasts. Anticancer Res. 13:167-171. [PubMed] [Google Scholar]
- 9.Chan, J. Y., X. L. Han, and Y. W. Kan. 1993. Cloning of Nrf1, an NF-E2-related transcription factor, by genetic selection in yeast. Proc. Natl. Acad. Sci. USA 90:11371-11375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chan, K., and Y. W. Kan. 1999. Nrf2 is essential for protection against acute pulmonary injury in mice. Proc. Natl. Acad. Sci. USA 96:12731-12736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chong, K. L., L. Feng, K. Schappert, E. Meurs, T. F. Donahue, J. D. Friesen, A. G. Hovanessian, and B. R. Williams. 1992. Human p68 kinase exhibits growth suppression in yeast and homology to the translational regulator GCN2. EMBO J. 11:1553-1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dhakishinamoorthy, S., and A. K. Jaiswal. 2000. Small Maf (MafG and MafK) proteins negatively regulate antioxidant response element-mediated expression and antioxidant induction of the NAD(P)H:quinone oxidoreductase1 gene. J. Biol. Chem. 275:40134-40141. [DOI] [PubMed] [Google Scholar]
- 13.Dinkova-Kostova, A. T., M. A. Massiah, R. E. Bozak, R. J. Hicks, and P. Talalay. 2001. Potency of Michael reaction acceptors as inducers of enzymes that protect against carcinogenesis depends on their reactivity with sulfhydryl groups. Proc. Natl. Acad. Sci. USA 98:3404-3409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dinkova-Kostova, A. T., W. D. Holtzclaw, R. N. Cole, K. Itoh, N. K. Y. Wakabayashi, M. Yamamoto, and P. Talalay. 2002. Direct evidence that sulfhydryl groups of Keap1 are the sensors regulating induction of phase 2 enzymes that protect against carcinogens and oxidants. Proc. Natl. Acad. Sci. USA 99:11908-11913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fernandez, J., B. Bode, A. Koromilas, J. A. Diehl, I. Krukovets, M. D. Snider, and M. Hatzoglou. 2002. Translation mediated by the internal ribosome entry site of the cat-1 mRNA is regulated by glucose availability in a PERK kinase-dependent manner. J. Biol. Chem. 277:11780-11787. [DOI] [PubMed] [Google Scholar]
- 16.Gething, M. J., and J. Sambrook. 1992. Protein folding in the cell. Nature 355:33-45. [DOI] [PubMed] [Google Scholar]
- 17.Gong, P., B. Hu, D. Stewart, M. Ellerbe, Y. G. Figueroa, V. Blank, B. S. Beckman, and J. Alam. 2001. Cobalt induces heme oxygenase-1 expression by a hypoxia-inducible factor-independent mechanism in Chinese hamster ovary cells: regulation by Nrf2 and MafG transcription factors. J. Biol. Chem. 276:27018-27025. [DOI] [PubMed] [Google Scholar]
- 18.Harding, H. P., M. Calfon, F. Urano, I. Novoa, and D. Ron. 2002. Transcriptional and translational control in the mammalian unfolded protein response. Annu. Rev. Cell Dev. Biol. 18:575-599. [DOI] [PubMed] [Google Scholar]
- 19.Harding, H. P., I. Novoa, Y. Zhang, H. Zeng, R. Wek, M. Schapira, and D. Ron. 2000. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol. Cell 5:1099-1108. [DOI] [PubMed] [Google Scholar]
- 20.Harding, H. P., H. Zeng, Y. Zhang, R. Jungries, P. Chung, H. Plesken, D. D. Sabatini, and D. Ron. 2001. Diabetes mellitus and exocrine pancreatic dysfunction in Perk-/- mice reveals a role for translational control in secretory cell survival. Mol. Cell 7:1153-1163. [DOI] [PubMed] [Google Scholar]
- 21.Harding, H. P., Y. Zhang, A. Bertolotti, H. Zeng, and D. Ron. 2000. Perk is essential for translational regulation and cell survival during the unfolded protein response. Mol. Cell 5:897-904. [DOI] [PubMed] [Google Scholar]
- 22.Harding, H. P., Y. Zhang, and D. Ron. 1999. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature 397:271-274. [DOI] [PubMed] [Google Scholar]
- 23.Harding, H. P., Y. Zhang, H. Zeng, I. Novoa, P. D. Lu, M. Calfon, N. Sadri, C. Yun, B. Popko, R. Paules, D. F. Stojdl, J. C. Bell, T. Hettmann, J. M. Leiden, and D. Ron. 2003. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol. Cell 11:619-633. [DOI] [PubMed] [Google Scholar]
- 24.Hayes, J. D., E. M. Ellis, G. E. Neal, D. J. Harrison, and M. M. Manson. 1999. Cellular response to cancer chemopreventive agents: contribution of the antioxidant responsive element to the adaptive response to oxidative and chemical stress. Biochem. Soc. Symp. 64:141-168. [PubMed] [Google Scholar]
- 25.Hayes, J. D., and M. McMahon. 2001. Molecular basis for the contribution of the antioxidant responsive element to cancer chemoprevention. Cancer Lett. 174:103-113. [DOI] [PubMed] [Google Scholar]
- 26.Hayes, J. D., and D. J. Pulford. 1995. The glutathione S-transferase supergene family: regulation of GST and the contribution of the isoenzymes to cancer chemoprotection and drug resistance. Crit. Rev. Biochem. Mol. Biol. 30:446-600. [DOI] [PubMed] [Google Scholar]
- 27.He, C. H., P. Gong, B. Hu, D. Stewart, M. E. Choi, A. M. K. Choi, and J. Alam. 2001. Identification of the activating transcription factor (ATF4) as an Nrf2-interacting protein. J. Biol. Chem. 276:20858-20865. [DOI] [PubMed] [Google Scholar]
- 28.Huang, H.-C., T. Nguyen, and C. B. Pickett. 2000. Regulation of the antioxidant response element by protein kinase C-mediated phosphorylation of NF-E2-related factor 2. Proc. Natl. Acad. Sci. USA 97:12475-12480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Itoh, K., T. Chiba, S. Takahashi, T. Ishii, K. Igarashi, Y. Katoh, T. Oyake, N. Hayahsi, K. Satoh, I. Hatayama, M. Yamamoto, and Y. Nabeshima. 1997. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem. Biophys. Res. Commun. 2236:313-322. [DOI] [PubMed] [Google Scholar]
- 30.Itoh, K., N. Wakabayashi, Y. Kotoh, T. Ishii, K. Igarashi, J. D. Engel, and M. Yamamoto. 1999. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev. 13:76-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kaufman, R. J. 1999. Stress signaling from the lumen of the endoplasmic reticulum: coordination of gene transcriptional and translational controls. Genes Dev. 13:1211-1233. [DOI] [PubMed] [Google Scholar]
- 32.Kozutsumi, Y., M. Segal, K. Normington, M. J. Gething, and J. Sambrook. 1988. The presence of malfolded proteins in the endoplasmic reticulum signals the induction of glucose-regulated proteins. Nature 332:462-464. [DOI] [PubMed] [Google Scholar]
- 33.McCullough, K. D., J. L. Martindale, L.-O. Klotz, T.-Y. Aw, and N. J. Holbrook. 2001. Gadd153 sensitizes cells to endoplasmic reticulum stress by down-regulating BclII and perturbing the cellular redox state. Mol. Cell. Biol. 21:1249-1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moi, P., K. Chan, I. Asunis, A. Cao, and Y. W. Kan. 1994. Isolation of NF-E2-related factor 2 (Nrf2), a NF-E2-like basic leucine zipper transcriptional activator that binds to the tandem NF-E2/AP1 repeat of the beta-globin locus control region. Proc. Natl. Acad. Sci. USA 91:9926-9930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nguyen, T., H. C. Huang, and C. B. Pickett. 2000. Transcriptional regulation of the antioxidant response element. Activation by Nrf2 and repression by MafK. J. Biol. Chem. 275:15466-15473. [DOI] [PubMed] [Google Scholar]
- 36.Rushmore, T. H., M. R. Morton, and C. B. Pickett. 1991. The antioxidant responsive element. Activation by oxidative stress and identification of the DNA consensus sequence required for functional activity. J. Biol. Chem. 266:11632-11639. [PubMed] [Google Scholar]
- 37.Scheuner, D., B. Song, E. McEwen, C. Liu, R. Laybutt, P. Gillespie, T. Saunders, S. Bonner-Weir, and R. J. Kaufman. 2001. Translational control is required for the unfolded protein response and in vivo glucose homeostasis. Mol. Cell 7:1165-1176. [DOI] [PubMed] [Google Scholar]
- 38.Shi, Y., K. M. Vattem, R. Sood, J. An, J. Liang, L. Stramm, and R. C. Wek. 1998. Identification and characterization of pancreatic eukaryotic initiation factor 2-subunit kinase, PEK, involved in translational control. Mol. Cell. Biol. 18:7499-7509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sood, R., A. C. Porter, D. Olsen, D. R. Cavener, and R. C. Wek. 2000. A mammalian homologue of GCN2 protein kinase important for translational control by phosphorylation of eukaryotic initiation factor-2α. Genetics 154:787-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tirasophon, W., A. A. Welihinda, and R. J. Kaufman. 1998. A stress response pathway from the endoplasmic reticulum to the nucleus requires a novel bifunctional protein kinase/endoribonuclease (Ire1p) in mammalian cells. Genes Dev. 12:2416-2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Todaro, G. J., and H. Green. 1963. Quantitative studies of the growth of mouse embryo cells in culture and their development into established lines. J. Cell Biol. 17:299-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang, X. Z., B. Lawson, J. W. Brewer, H. Zinszner, A. Sanjay, L. J. Mi, R. Boorstein, G. Kreibich, L. M. Hendershot, and D. Ron. 1996. Signals from the stressed endoplasmic reticulum induce C/EBP-homologous protein (CHOP/GADD153). Mol. Cell. Biol. 16:4273-4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu, R., C. Chen, Y. Y. Mo, V. Hebbar, E. D. Owuor, T. H. Tan, and A. N. Kong. 2000. Activation of mitogen-activated protein kinase pathways induces antioxidant response element-mediated gene expression via a Nrf2-dependent mechanism. J. Biol. Chem. 275:39907-39913. [DOI] [PubMed] [Google Scholar]
- 44.Zhang, P., B. McGrath, S. Li, A. Frank, F. Zambito, J. Reinert, M. Gannon, K. Ma, K. McNaughton, and D. R. Caverner. 2002. The PERK eukaryotic initiation factor 2α kinase is required for the development of the skeletal system, postnatal growth, and the function and viability of the pancreas. Mol. Cell. Biol. 22:3864-3874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zipper, L. M., and R. T. Mulcahy. 2000. Inhibition of Erk and p38 MAP kinases inhibits binding of Nrf2 and induction of GCS genes. Biochem. Biophys. Res. Commun. 278:484-492. [DOI] [PubMed] [Google Scholar]
- 46.Zipper, L. M., and R. T. Mulcahy. 2002. The Keap1 BTB/POZ dimerization function is required to sequester Nrf2 in cytoplasm. J. Biol. Chem. 277:36544-36552. [DOI] [PubMed] [Google Scholar]