Abstract
CEACAM1 is an intercellular adhesion glycoprotein. As CEACAM1 plays an important role in epithelial cell signaling and functions, we have examined its localization in epithelial cells. We have observed that distribution at cell contacts is not always seen in these cells, suggesting that CEACAM1 localization might be regulated. In Swiss 3T3 cells, the targeting of CEACAM1 at cell-cell boundaries is regulated by the Rho GTPases. In the present study, we have used the MDCK epithelial cells to characterize the effects of the Rho GTPases and their effectors on CEACAM1 intercellular targeting. Activated Cdc42 and Rac1 or their downstream effector PAK1 targeted CEACAM1 to sites of cell-cell contacts. On the other hand, neither activated RhoA nor activated Rho kinase directed CEACAM1 to cell boundaries, resulting in a condensed distribution of CEACAM1 at the cell surface. Interestingly, inhibition of this pathway resulted in CEACAM1 intercellular localization suggesting that a tightly regulated balance of Rho GTPase activities is necessary to target CEACAM1 at cell-cell boundaries. In addition, using CEACAM1 mutants and chimeric fusion constructs containing domains of the colony-stimulating factor receptor, we have shown that the transmembrane domain of CEACAM1 is responsible for the Cdc42-induced targeting at cell-cell contacts.
In epithelial tissues, cell compendiums are held together by a number of protein complexes such as occludin and the ZO proteins in tight junctions, the cadherins in adherens junctions, and the desmogleins and desmocollins in desmosomes. In addition, integrins are linked to the extracellular matrix and form dynamic adhesion complexes with cellular kinases, phosphatases, and adaptor proteins. However, under different conditions, such as organogenesis, shear force, stress, wounding, or epithelial-mesenchymal transitions, epithelial cells change their morphology, dissociate from one another, and undergo cell migration. Therefore, the adhesion complexes are not static and can be subjected to dynamic changes.
Members of the Rho-like GTPase family of small GTP-binding proteins, including Cdc42, Rac1, and RhoA, actively participate in tissue remodeling by regulating the organization of the actin cytoskeleton (for a recent review, see reference 16). The Rho-like GTPases cycle between GDP- and GTP-bound forms, and their activities are modulated by a number of binding proteins. Changes in cytoskeletal structures were initially noticed in Swiss 3T3 fibroblasts, where the activation of RhoA leads to the development of stress fibers and focal adhesion complexes (57). Activation of Rac1 induces the polymerization of actin at the cell membrane, giving rise to lamellipodia and membrane ruffles (56), whereas activation of Cdc42 initiates fine peripheral cell extensions such as filopodia and microspikes (33, 48). These GTPases act in conjunction with each other, signaling by cross talk. They play a major role in epithelial morphogenic processes (for a review, see reference 72), such as the regulation of adherens and tight junction formation. It has been proposed, for instance, that Rac1 may mediate the reorganization of the actin cytoskeleton necessary to stabilize cadherin receptors at cell-cell contact sites. Interestingly, these effects depend on the cell type considered and Rac1 can display opposite effects in keratinocytes (6) and MDCK cells (23, 62).
Rho GTPases have a number of downstream effector proteins (for a review, see reference 3). p21-associated kinase (PAK) and Rho-associated kinase (ROK) are two such effectors that respond to activation of either Cdc42 and Rac1 or RhoA, respectively. PAK, a Ser/Thr kinase protein, plays a pivotal role in actin cytoskeleton dynamics and cell adhesion (41). The Rho-specific pathways depend on the activity of a number of effectors, including ROK, such as p164ROKα and p160ROKβ, or ROCK (27, 38, 43) as well as Dia1 and Dia2 (75).
CEACAM1, formerly known as biliary glycoprotein, CD66a, or C-CAM, is a homophilic intercellular adhesion molecule of the immunoglobulin (Ig) superfamily (2). Its cell-adhesive properties are mediated by its first variable-like Ig domain (28, 76). The CEACAM1 primary transcript is subjected to alternative splicing, producing four different evolutionarily conserved isoforms with either two or four extracellular Ig domains and either a short 10-amino-acid (CEACAM1-S) or a long 73-amino-acid (CEACAM1-L) intracytoplasmic domain. In addition to its intercellular adhesion properties, CEACAM1 functions as a signal regulatory molecule (50) and negatively controls epithelial tumor cell growth (24, 35) as well as T-cell (5, 29, 46, 47) and B-cell (8) proliferation and functions. CEACAM1 also regulates early maturation and activation of dendritic cells (30). In the liver, Tyr- and Ser-phosphorylated CEACAM1 is associated with insulin receptor endocytosis and degradation. Transgenic mice overexpressing a dominant-negative CEACAM1 mutant in the liver develop hyperinsulinemia from defective insulin clearance (54). Furthermore, CEACAM1is an angiogenic factor and an effector of vascular endothelial growth factor in endothelia (15). In addition to its role in cell physiology, CEACAM1 has also been subverted by a number of pathogens. In mice, CEACAM1 is the receptor for mouse hepatitis virus (4), whereas in humans, it binds to pathogenic Neisseria (21, 74), Haemophilus influenzae (73), Escherichia coli, and Salmonella (39). All of these functions require the expression of CEACAM1 at the cell surface.
It has been previously shown that, in Swiss 3T3 fibroblasts, CEACAM1 cell-cell contact localization is dependent upon activation of Rho-like GTPases (60). Recently, Schumann et al. have shown that CEACAM1-S binds directly to F-actin, whereas both CEACAM1 isoforms associate directly with G-actin and tropomyosin (64).
As CEACAM1 plays an important role in epithelial cell signaling and functions, we first questioned its localization at the surface of two different epithelial models and have examined the Rho GTPase-mediated mechanisms modulating its localization at cell contacts of MDCK cells. In addition, we have studied the contribution of the different CEACAM1 domains mediating cell contact localization triggered by activated Cdc42. We conclude that CEACAM1 epithelial cell contact localization responds to a balance in the activity of Cdc42/Rac1 and RhoA via a process involving the CEACAM1 transmembrane domain.
MATERIALS AND METHODS
Cell culture and microinjection.
Primary rat hepatocytes were prepared as previously described (32). Cells were incubated for 24 h in Dulbecco's modified Eagle-F12 medium (DMEM) containing 10% fetal bovine serum (FBS) (Invitrogen, Burlington, Ontario, Canada) containing 10 mM HEPES (pH 8.0), 20 mM NaHCO3, 500 U of penicillin/ml, and 500 μg of streptomycin/ml. Cells were plated on collagen matrix (Vitrogen-100)-coated microscope slides at 50 to 80% confluence. Mouse rectal carcinoma cells (CMT-93) (ATCC CCL 223) and Madin-Darby canine kidney (MDCK) epithelial cells were grown in DMEM containing 50 U of penicillin/ml and 50 μg of streptomycin/ml. The culture medium was supplemented with 10% FBS. For microinjections, 5 × 104 to 20 × 104 cells were seeded on glass coverslips and grown for either 48 or 24 h, respectively. Plasmid DNA (30 or 60 μg/ml, diluted in phosphate-buffered saline [PBS]) was microinjected (Eppendorf microinjection system) into the nucleus of 100 cells maintained in culture medium with FBS. The cells were incubated for 2 h for protein expression, except those expressing the L107FPAK1 mutant, which were incubated for 1 h only, due to its toxic effects. They were then fixed and subjected to immunofluorescence. The Rho-kinase inhibitor Y27632 (Yoshitomi Pharmaceutical Industries) (10 μM) was added directly to the medium 30 min prior to fixation.
cDNA constructs used.
The Ceacam1 cDNA constructs were cloned into the pRK5 vector. The CEACAM1-S and S449, S452A, S449,452A, or Δcyto CEACAM1-S membrane-proximal mutants were prepared by using a previously described overlap PCR method with specific mutating oligonucleotides (25) and cloned into the pRK5 vector. For S449,452A mutations, the sense oligo used was 5′-CTCTATGCCAGGAAGGCTGGCGG and the antisense oligo was 5′-CCGCCAGCCTTCCTGGCATAGAG (nucleotides in bold correspond to the mutations). The S452A mutation was introduced by using the sense oligo 5′-TCCAGGAAGGCTGGCGG and the antisense oligo 5′-CCGCCAGCCTTCCTGGA. Deletion of the CEACAM1 cytoplasmic domain was performed by inserting a stop codon at S449 with the sense oligo 5′-TATTCCCTCTATTGAAGGAAGTCT and the antisense oligo 5′-AGACTTCCTTCAATAGAGGAAATA. All new mutants created were subjected to DNA dideoxy sequencing prior to use.
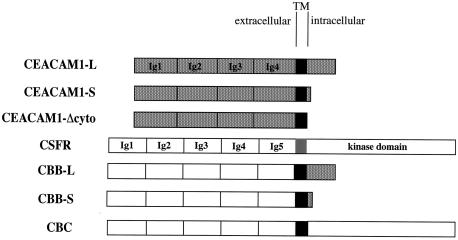
Chimeric proteins expressing the five extracellular Ig-like domains of the colony-stimulating factor receptor (CSFR) (10) fused to the transmembrane and cytoplasmic domains of CEACAM1-S or -L were generated. The wild-type CSFR cDNA was a kind gift of Martine Roussel (St. Jude Children's Research Hospital, Memphis, Tenn.) and was retrieved as a BamHI insert for cloning into the pRK5 vector. The chimeras were assembled from overlapping PCR fragments. These included a 479-bp CSFR fragment from a HindIII site at nucleotide (nt) 1348 (5′-GAGCCCAAGCTTGCTAA) and sequences upstream of the encoded transmembrane domain at nt 1827 with the reverse oligo 5′-GGGATCCGTGTGGAGGCCTGC containing an engineered StuI site. This fragment was joined to the CEACAM1 fragments encompassing the transmembrane and cytoplasmic domains of either CEACAM1-S or -L that were amplified by PCR. The sense oligo was 5′-ACACAAAGGAGGCCTCTCAGAT and contained an endogenous StuI site. The reverse oligo was from the 3′ untranslated region of the Ceacam1 cDNAs (5′-CATCACTGGTGCAGCC). The overlapping fragments of the chimeras were connected to the 5′ end of the CSFR cDNA at its endogenous HindIII site. The chimeras were cloned into the pRK5 vector for cell expression. These constructs have been named CBB-S and CBB-L for CSFR (extra)-B (Bgp TM)-B (Bgp intra) (see Fig. 5 for a graphic description). All cDNAs generated by PCR amplification were subjected to DNA sequencing prior to use.
FIG. 5.
Graphic representation of the CEACAM1 and chimeric constructs used to define the CEACAM1 domain involved in cellular targeting. The stippled boxes represent the CEACAM1 extracellular Ig domains linked by its transmembrane domain (black box) to a 73-amino-acid long (L) or 10-amino-acid short (S) cytoplasmic domain (stippled box). A tailless CEACAM1 protein was generated by inserting a stop codon in the cDNA at nucleotides corresponding to S449 (CEACAM1-Δcyto). The CSFR contains five extracellular Ig domains (white boxes) linked by its transmembrane domain (grey box) to a cytoplasmic kinase domain (white box). Chimeric proteins expressing the CSFR Ig domains (white boxes) linked via the CEACAM1 transmembrane domain (black box) to either the long (CBB-L) or short (CBB-S) CEACAM1 cytoplasmic domains (stippled boxes) or to the CSFR (CBC) cytoplasmic kinase domain were also produced and tested.
The CBC chimera consists of the extracytoplasmic domain of the CSFR, the transmembrane domain of CEACAM1, and the cytoplasmic domain of the CSFR (see Fig. 5). This construct was created by overlap PCR of three DNA products. The first PCR fragment was obtained by using the following combination of primers. The forward primer NB5 (5′-GAGCCCAAGCTTGCTAA-3′) corresponds to nt 1350 to 1367 in the CSFR extracytoplasmic domain cDNA sequence and contains an endogenous HindIII restriction site. The reverse primer RBF5 (5′-GATGCCAGCAATGGCGCCCTCATCCGGGGGATGCGTGTG-3′) was designed to overlap the CSFR extracytoplasmic domain (nt 1816 to 1837) and the CEACAM1 transmembrane domain sequences (nt 1372 to 1390). These primers were used in combination with the CSFR cDNA template. The second PCR product was amplified on the Ceacam1 cDNA template by using a primer corresponding to the reverse sequence of the primer RBF5 (primer BF5, 5′-CACACGCATCCCCCGGATGAGGGCGCCATTGCTGGCATC-3′) and the reverse primer overlapping the transmembrane domain sequence of CEACAM1 (nt 1426 to 1447) and the cytoplasmic domain sequence of the CSFR (nt 1911 to 1932) (primer RBF6, 5′-GGGCTTCTGCTTATACTTGTAATAGAGGAAATATGCCAGCCC-3′). The third PCR product was produced on the CSFR cDNA template by using the forward primer corresponding to the reverse sequence of the primer RBF6 (primer BF6, 5′-GGGCTGGCATATTTCCTCTATTACAAGTATAAGCAGAAGCCC-3′) and the reverse primercorresponding to a sequence in the CSFR cytoplasmic domain containing an endogenous SalI restriction site (nt 2481 to 2402) (primer RSalI), (5′-TTCTTATAGTCGACGCCTCCC-3′). The first and the second PCR products were then joined together by overlap PCR and then combined with the third PCR product to generate the CBC overlap that was cloned in the pGEM-T vector (Promega) and subjected to DNA sequencing. The 3′ end of the CSFR corresponding to a SalI (at position 2389)-HindIII (from the pRK5 vector) fragment was excised from the pRK5-CSFR vector. The 5′ end was excised as a BamHI-HindIII fragment from the pBABEpuro-CSFR vector. The HindIII-SalI CBC overlap was removed from the pGEM-T vector. These 3 fragments were cloned in the pBluescript SK(+) vector, and the complete CBC chimera was finally cloned in the pRK5 vector in a SmaI site.
The pRK5-Myc vectors encoding the constitutively activated Rho GTPase mutants L61Cdc42, L61Rac1, and L63RhoA (7) and the activated L61Cdc42 mutants bearing either the F37A or the Y40C amino acid substitution (37) have previously been described. pRK5-Myc encoding either the constitutively activated Rho kinase catalytic domain (amino acid 5 to 542) or a dominant-negative Rho kinase containing the Rho binding (RB) domain (amino acid 950 to 1069) were a kind gift of David Drechsel and Alan Hall (London, United Kingdom). These constructs were generated from the ROK-α cDNA (43). The activated L107FPAK1 was also used. The pCDB-PAKR mutant was a kind gift from Onyx Pharmaceuticals.
Transient transfections of MDCK cells.
MDCK cells were seeded at a concentration of 106 cells per 60-mm-diameter tissue culture dishes 24 h prior to transfection. Liposomes were prepared by a 30-min incubation at 20°C of 2 or 6 μl of Lipofectamine 2000 reagent (Invitrogen) together with 1 μg of pRK5-CEACAM1-S or -L plasmid and 3.0 μg of pRK5-Myc plasmid encoding the various Rho GTPase mutants, respectively, in a total volume of 600 μl of OptiMEM1 medium (Invitrogen). Transient transfections of the CSFR chimeric constructs was performed in the same way with 5 μg of the pRK5 empty vector, the pRK5-CSFR or pRK5-CBC construct, and 2.0 μg of the pRK5-CBB-S or -L construct. The liposome-DNA mix was incubated with the cells for 18 h, and the cells were then collected and processed for analyses of expression of the various proteins by immunoblotting.
Immunoblotting.
Expression of the cDNA constructs used in this study was confirmed by transient transfection of the constructs in MDCK cells as described above. Eighteen hours after the transfections, the cells were collected by scraping in lysis buffer (10 mM Tris-Cl [pH 7.4], 1% Triton X-100, 1 mM EDTA, 50 mM NaCl, complete protease inhibitors [Roche]). Protein concentrations were determined by using a bicinchoninic acid protein assay kit (Pierce Chemicals). Total lysate proteins were separated on sodium dodecyl sulfate (SDS)-8% polyacrylamide gel electrophoresis (PAGE) gels and transferred to Immobilon membranes. The membranes were probed with either a rabbit polyclonal anti-mouse CEACAM1 antibody (antibody [Ab] 2456, 1/1,000) (28), a mouse monoclonal anti-Myc antibody (monoclonal Ab [MAb] 9E10, 1/200; Calbiochem) or a rabbit polyclonal anti-human c-Fms antibody (1/1,000; Upstate Biotechnology). Immune complexes were detected by horseradish peroxidase-conjugated secondary antibodies and an enhanced chemiluminescence (ECL) kit (Amersham). Blots were exposed to X-ray films (Kodak).
Immunofluorescence.
Cells were fixed in 1.8% paraformaldehyde for 20 min, and nonspecific sites were saturated by incubation with a 10% solution of normal goat serum (Jackson ImmunoResearch Laboratories) diluted in PBS. CEACAM1protein expression was detected with the Ab 2456 (1/800). Alternatively, CMT-93 cells were stained with a mouse anti-mouse CEACAM1 monoclonal antibody (MAb CC1, 1/200), a kind gift of K.V. Holmes, University of Colorado (4). The wild-type CSFR or the chimeric CBB-S, CBB-L, and CBC proteins were detected by using a rat anti-human CSFR monoclonal antibody (1/200; Calbiochem). Alternatively, the Rho GTPases or their effectors were detected after permeabilization of the cells by treatment with Triton X-100 for 10 min before incubation with anti-Myc Ab 9E10 (1/200) for 45-min incubation periods. The antibodies were diluted in PBS containing 5% normal goat serum. After three washes in PBS, the cells were incubated for 45 min with a fluorescein isothiocyanate-conjugated anti-rabbit IgG antibody (1/400; ICN) or a Cy3-conjugated anti-mouse IgG antibody (1/400; Jackson ImmunoResearch Laboratories) or a fluorescein isothiocyanate-conjugated anti-rat IgG antibody (1/200) diluted in a PBS solution of 5% normal goat serum. The cells were washed three times in PBS. Polymerized actin was detected by incubating the slides for 45 min with tetramethyl rhodamine isothiocyanate (TRITC)-conjugated phalloidin (1/1,000; Sigma) after permeabilization in a PBS solution containing 0.2% Triton X-100 for 10 min. Coverslips were mounted with moviol containing p-phenylenediamine (1 mg/ml). The cells were examined by using either a Zeiss Axiophot fluorescence microscope (Thornwood, N.Y.) or a Zeiss Axiovert 510 confocal microscope (Hercules, Calif.). Images were collected with the LSM 510 software and processed in Adobe Photoshop.
Quantification of CEACAM1 at cell-cell contacts.
The percentage of cells showing CEACAM1 present at cell-cell contacts was calculated on an average of 50 to 60 CEACAM1-positive cells in several independent experiments.
RESULTS
CEACAM1 localization in primary hepatocytes and in a transformed epithelial cell line.
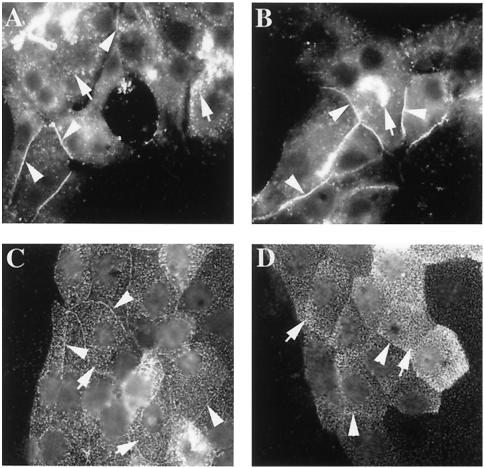
CEACAM1 is a cell surface molecule expressed in epithelial, endothelial, and hemopoietic cells (2). Immunolocalization of the protein in several cell lines indicated that its expression at cell-cell contacts was possibly a regulated event in epithelial cells. We investigated CEACAM1 localization in two different models. One model represents normal cells (rat primary hepatocytes), whereas the second model is an established transformed cell line (CMT-93, isolated from a mouse rectal carcinoma). Primary hepatocytes express both the CEACAM1-S and -L isoforms, and the CMT-93 cells predominantly express the CEACAM1-S isoform (58). Several fields of rat primary hepatocytes plated on collagen-coated microscope slides were examined after fixation and immunostaining with a polyclonal antibody specific to CEACAM1 (Ab 2456). The cells were not permeabilized prior to immunostaining. As seen in Fig. 1A and B, CEACAM1 staining was found over the entire surface of all observed cells in a punctate pattern and was also localized at some cell-cell contacts. A similar distribution was also observed in CMT-93 cells immunostained with anti-CEACAM1 MAb CC1, showing both punctate, evenly distributed cell surface expression and intercellular contact localization (Fig. 1C). Interestingly, as observed with primary hepatocytes, some cell contacts were negative for CEACAM1 (Fig. 1D). These results are similar to what has been observed in NbE rat normal prostatic epithelial cells or in NBT-II rat bladder carcinoma cells, where the expression of the endogenous CEACAM1 isoforms was detected both over the entire surface of the cells and at cell-cell contacts in subconfluent cells. In contrast, the endogenous protein was found mainly at cell-cell contacts as the cells entered into quiescence and underwent polarization (65). These results indicate that in epithelial cells, CEACAM1 is distributed heterogeneously at cell-cell contacts and at the cell surface, independently of the fact that these cells are normal or transformed.
FIG. 1.
Endogenous expression of CEACAM1 in primary rat hepatocytes and CMT-93 mouse rectal carcinoma cells. Primary rat hepatocytes were prepared as described in Materials and Methods and plated on collagen-coated microscope slides. CMT-93 cells were grown on microscope slides in DMEM containing 10% FBS to 50 to 80% confluence. CEACAM1 protein expression in primary hepatocytes (A and B) was detected by indirect immunofluorescence with a rabbit anti-mouse CEACAM1 antibody (Ab 2456; 1/800). Alternatively, CMT-93 cells (C and D) were stained with a mouse anti-mouse CEACAM1 monoclonal antibody (MAb CC1; 1/200). Arrowheads indicate significant localization of CEACAM1 at cell-cell contacts, and arrows indicate punctate dispersed expression of CEACAM1 at the cell surface.
CEACAM1 expression is directed to cell-cell contacts by activated Rac1 and Cdc42 GTPases but not by RhoA in MDCK cells.
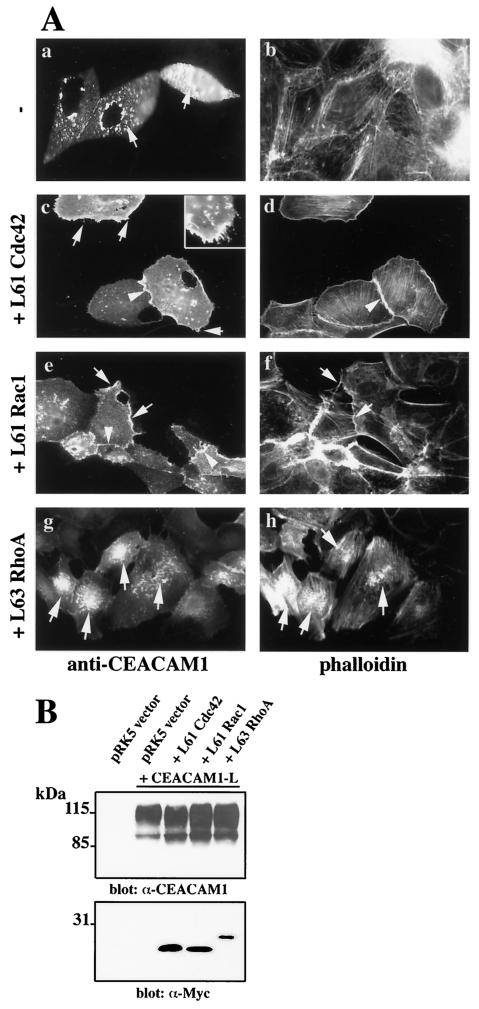
It was previously demonstrated that coinjection of CEACAM1-L with activated mutants of the Rho GTPases led to its expression at cell-cell contacts in Swiss 3T3 cells (60). It was therefore probable that Rho GTPase activity might be responsible for the modulation of CEACAM1 localization observed in hepatocytes and CMT-93 cells. We chose the MDCK cell line as a model for the study of CEACAM1 localization in epithelia, since CEACAM1 is expressed in the renal collecting tubular epithelium (55). The MDCK cells are amenable to direct delivery of cDNA constructs in the cell nucleus by microinjection. Many of the activated or dominant-negative Rho GTPase mutants have detrimental effects on cell morphology and survival if expressed over long periods of time. Therefore, microinjection of cDNA constructs expressed for shorter periods can obviate these deleterious effects. Cells were injected at 50 to 80% confluence to allow the cells to form membrane extensions, such as filopodia and membrane ruffles, or eventually to spread in response to Rho GTPase activity. These conditions are different from those of MDCK cells grown as polarized sheets. The Ceacam1-L cDNA was therefore microinjected either alone (Fig. 2A -a and b) or in combination with activating mutant cDNAs of Cdc42, Rac1, and RhoA (Fig. 2A-c to h) in subconfluent MDCK cells. When the Ceacam1-L cDNA was expressed alone, the encoded protein was distributed over the entire surface of the cell in a punctate pattern (Fig. 2A-a). This was similar to the distribution of CEACAM1 observed in some hepatocytes and CMT-93 cells. Phalloidin staining of these cells indicated that the majority of filamentous actin was concentrated at the cell periphery (Fig. 2A-b). Coinjection of Ceacam1-L cDNA with activated Cdc42 or Rac1 into the cell nucleus led to CEACAM1 cell-cell contact expression (Fig. 2A-c and e, respectively). Cells expressing the activated mutants of Cdc42 and Rac1 showed characteristic filopodia (Fig. 2A-c, inset) and lamellipodia actin structures, respectively (Fig. 2A-f).
FIG. 2.
(A) CEACAM1 is targeted to cell-cell contacts, and typical actin structures are apparent upon Cdc42 and Rac1 activation. The Ceacam1-L cDNA was microinjected into the nucleus of subconfluent MDCK cells either alone (a and b) or together with an activated Myc-tagged L61Cdc42 mutant (c and d), a Myc-tagged L61Rac1 (e and f), or a Myc-tagged L63RhoA mutant (g and h). CEACAM1 expression was detected by indirect immunofluorescence with an anti-CEACAM1 (α-CEACAM1) antibody (Ab 2456), and polymerized actin was revealed by TRITC-coupled phalloidin. Expression of Rho GTPases was detected by immunofluorescence with a monoclonal anti-Myc antibody (Ab 9E10) (data notshown). The arrows in panel a indicate the punctate localization of CEACAM1-L. The arrows in panels c to h coincide with either filopodia/microspike (c) or lamellipodial (e and f) CEACAM1 expression. The arrowheads indicate CEACAM1 localization at cell-cell contacts (c and e). Either 0% (a), 82% (c), or 81% (e) of CEACAM1-positive cells expressed CEACAM1 at cell junctions, respectively. Coinjections of the Ceacam1-L cDNA with an activated L63RhoA mutant did not result in targeting to cell-cell contacts, as none of the CEACAM1-positive cells expressed this protein at cell junctions. Instead, the protein was present in a punctate pattern at the cell surface (arrows in panels g and h). Stress fibers were consistently seen in these cells. (B) Expression levels of the CEACAM1-L protein upon coinjection of the activated Rho-GTPases. MDCK cells were transiently transfected with Lipofectamine 2000 liposomes with either the empty vector alone or the vector expressing the Ceacam1-L cDNA alone and together with Myc-tagged L61Cdc42 or L61Rac1 or L63RhoA mutants. After 18 h of expression, the cells were collected and lysed. Equivalent amounts of total cell lysate proteins were separated by SDS-PAGE. The CEACAM1-L protein was detected with a polyclonal antibody (Ab 2456) and the activated Rho-GTPase mutants were revealed with a monoclonal anti-Myc (α-Myc) antibody (Ab 9E10). Immune complexes were detected with the ECL system.
Coinjection of the activating L63RhoA mutant with CEACAM1-L did not lead to cell-cell contact localization of the glycoprotein. Instead, CEACAM1 staining displayed a punctate pattern present at the cell surface (Fig. 2A-g). The structures expressing CEACAM1 under these conditions appear more condensed (Fig. 2A-g) than when CEACAM1 is expressed alone (Fig. 2A-a). It is presently unclear what these structures represent. Stress fibers were present in these cells, as expected for RhoA activity (Fig. 2A-h). Since the CEACAM1 protein is detected on nonpermeabilized cells, the protein is essentially localized at the cell surface, as confirmed by confocal microscopy sectioning (data not shown). These results suggest that, in epithelial cells, CEACAM1 is relocalized upon signaling from activated Cdc42, Rac1, and RhoA.
To examine whether the different patterns of CEACAM1 cellular localization observed upon Rho GTPase mutant expression resulted from a change of CEACAM1 cellular distribution and not from different levels of protein expression, we evaluated the expression of CEACAM1-L in the presence of Rho GTPases by immunoblotting. MDCK cells were transiently transfected with the CEACAM1-L construct alone or together with the activated Myc-tagged L61Cdc42, L61Rac1, or L63RhoA construct. After 18 h of expression, the cells were collected and total cell proteins were prepared and separated by SDS-PAGE. As demonstrated in Fig. 2B, MDCK cells transfected with the empty vector did not express the CEACAM1 isoforms (Fig. 2B, lane pRK5). CEACAM1 is a highly glycosylated protein, and expression of the CEACAM1-L transfected into MDCK cells was detected as a large smear of approximately 120 kDa (Fig. 2B, lane pRK5 + CEACAM1-L). Coexpression of the activated Rho GTPase mutants (Fig. 2B, lanes +L61Cdc42, +L61Rac1, and +L63RhoA), as gauged in the bottom panel detected with an anti-Myc antibody, did not significantly alter the expression of CEACAM1-L.
Activated PAK targets CEACAM1 to cell-cell contacts.
Both Cdc42 and Rac1 influence cellular activities through functional modulation of a number of key effectors, some of which are common to Cdc42 and Rac1. PAK is a Ser/Thr kinase acting downstream of Rac1 and Cdc42 (41). It is intricately involved in cell motility through phosphorylation and inactivation of myosin light-chain kinase, causing disassembly of stress fibers and focal adhesions (63). PAK also phosphorylates LIM kinase, leading to its enhanced activity towards the actin-depolymerizing protein cofilin. This results in membrane ruffling, a hallmark of motile cells (1, 14, 78). Furthermore, formation of a paxillin/GIT1/PIX/PAK/Nck complex is necessary for localization of PAK to focal complexes within lamellipodia and filopodia (70).
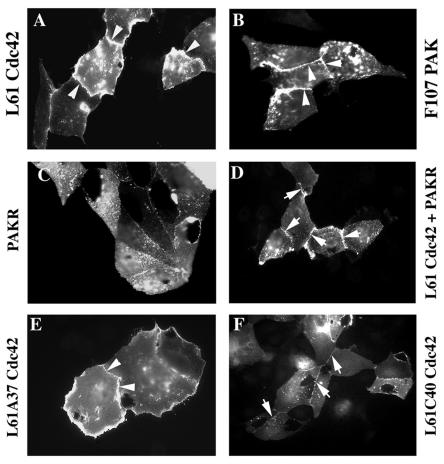
As CEACAM1 localization responds to Rac1 and Cdc42 activation and because CEACAM1-L has been shown to associate with paxillin (12), we investigated whether their effector PAK was involved. Similar to results obtained with the activated Cdc42 mutant (Fig. 3A), coinjection of a constitutively active PAK cDNA (L107FPAK1) resulted in CEACAM1-L targeting at intercellular contacts (Fig. 3B), showing its active role in the relocalization of CEACAM1. To assess the contribution of PAK in Cdc42-induced CEACAM1 targeting, we coexpressed the N-terminal regulatory domain (amino acids 1 to 225) of PAK2 (PAKR) (42, 44) with CEACAM1-L and activated L61Cdc42. PAKR includes an autoinhibitory region that blocks the kinase activity of PAK in vitro and in vivo (69, 80). Coinjection of CEACAM1-L and PAKR revealed a punctate localization of CEACAM1-L (Fig. 3C) similar to results obtained with CEACAM1-L alone, showing that this construct has no effect by itself on CEACAM1 localization. When PAKR was coinjected with L61Cdc42, a weak intercellular CEACAM1 staining was observed (Fig. 3D), suggesting that inhibition of the Cdc42-induced PAK activation decreases CEACAM1 targeting at cell contacts but does not completely abolish it. Therefore, other Cdc42 effector(s) may contribute to CEACAM1 localization at cell-cell contacts.
FIG. 3.
Activated PAK targets CEACAM1 to cell-cell contacts. The cDNA encoding CEACAM1-L was microinjected with constructs encoding either Myc-tagged L61Cdc42 (A and D) or L107FPAK1 (B), PAKR (C and D), L61A37Cdc42 (E), and L61C40Cdc42 (F). Expression of CEACAM1-L was detected in microinjected MDCK cells by immunofluorescence with an anti-CEACAM1 antibody (Ab 2456), and Myc-tagged proteins in injected cells were detected by immunofluorescence with an anti-Myc antibody (Ab 9E10) (data not shown). The arrowheads indicate significant localization of CEACAM1 at cell-cell contacts, and the arrows indicate faint detection of CEACAM1 at cell-cell contacts. The percentages of CEACAM1-positive cells expressing CEACAM1 at cell junctions were 82% (A), 67% (B), 0% (C), 71% (D), and 90% (E and F).
To corroborate the partial contribution of PAK in CEACAM1 targeting, we used the L61C40Cdc42 effector mutant that is unable to interact with PAK and that eliminates both PAK activation and the JNK/SAPK mitogen-activated protein kinase pathway (37). As a control, we used the L61A37Cdc42 mutant that retains these functions. Coinjection of CEACAM1-L with the L61A37Cdc42 mutant resulted in CEACAM1 localization at cell-cell contacts (Fig. 3E), which was comparable to effects observed with the L61Cdc42 mutant. On the other hand, the L61C40Cdc42 mutant induced CEACAM1 relocalization, but the staining observed at cell contacts in this case is notably weaker (Fig. 3F) than the L61Cdc42 results. Therefore, these results suggest that PAK is not the only effector involved in the Cdc42-induced localization of CEACAM1, but it largely contributes to this effect. Hence, activated Cdc42 may also mediate CEACAM1 relocalization via PAK-independent signaling pathways.
Down-regulation of Rho pathway activity leads to CEACAM1at cell-cell boundaries.
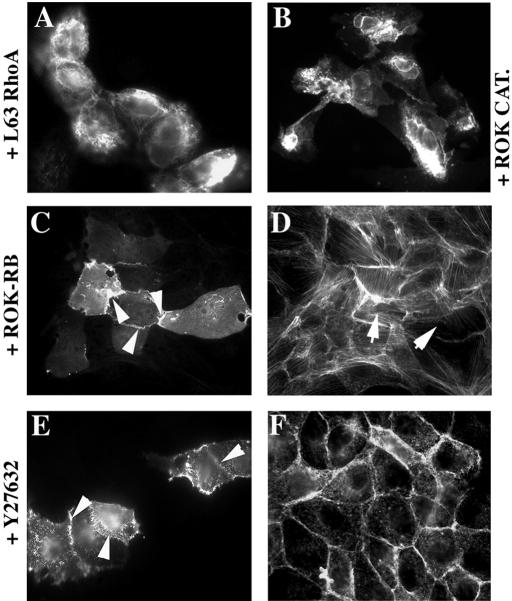
Contrary to activated Cdc42 and Rac1, the activated L63RhoA mutant did not influence cell-cell contact localization of CEACAM1 in MDCK cells (Fig. 2A-g and Fig. 4A). Upon activated RhoA coexpression, the CEACAM1 protein was condensed at the cell surface. RhoA binds to the Rho-associated Ser/Thr kinase isoforms (ROKα, p160ROCK), which then localize to the cell membrane and act as RhoA effectors (11, 27, 38, 43). We therefore tested whether the effect seen with RhoA was mediated by ROK. Coinjection of a constitutively activated ROK (ROK catalytic domain, or ROK CAT) with CEACAM1-L led to a localization pattern (Fig. 4B) similar to what had been noticed with activated RhoA. In contrast to results obtained with ROK CAT, when the C-terminal portion of ROK encompassing the RB domain (ROK-RB), which acts as a dominant-negative mutant, was coinjected with CEACAM1-L, the glycoprotein was found at sites of cell-cell contacts (Fig. 4C). Moreover, when cells were treated with the Y27632 ROK inhibitor, competing with ATP for binding to the ROK domain (71), CEACAM1-L was again localized at intercellular contacts (Fig. 4E). As expected, loss of stress fibers was observed in cells injected with ROK-RB (Fig. 4D) or in cells treated with the Y27632 compound (Fig. 4F). We conclude that the effect of RhoA on CEACAM1 localization is mediated by ROK. Moreover, inhibition of this signaling pathway reversed the effect on CEACAM1 localization, conferring a similar pattern to that observed with activated Cdc42 or Rac1.
FIG. 4.
The abolition of Rho-mediated signaling targets CEACAM1 to cell-cell contacts. pRK5-CEACAM1-L was coinjected with either (A) Myc-L63RhoA, Myc-ROK CAT (B), or Myc-ROK-RB (C and D) into subconfluent MDCK cells. (E and F) Cells microinjected with the CEACAM1-L construct were treated with 10 μM Y27632 ROK inhibitor for 30 min prior to fixation. Anti-CEACAM1 antibody (Ab 2456) staining indicated that CEACAM1-L was expressed diffusely at the cell surface (no CEACAM1 at cell junctions in panels A and B) or in cell-cell contacts (84 and 80% of CEACAM1-positive cells had CEACAM1 at cell junctions, arrowheads in panels C and E). TRITC-coupled phalloidin staining (D and F) showed that stress fibers were absent from injected cells (arrows in panel D) or in Y27632-treated cells (F). The expression of Myc-tagged proteins was detected with an anti-Myc antibody (Ab 9E10) (data not shown).
The CEACAM1 intracellular and extracellular domains are dispensable for intercellular localization.
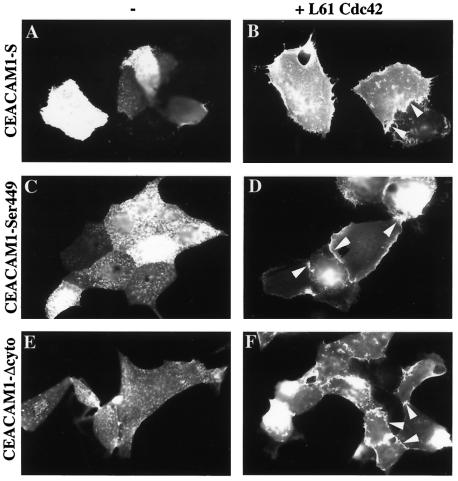
The long intracytoplasmic domain present in CEACAM1-L is essential for many CEACAM1-L functions such as tumor growth control (28). We questioned whether, in MDCK epithelial cells, the CEACAM1-S and CEACAM1-L splice isoforms (Fig. 5) localized at intercellular contacts when activated forms of Rac1 or Cdc42 were coinjected. CEACAM1-S encompasses only a 10 amino acid cytoplasmic domain (Fig. 5), but it behaves like CEACAM1-L relative to its targeting at epithelial cell-cell contacts in the presence of activated Cdc42 (Fig. 6B). Several Ser residues are found within the short cytoplasmic tail, one or several of which are phosphorylated by protein kinase C (13). We replaced Ser449, Ser452, and Ser449,452 with nonphophorylatable Ala residues to verify whether Ser phosphorylation of these sites interfered with the localization of CEACAM1-S. None of these amino acid substitutions produced any significant differences on CEACAM1-S intercellular location in the absence or presence of activated Cdc42 (Fig. 6C and D and data not shown). We then completely truncated the cytoplasmic domain by introducing a stop codon at Ser449, producing the mutant CEACAM1-Δcyto. Eliminating the CEACAM1 cytoplasmic domain did not alter its intercellular junction localization in the presence of activated Cdc42 (Fig. 6F). These results strongly suggest that the cytoplasmic domain of CEACAM1 is not required for its proper localization to cell-cell contacts.
FIG. 6.
The cytoplasmic domain of CEACAM1 is not involved in intercellular targeting. CEACAM1 mutant constructs were microinjected either alone (A, C, and E) or together with activated L61Cdc42 (B, D, and F) into subconfluent MDCK cells. Cells were stained with an anti-CEACAM1 antibody (Ab 2456) recognizing extracellular epitopes. Expression at cell-cell contacts is denoted by the presence of arrowheads. (A and B) CEACAM1-S; (C and D) CEACAM1-S-Ser449 mutant; (E and F) CEACAM1-Δcyto mutant. Eighty-four and eighty percent of CEACAM1-positive cells expressed CEACAM1 at cell junctions in panels D and F, respectively.
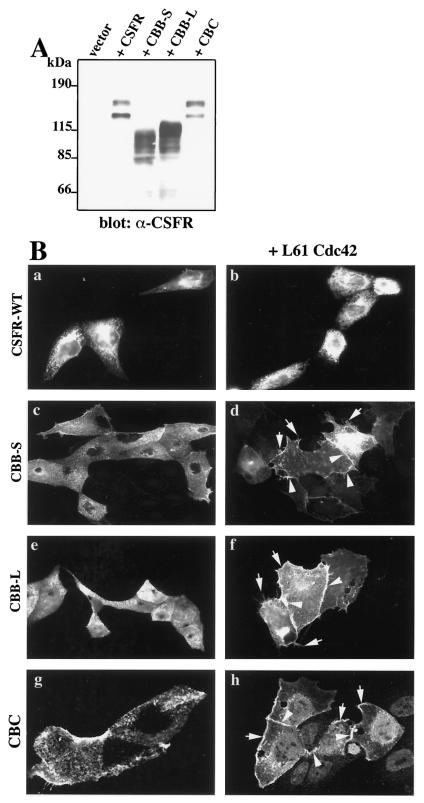
To determine the role of the CEACAM1 extracellular domain in cell-cell contact localization, we substituted the 4 CEACAM1 Ig-like domains with the 5 extracellular Ig domains of the CSFR (10) (a kind gift of Martine Roussel) (Fig. 5). The chimeric proteins were anchored to the cell membrane via the CEACAM1 transmembrane domain and contained either the short (CBB-S) or long (CBB-L) CEACAM1 cytoplasmic domains (Fig. 5). We first verified whether the chimeric proteins were properly synthesized in MDCK cells by transient transfection of the respective constructs and detection by immunoblotting after migration on SDS-PAGE. As seen in Fig. 7A, transfection of the CSFR construct generated a protein of approximately 145 kDa corresponding to the known molecular mass of CSFR (10). A second band of lower molecular mass was also detected on the immunoblot and corresponds to a degradation product (as discussed in the technical brochure from Upstate Biotechnology). The CBB-S and CBB-L constructs produced proteins of approximately 115 and 120 kDa, consistent with their predicted molecular masses.
FIG. 7.
The CEACAM1 transmembrane domain mediates CEACAM1 localization to cell contacts in response to L61Cdc42. (A) Expression of the chimeric constructs in transiently transfected MDCK cells. MDCK cells were transiently transfected with Lipofectamine 2000 liposomes with either the empty vector alone or the vector expressing either the wild-type CSFR cDNA or those encompassing the CBB-S, CBB-L, or CBC chimeric constructs. After 24 h of expression, the cells were collected and lysed. Equivalent amounts of total cell lysate proteins were separated by SDS-PAGE. The chimeric proteins were detected with a rabbit polyclonal antibody recognizing the human c-Fms protein. Immune complexeswere detected by horseradish peroxidase-conjugated secondary antibodies and an ECL kit. (B) Expression of chimeric constructs in microinjected MDCK cells. The wild-type (WT) CSFR or chimeric constructs were microinjected either alone (a, c, e, and g) or together (b, d, f, and h) with the activated L61Cdc42 mutant into subconfluent MDCK cells. Cells were stained with an anti-CSFR antibody. Wild-type CSFR (a and b), CBB-S (c and d), and CBB-L (e and f), and CBC (g and h) constructs were used. Arrowheads indicate localization to cell-cell contacts. Arrows point to filopodial structures induced by the activated Cdc42 activity. Seventy-seven, eighty-one, and fifty-three percent of positive cells expressed the protein at cell junctions in panels d, f, and h, respectively. Photographs in panels g and h are from 0.25-μm-thick confocal sections (B).
Injection of the wild-type CSFR construct in MDCK cells with or without activated Cdc42 revealed a dispersed localization after staining with the anti-CSFR antibody (Fig. 7B-a and b), demonstrating that this transmembrane protein structurally related to CEACAM1-L does not respond to Cdc42 activation in the same way as CEACAM1-L. When the chimeric CBB-S or CBB-L proteins were injected alone into MDCK cells, they remained disperse in a punctate pattern as detected with an anti-CSFR antibody (Fig. 7B-c and e, respectively). However, when coexpressed with activated Cdc42, the chimeric proteins were again found at intercellular contacts (Fig. 7B-d and f, respectively), similar to the wild-type CEACAM1 cell expression pattern. Filopodia can be seen extending from the cell surface. Therefore, these results suggest that neither the truncation of the CEACAM1 cytoplasmic domain nor the replacement of its extracellular domain changed the cell-cell contact targeting of the protein, thereby identifying the transmembrane domain as most likely essential.
The CEACAM1 transmembrane domain is sufficient for targeting to cell-cell contacts.
To confirm this hypothesis, a chimeric protein where the transmembrane domain of CSFR was replaced by that of CEACAM1 (CBC) was produced. When expressed in MDCK by transient transfection, the CBC protein behaved like the CSFR protein upon SDS-PAGE (Fig. 7A). When injected alone into MDCK cells, CBC was localized at the cell surface in a punctate pattern (Fig. 7B-g). When CBC was injected with the activated L61Cdc42, the chimeric CBC protein was detected at cell-cell contacts (Fig. 7B-h) and in lamellipodia and membrane ruffles (Fig, 7B-h). We conclude that introducing the CEACAM1 transmembrane domain within the CSFR construct is sufficient to confer to this protein the cell localization properties observed with CEACAM1 upon Cdc42 activation. Therefore, the transmembrane domain of CEACAM1 plays a crucial role in CEACAM1 targeting to cell-cell contacts.
DISCUSSION
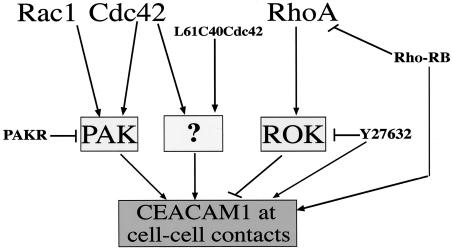
In this work, we show that activated Cdc42, Rac1, and their common effector PAK can direct CEACAM1 to intercellular contacts, as depicted by the model in Fig. 8. Our results have highlighted that unknown effector(s) are also likely targeting CEACAM1 to intercellular boundaries. Indeed, inhibiting PAK activity through a dominant-negative PAKR protein or disengaging PAK from Cdc42 interactions and eliminating PAK activation resulted in consistently present, albeit weak, CEACAM1 staining at cell-cell contacts. In contrast to activated Rac1, Cdc42, and PAK, activated RhoA or ROK CAT coexpression does not result in CEACAM1 intercellular targeting. Moreover, inhibition of the Rho pathway led to the addressing of CEACAM1 to sites of cell-cell contacts. These results suggest that a fine balance between the Rho GTPase pathways must be established for proper CEACAM1 localization to occur. According to this hypothesis, Cdc42 and Rac1, on one hand, and RhoA, on the other hand, would induce two different pathways, leading to opposite effects by either targeting CEACAM1 at cell-cell contacts or concentrating it at the top of the cells, respectively. At an equilibrium between the two pathways, CEACAM1 would be homogeneously diffuse at the cell membrane. Upon Rho pathway inactivation, the Cdc42 and Rac1 pathways, involving at least the PAK effector, would predominate and CEACAM1 would be targeted to cell contacts. This situation is reminiscent of other reports demonstrating the opposite effects of PAK and ROK in the hepatocyte growth factor-induced epithelial-mesenchymal transition (59) or the different effects of the Rho GTPases in neuronal growth (34). Similarly, Rac-mediated down-regulation of Rho in NIH 3T3 fibroblasts is another example of the antagonistic effects of Rho GTPase pathways (52). The model proposed in this study of MDCK cells may correspond to the regulation of CEACAM1 expression in kidney tissue, a tissue that expresses CEACAM1 endogenously. Accordingly, this model could explain the heterogeneity observed in the localization of endogenous CEACAM1 in primary hepatocytes or CMT-93 cells, where CEACAM1 is dispersed at the cell surface in most cells and present at cell contacts in only a few cells. Attempts to pharmacologically activate the Rho GTPases or to express either activated Rho GTPase proteins or cDNA constructs in the CMT-93 cells or primary hepatocytes have been unsuccessful at provoking alterations in cytoskeletal structures or CEACAM1 relocalization. Whether the model defined in MDCK cells would also prevail in primary or transformed epithelial cells will need to be clarified in the future. In this case, more studies will be needed to verify whether the CEACAM1 localization at cell contacts is correlated with activation of Cdc42 or Rac1 and inhibition of RhoA in these particular cells.
FIG. 8.
Model of pathways involved in targeting CEACAM1 to intercellular contacts. Both Rac1 and Cdc42 activities lead to CEACAM1 cell-cell contact targeting. Since PAK activity reproduces this effect, we propose that the pathway involved in CEACAM1 relocalization includes this kinase (arrows). However, inhibiting the PAK activity with a PAKR dominant-negative mutant (bar) or disengaging PAK from Ccd42 binding with the L61C40Cdc42 mutant (arrow) still resulted in faint expression of CEACAM1 at sites of cell-cell contacts. This suggests that alternate and as yet unknown effector(s) (question mark) might be activated in these circumstances. Activated RhoA functions through its effector ROK (arrow) to inhibit CEACAM1 targeting to cell-cell contacts (bar). Inhibition of RhoA activity by expression of the RB domain of ROK (bar) or inhibition of ROK activity by the Y27632 inhibitor (bar) both restore CEACAM1 targeting to cell-cell contacts.
In E-cadherin-mediated cell adhesion, Rac1 and Cdc42 both induce the accumulation of E-cadherin, β-catenin, and actin filaments to sites of cell-cell contacts in MDCKII cells (31, 36, 68). As discussed by Fukata and Kaibuchi (19), Rac1 and Cdc42 positively regulate E-cadherin-mediated adhesion by modulating the cadherin-catenin complex, whereas RhoA alters adhesion through changes in the actin cytoskeleton or other components. However, Sahai and Marshall (61) have recently shown that inhibition of RhoA activity in MDCK cells, but not that of its effector ROK, led to reduced expression of β-catenin at sites of cell-cell contacts and this severely disturbed the columnar organization of these cells. They further demonstrated that Dia1, another Rho effector, was involved in stabilizing the adherens junctions at the cell periphery, whereas ROK disrupted them by establishing contractile forces (61). We do not know what role Dia1 might play in CEACAM1 cell-cell contact targeting and adhesion and this will need to be further investigated. On the other hand, it has been shown that cadherins trigger signaling that regulates Rho GTPase activity (79). As shown by Noren et al., cadherin-elicited adhesion activates Rac1 and Cdc42 and inhibits RhoA activation (49). It is possible that within an epithelial layer, cadherin engagement and its consequent effects on Rho GTPase activity lead to CEACAM1 localization at cell contacts.
CEACAM1 functions as an intercellular adhesion molecule through its first Ig domain (28, 58, 76, 77). We anticipate that the localization of CEACAM1 at cell contacts triggered by activation of Cdc42 and/or Rac1 in MDCK cells is crucial for the CEACAM1 adhesive function. Unfortunately, it is difficult to evaluate the effect of Rho GTPase activity on CEACAM1-mediated cell adhesion in this cellular model. In these cells, the E-cadherin intercellular adhesion system is predominant and elimination of this adhesion activity by reduction of the calcium concentration or antibody inhibition leads to complete disorganization of cell morphology and multicellular organization (19). However, there are two major lines of evidence that currently validate the adhesive activities of CEACAM1 at cell contacts. First, in fibroblast cells, where there is no interference from the E-cadherin adhesion complex, targeting of CEACAM1-L at Swiss 3T3 cell contacts provoked its engagement in homophilic interactions (60) and induced cell aggregation in NIH 3T3 cells (28). Second, the role of CEACAM1 as a cell adhesion molecule in epithelial cells has been studied by Sundberg and Öbrink in fully polarized MDCK cells transfected with CEACAM1-L (67). The localization of this protein in the lateral membrane with consequent blockage of the N-epitope to antibody recognition represented the state of homophilic, antiparallel binding observed when CEACAM1 is engaged in adhesion. Hyperosmotic treatment provoked shrinkage and retraction of the cells, thereby freeing the N epitope for antibody recognition (67). The same study showed that in polarized MDCK cells, CEACAM1-L is localized at the apical and lateral surfaces where it engages in intercellular adhesion, whereas CEACAM1-S is expressed exclusively at the apical membrane (67), suggesting a discrepancy in the ability of the two isoforms to trigger cell adhesion. In our study, we did not notice any differences in the localization of CEACAM1-S versus CEACAM1-L, indicating that both of them relocalize upon Rho-GTPase mutant coexpression. This can be explained by the fact that we have investigated CEACAM1 localization in cells organized as individual colonies (50% confluence). In these conditions, cells are not polarized and the two compartments, basolateral and apical, are not fully differentiated. However, it is likely that the presence of CEACAM1 at cell-cell contacts still favors intercellular adhesion in nonpolarized cells, as seen in fibroblasts.
In addition to its accumulation at cell contacts, CEACAM1 is also relocalized in either filopodia or lamellipodia, depending on the activation of Cdc42 or Rac1, respectively. This suggests another functional property, in addition to trafficking to intercellular contacts for adhesion purposes. In filopodia, CEACAM1 is at the very tip of the protrusion. Filopodia are dynamic structures, extending and retracting constantly. They adhere to the substratum, pulling the motile cell forward and acting in concert with lamellipodia that interdigitate between protrusions (45, 66). The localization of CEACAM1 within these structures suggests a potential role for CEACAM1 in cell motility. Consistent with these observations, CEACAM1-L expression enhances the chemotaxis and formation of capillary tubes of a human microvascular endothelial cell line (15) and the migration of colon carcinoma cells (B. Fournès, S. Testay, C. Turbide, and N. Beauchemin, unpublished data).
As the two isoforms behave similarly in their targeting to cell-cell contacts, we investigated which CEACAM1 structural elements might be involved in this phenotype. Our results demonstrate that none of the phosphorylated Ser residues in the short cytoplasmic domain of CEACAM1 participate in the targeting of CEACAM1. A deletion construct, completely eliminating the CEACAM1 cytoplasmic domain, confirmed that the site of intercellular targeting does not lie in this domain. We therefore tested the role of the extracellular domain, since it is the main player in cell adhesion. The extracellular region of CEACAM1 did not apparently contain any specific targeting signals, as its substitution with that of CSFR (CBB-S and CBB-L chimeras) does not disturb the cell-cell contact targeting upon Cdc42 activation. We then tested the role of the transmembrane domain. Interestingly, the replacement of the CSFR transmembrane domain with that of CEACAM1 (CBC chimera) demonstrated that this amino acid sequence was sufficient for localization of the chimera to intercellular contacts upon Cdc42 activation. Therefore, we propose that the CEACAM1 transmembrane region is the target of Rho GTPase-triggered pathways.
Other investigators have described the involvement of transmembrane regions of cell surface proteins in many signaling functions. For instance, the transmembrane domain of the adhesion molecule P-selectin, within the context of the P-selectin cytoplasmic domain, mediates its granular targeting in rat insulinoma cells (18). In addition, the juxtamembrane region of the erythropoietin receptor contains three hydrophobic motifs that are crucial for JAK2 activation and downstream signaling (9). Finally, the adhesive functions of E-cadherin are preferentially mediated by its cis-dimerization, and its transmembrane domain is required for this lateral association (52). As CEACAM1 also forms dimers (26), it is possible that its transmembrane domain plays a significant role in maintaining the cis-dimers in an adhesive configuration and then stabilizing the protein at cell contacts.
Moreover, there is some evidence suggesting that Rho- GTPases are related to membrane microdomains. For instance, Cdc42 is localized in the caveolae-enriched domains in endothelial cells (20), and Field et al. proposed that Cdc42 plays a role in lipid raft biosynthesis in mast cells (17). In addition, membrane raft microdomains mediate the front-rear polarity in migratory cells. Upon induction of MCF-7 cell migration, lipid raft domains cluster and accumulate at the cell front in structures such as pseudopodia and lamellipodia (40). Since Cdc42 and Rac1 stimulate the formation of these structures in fibroblasts and epithelial cells (22) and since CEACAM1, via its transmembrane domain, is enriched in these regions, we propose the following model: Rac1- or Cdc42-induced modifications in the organization of the lipid bilayer would result in the formation of microdomains at specific sites of the cell surface such as cell contacts, filopodia, and lamellipodia. The CEACAM1 transmembrane domain would target the protein to these membrane microdomains or lipid rafts, where CEACAM1 can function as a cell adhesion molecule or as a mediator of cell migration. Such relocalization has been observed with another member of the Ig superfamily, CD44, which gets targeted to basolateral lipid rafts of EpH4 mammary epithelial cells (51) by its transmembrane domain (53), whereupon it interacts with the actin cytoskeleton through its association with the ezrin-radixin-moesin complex (51). Finally, under conditions where MDCK cells become fully polarized, the CEACAM1-L cytoplasmic domain would play a significant role in partitioning some of the CEACAM1-L protein to the lateral membrane, where it would engage in intercellular adhesion, and some of the CEACAM1-S isoform to the apical domain, where its function is still unclear.
Given the importance of CEACAM1 in a number of physiological processes, the proper localization of CEACAM1 at sites of cell-cell contacts and membrane extensions in response to the Rho GTPase balance ensures proper interconnections within intracellular signaling pathways. Our findings provide new insight into understanding the molecular mechanisms underlying CEACAM1 localization at specific sites of cell-cell boundaries.
Acknowledgments
We thank Victor Dumas and Barry I. Posner as well as Alan Cheng and Michel L. Tremblay from McGill University for providing us with rat and mouse primary hepatocytes, respectively. We are greatly indebted to Morag Park (Montreal, Quebec, Canada) and Isabelle Royal (Montreal, Quebec, Canada) for constructs and discussions. We also acknowledge the contribution by Onyx Pharmaceutical of the pCDB-PAKR plasmid. We are also greatly indebted to Martine Roussel for the wild-type CSFR cDNA, to Yoshitomi Pharmaceutical Industries for the Y27632 ROK inhibitor, and to David Drechsel and Alan Hall (London, United Kingdom) for plasmids encoding the ROK-RB and the ROK CAT.
B. Fournès and N. Beauchemin are funded, respectively, by a postdoctoral fellowship and a “Chercheur National” senior scholarship award from the Fonds de la Recherche en Santé du Québec. J. Farrah is a recipient of Canderel and MUHC studentship awards. N. Lamarche-Vane is a CIHR New Investigator scholar. This work was funded by the Canadian Institutes of Health Research.
REFERENCES
- 1.Arber, S., F. A. Barbayannis, H. Hanser, C. Schneider, C. A. Stanyon, O. Bernard, and P. Caroni. 1998. Regulation of actin dynamics through phosphorylation of cofilin by LIM-kinase. Nature 393:805-809. [DOI] [PubMed] [Google Scholar]
- 2.Beauchemin, N., and S. H. Lin. 1998. Role of C-CAM as a tumor suppressor, p. 155-175. In C. P. Stanners (ed.), Cell adhesion and communication mediated by the CEA family. Harwood Academic Publishers, Amsterdam, The Netherlands.
- 3.Bishop, A. L., and A. Hall. 2000. Rho GTPases and their effector proteins. Biochem. J. 348:241-255. [PMC free article] [PubMed] [Google Scholar]
- 4.Blau, D. M., C. Turbide, M. Tremblay, M. Olson, S. Letourneau, E. Michaliszyn, S. Jothy, K. V. Holmes, and N. Beauchemin. 2001. Targeted disruption of the Ceacam1 (MHVR) gene leads to reduced susceptibility of mice to mouse hepatitis virus infection. J. Virol. 75:8173-8186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boulton, I. C., and S. D. Gray-Owen. 2002. Neisserial binding to CEACAM1 arrests the activation and proliferation of CD4+ T lymphocytes. Nat. Immunol. 3:229-236. [DOI] [PubMed] [Google Scholar]
- 6.Braga, V. M., M. Betson, X. Li, and N. Lamarche-Vane. 2000. Activation of the small GTPase Rac is sufficient to disrupt cadherin-dependent cell-cell adhesion in normal human keratinocytes. Mol. Biol. Cell. 11:3703-3721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caron, E., and A. Hall. 1998. Identification of two distinct mechanisms of phagocytosis controlled by different Rho GTPases. Science 282:1717-1721. [DOI] [PubMed] [Google Scholar]
- 8.Chen, T., W. Zimmermann, J. Parker, I. Chen, A. Maeda, and S. Bolland. 2001. Biliary glycoprotein (BGPa, CD66a, CEACAM1) mediates inhibitory signals. J. Leukoc. Biol. 70:335-340. [PubMed] [Google Scholar]
- 9.Constantinescu, S. N., L. J. Huang, H. Nam, and H. F. Lodish. 2001. The erythropoietin receptor cytosolic juxtamembrane domain contains an essential, precisely oriented, hydrophobic motif. Mol. Cell 7:377-385. [DOI] [PubMed] [Google Scholar]
- 10.Coussens, L., C. Van Beveren, D. Smith, E. Chen, R. L. Mitchell, C. M. Isacke, I. M. Verma, and A. Ullrich. 1986. Structural alteration of viral homologue of receptor proto-oncogene fms at carboxyl terminus. Nature 320:277-280. [DOI] [PubMed] [Google Scholar]
- 11.Dharmawardhane, S., and G. M. Bokoch. 1997. Rho GTPases and leukocyte cytoskeletal regulation. Curr. Opin. Hematol. 4:12-18. [DOI] [PubMed] [Google Scholar]
- 12.Ebrahimnejad, A., R. Flayeh, G. Unteregger, C. Wagener, and J. Brummer. 2000. Cell adhesion molecule CEACAM1 associates with paxillin in granulocytes and epithelial and endothelial cells. Exp. Cell Res. 260:365-373. [DOI] [PubMed] [Google Scholar]
- 13.Edlund, M., K. Wikstrom, R. Toomik, P. Ek, and B. Obrink. 1998. Characterization of protein kinase C-mediated phosphorylation of the short cytoplasmic domain isoform of C-CAM. FEBS Lett. 425:166-170. [DOI] [PubMed] [Google Scholar]
- 14.Edwards, D. C., L. C. Sanders, G. M. Bokoch, and G. N. Gill. 1999. Activation of LIM-kinase by Pak1 couples Rac/Cdc42 GTPase signalling to actin cytoskeletal dynamics. Nat. Cell Biol. 1:253-259. [DOI] [PubMed] [Google Scholar]
- 15.Ergün, S., N. Kilic, G. Ziegeler, A. Hansen, P. Nollau, J. Götze, J. H. Wurmbach, A. Horst, J. Weil, M. Fernando, and C. Wagener. 2000. CEA-related cell adhesion molecule 1 (CEACAM1): a potent angiogenic factor and a major effector of vascular endothelial growth factor (VEGF). Mol. Cell 5:311-320. [DOI] [PubMed] [Google Scholar]
- 16.Etienne-Manneville, S., and A. Hall. 2002. Rho GTPases in cell biology. Nature 420:629-635. [DOI] [PubMed] [Google Scholar]
- 17.Field, K. A., J. R. Apgar, E. Hong-Geller, R. P. Siraganian, B. Baird, and D. Holowka. 2000. Mutant RBL mast cells defective in FcɛR1 signaling and lipid raft biosynthesis are reconstituted by activated Rho-family GTPases. Mol. Biol. Cell 11:3661-3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fleming, J. C., G. Berger, J. Guichard, E. M. Cramer, and D. D. Wagner. 1998. The transmembrane domain enhances granular targeting of P-selectin. Eur. J. Cell Biol. 75:331-343. [DOI] [PubMed] [Google Scholar]
- 19.Fukata, M., and K. Kaibuchi. 2001. Rho-family GTPases in cadherin-mediated cell-cell adhesion. Nat. Rev. Mol. Cell Biol. 2:887-897. [DOI] [PubMed] [Google Scholar]
- 20.Gingras, D., F. Gauthier, S. Lamy, R. Desrosiers, and R. Béliveau. 1998. Localisation of RhoA GTPase to endothelial caveolae-enriched membrane domains. Biochem. Biophys. Res. Commun. 247:888-893. [DOI] [PubMed] [Google Scholar]
- 21.Gray-Owen, S. D., C. Dehio, A. Haude, F. Grunert, and T. F. Meyer. 1997. CD66 carcinoembryonic antigens mediate interactions between Opa-expressing Neisseria gonorrhoeae and human polymorphonuclear phagocytes. EMBO J. 16:3435-3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hall, A. 1998. Rho GTPases and the actin cytoskeleton. Science 279:509-514. [DOI] [PubMed] [Google Scholar]
- 23.Hordijk, P. L., J. P. ten Klooster, R. A. van der Kammen, F. Michiels, L. C. Oomen, and J. G. Collard. 1997. Inhibition of invasion of epithelial cells by Tiam1-Rac signaling. Science 278:1464-1466. [DOI] [PubMed] [Google Scholar]
- 24.Hsieh, J.-T., W. Luo, W. Song, Y. Wang, D. I. Kleinerman, N. T. Van, and S. H. Lin. 1995. Tumor suppressive role of an androgen-regulated epithelial cell adhesion molecule (C-CAM) in prostate carcinoma cell revealed by sense and antisense approaches. Cancer Res. 55:190-197. [PubMed] [Google Scholar]
- 25.Huber, M., L. Izzi, P. Grondin, C. Houde, T. Kunath, A. Veillette, and N. Beauchemin. 1999. The carboxyl-terminal region of biliary glycoprotein controls its tyrosine phosphorylation and association with protein-tyrosine phosphatases SHP-1 and SHP-2 in epithelial cells. J. Biol. Chem. 274:335-344. [DOI] [PubMed] [Google Scholar]
- 26.Hunter, I., H. Sawa, M. Edlund, and B. Obrink. 1996. Evidence for regulated dimerization of cell-cell adhesion molecule (C-CAM) in epithelial cells. Biochem. J. 320:847-853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ishizaki, T., M. Maekawa, K. Fujisawa, K. Okawa, A. Iwamatsu, A. Fujita, N. Watanabe, Y. Saito, A. Kakizuka, N. Morii, and S. Narumiya. 1996. The small GTP-binding protein Rho binds to and activates a 160 kDa Ser/Thr protein kinase homologous to myotonic dystrophy kinase. EMBO J. 15:1885-1893. [PMC free article] [PubMed] [Google Scholar]
- 28.Izzi, L., C. Turbide, C. Houde, T. Kunath, and N. Beauchemin. 1999. cis-Determinants in the cytoplasmic domain of CEACAM1 responsible for its tumor inhibitory function. Oncogene 18:5563-5572. [DOI] [PubMed] [Google Scholar]
- 29.Kammerer, R., S. Hahn, B. B. Singer, J. S. Luo, and S. von Kleist. 1998. Biliary glycoprotein (CD66a), a cell adhesion molecule of the immunoglobulin superfamily, on human lymphocytes: structure, expression and involvement in T cell activation. Eur. J. Immunol. 28:3664-3674. [DOI] [PubMed] [Google Scholar]
- 30.Kammerer, R., D. Stober, B. B. Singer, B. Obrink, and J. Reimann. 2001. Carcinoembryonic antigen-related cell adhesion molecule 1 on murine dendritic cells is a potent regulator of T cell stimulation. J. Immunol. 166:6537-6544. [DOI] [PubMed] [Google Scholar]
- 31.Kodama, A., K. Takaishi, K. Nakano, H. Nishioka, and Y. Takai. 1999. Involvement of Cdc42 small G protein in cell-cell adhesion, migration and morphology of MDCK cells. Oncogene 1 8:3996-4006. [DOI] [PubMed] [Google Scholar]
- 32.Kong, M., C. Mounier, V. Dumas, and B. I. Posner. 2003. Epidermal growth factor-induced DNA synthesis. Key role for Src phosphorylation of the docking protein Gab2. J. Biol. Chem. 278:5837-5844. [DOI] [PubMed] [Google Scholar]
- 33.Kozma, R., S. Ahmed, A. Best, and L. Lim. 1995. The Ras-related protein Cdc42Hs and bradykinin promote formation of peripheral actin microspikes and filopodia in Swiss 3T3 fibroblasts. Mol. Cell. Biol. 4:1942-1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kozma, R., S. Sarner, S. Ahmed, and L. Lim. 1997. Rho family GTPases and neuronal growth cone remodelling: relationship between increased complexity induced by Cdc42Hs, Rac1, and acetylcholine and collapse induced by RhoA and lysophosphatidic acid. Mol. Cell. Biol. 17:1201-1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kunath, T., C. Ordonez-Garcia, C. Turbide, and N. Beauchemin. 1995. Inhibition of colonic tumor cell growth by biliary glycoprotein. Oncogene 11:2375-2382. [PubMed] [Google Scholar]
- 36.Kuroda, S., M. Fukata, K. Fujii, T. Nakamura, I. Izawa, and K. Kaibuchi. 1997. Regulation of cell-cell adhesion of MDCK cells by Cdc42 and Rac1 small GTPases. Biochem. Biophys. Res. Commun. 240:430-435. [DOI] [PubMed] [Google Scholar]
- 37.Lamarche, N., N. Tapon, L. Stowers, P. D. Burbelo, P. Aspenstrom, T. Bridges, J. Chant, and A. Hall. 1996. Rac and Cdc42 induce actin polymerization and G1 cell cycle progression independently of p65PAK and the JNK/SAPK MAP kinase pathway. Cell 87:519-529. [DOI] [PubMed] [Google Scholar]
- 38.Leung, T., E. Manser, L. Tan, and L. Lim. 1995. A novel serine/threonine kinase binding the Ras-related RhoA GTPase which translocates the kinase to peripheral membranes. J. Biol. Chem. 270:29051-29054. [DOI] [PubMed] [Google Scholar]
- 39.Leusch, H. G., Z. Drzeniek, Z. Markos-Pusztai, and C. Wagener. 1991. Binding of Escherichia coli and Salmonella strains to members of the carcinoembryonic antigen family: differential binding inhibition by aromatic alpha-glycosides of mannose. Infect. Immun. 59:2051-2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mañes, S., E. Mira, C. Gomez-Mouton, R. A. Lacalle, P. Keller, J. P. Labrador, and C. Martinez-A. 1999. Membrane raft microdomains mediate front-rear polarity in migrating cells. EMBO J. 18:6211-6220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Manser, E., T. Leung, H. Salihuddin, Z. S. Zhao, and L. Lim. 1994. A brain serine/threonine protein kinase activated by Cdc42 and Rac1. Nature 367:40-46. [DOI] [PubMed] [Google Scholar]
- 42.Martin, G. A., G. Bollag, F. McCormick, and A. Abo. 1995. A novel serine kinase activated by rac1/CDC42Hs-dependent autophosphorylation is related to PAK65 and STE20. EMBO J. 14:4385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Matsui, T., M. Amano, T. Yamamoto, K. Chihara, M. Nakafuku, M. Ito, T. Nakano, K. Okawa, A. Iwamatsu, and K. Kaibuchi. 1996. Rho-associated kinase, a novel serine/threonine kinase, as a putative target for small GTP binding protein Rho. EMBO J. 15:2208-2216. [PMC free article] [PubMed] [Google Scholar]
- 44.Minden, A., A. Lin, F. X. Claret, A. Abo, and M. Karin. 1995. Selective activation of the JNK signaling cascade and c-Jun transcriptional activity by the small GTPases Rac and Cdc42Hs. Cell 81:1147-1157. [DOI] [PubMed] [Google Scholar]
- 45.Mitchison, T. J., and L. P. Cramer. 1996. Actin-based cell motility and cell locomotion. Cell 84:371-379. [DOI] [PubMed] [Google Scholar]
- 46.Morales, V. M., A. Christ, S. M. Watt, H. S. Kim, K. W. Johnson, N. Utku, A. M. Texieira, A. Mizoguchi, E. Mizoguchi, G. J. Russell, S. E. Russell, A. K. Bhan, G. J. Freeman, and R. S. Blumberg. 1999. Regulation of human intestinal intraepithelial lymphocyte cytolytic function by biliary glycoprotein (CD66a). J. Immunol. 163:1363-1370. [PubMed] [Google Scholar]
- 47.Nakajima, A., H. Iijima, M. F. Neurath, T. Nagaishi, E. E. Nieuwenhuis, R. Raychowdhury, J. Glickman, D. M. Blau, S. Russell, K. V. Holmes, and R. S. Blumberg. 2002. Activation-induced expression of carcinoembryonic antigen-cell adhesion molecule 1 regulates mouse T lymphocyte function. J. Immunol. 168:1028-1035. [DOI] [PubMed] [Google Scholar]
- 48.Nobes, C. D., and A. Hall. 1995. Rho, Rac, and Cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell 81:53-62. [DOI] [PubMed] [Google Scholar]
- 49.Noren, N. K., C. M. Niessen, B. M. Gumbiner, and K. Burridge. 2001. Cadherin engagement regulates Rho family GTPases. J. Biol. Chem. 276:33305-33308. [DOI] [PubMed] [Google Scholar]
- 50.Öbrink, B. 1997. CEA adhesion molecules: multifunctional proteins with signal-regulatory properties. Curr. Opin. Cell Biol. 9:616-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Oliferenko, S., K. Paiha, T. Harder, V. Gerke, C. Schwärzler, H. Schwarz, H. Beug, U. Günthert, and L. A. Huber. 1999. Analysis of CD44-containing lipid rafts: recruitment of annexin II and stabilization by the actin cytoskeleton. J. Cell Biol. 146:843-854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ozawa, M. 2002. Lateral dimerization of the E-cadherin extracellular domain is necessary but not sufficient for adhesive activity. J. Biol. Chem. 277:19600-19608. [DOI] [PubMed] [Google Scholar]
- 53.Perschl, A., J. Lesley, N. English, R. Hyman, and I. S. Trowbridge. 1995. Transmembrane domain of CD44 is required for its detergent insolubility in fibroblasts. J. Cell Sci. 108:1033-1041. [DOI] [PubMed] [Google Scholar]
- 54.Poy, M. N., Y. Yang, K. Rezaei, M. A. Fernstrom, A. D. Lee, Y. Kido, S. K. Erickson, and S. M. Najjar. 2002. CEACAM1 regulates insulin clearance in liver. Nat. Genet. 30:270-276. [DOI] [PubMed] [Google Scholar]
- 55.Prall, F., P. Nollau, M. Neumaier, H. D. Haubeck, Z. Drzeniek, U. Helmchen, T. Loning, and C. Wagener. 1996. CD66a (BGP), an adhesion molecule of the carcinoembryonic antigen family, is expressed in epithelium, endothelium, and myeloid cells in a wide range of normal human tissues. J. Histochem. Cytochem. 44:35-41. [DOI] [PubMed] [Google Scholar]
- 56.Ridley, A. J., and A. Hall. 1992. Distinct patterns of actin organization regulated by the small GTP-binding proteins Rac and Rho. Cold Spring Harbor Symp. Quant. Biol. 57:661-671. [DOI] [PubMed] [Google Scholar]
- 57.Ridley, A. J., and A. Hall. 1992. The small GTP-binding protein Rho regulates the assembly of focal adhesions and actin stress fibers in response to growth factors. Cell 70:389-399. [DOI] [PubMed] [Google Scholar]
- 58.Rosenberg, M., P. Nédellec, S. Jothy, D. Fleiszer, C. Turbide, and N. Beauchemin. 1993. The expression of mouse biliary glycoprotein, a carcinoembryonic antigen-related gene, is down-regulated in malignant mouse tissues. Cancer Res. 55:4938-4945. [PubMed] [Google Scholar]
- 59.Royal, I., L. Lamorte, K. Kaibuchi, and M. Park. 2000. Activation of cdc42, rac, PAK, and rho-kinase in response to hepatocyte growth factor differentially regulates epithelial cell colony spreading and dissociation. Mol. Biol. Cell 11:1709-1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sadekova, S., N. Lamarche-Vane, X. Li, and N. Beauchemin. 2000. The CEACAM1-L glycoprotein associates with the actin cytoskeleton and localizes to cell-cell contact through activation of Rho-like GTPases. Mol. Biol. Cell 11:65-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sahai, E., and C. J. Marshall. 2002. ROCK and Dia have opposing effects on adherens junctions downstream of Rho. Nat. Cell Biol. 4:408-415. [DOI] [PubMed] [Google Scholar]
- 62.Sander, E. E., J. P. ten Klooster, S. van Delft, R. A. van der Kammen, and J. G. Collard. 1999. Rac downregulates Rho activity: reciprocal balance between both GTPases determines cellular morphology and migratory behavior. J. Cell Biol. 147:1009-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sanders, L. C., F. Matsumura, G. M. Bokoch, and P. de Lanerolle. 1999. Inhibition of myosin light chain kinase by p21-activated kinase. Science 283:2083-2085. [DOI] [PubMed] [Google Scholar]
- 64.Schumann, D., C. J. Chen, B. Kaplan, and J. E. Shively. 2001. Carcinoembryonic antigen cell adhesion molecule 1 directly associates with cytoskeleton proteins actin and tropomyosin. J. Biol. Chem. 276:47421-47433. [DOI] [PubMed] [Google Scholar]
- 65.Singer, B. B., I. Scheffrahn, and B. ÖBrink. 2000. The tumor growth-inhibiting cell adhesion molecule CEACAM1 (C-CAM) is differently expressed in proliferating and quiescent epithelial cells and regulates cell proliferation. Cancer Res. 60:1236-1244. [PubMed] [Google Scholar]
- 66.Small, J. V., T. Stradal, E. Vignal, and K. Rottner. 2002. The lamellipodium: where motility begins. Trends Cell Biol. 12:112-120. [DOI] [PubMed] [Google Scholar]
- 67.Sundberg, U., and B. Öbrink. 2002. CEACAM1 isoforms with different cytoplasmic domains show different localization, organization and adhesive properties in polarized epithelial cells. J. Cell Sci. 115:1273-1284. [DOI] [PubMed] [Google Scholar]
- 68.Takaishi, K., T. Sasaki, H. Kotani, H. Nishioka, and Y. Takai. 1997. Regulation of cell-cell adhesion by rac and rho small G proteins in MDCK cells. J. Cell Biol. 139:1047-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tu, H., and M. Wigler. 1999. Genetic evidence for Pak1 autoinhibition and its release by Cdc42. Mol. Cell. Biol. 19:602-611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Turner, C. E. 2000. Paxillin interactions. J. Cell Sci. 113:4139-4140. [DOI] [PubMed] [Google Scholar]
- 71.Uehata, M., T. Ishizaki, H. Satoh, T. Ono, T. Kawahara, T. Morishita, H. Tamakawa, K. Yamagami, J. Inui, M. Maekawa, and S. Narumiya. 1997. Calcium sensitization of smooth muscle mediated by a Rho-associated protein kinase in hypertension. Nature 389:990-994. [DOI] [PubMed] [Google Scholar]
- 72.Van Aelst, L., and M. Symons. 2002. Role of Rho family GTPases in epithelial morphogenesis. Genes Dev. 16:1032-1054. [DOI] [PubMed] [Google Scholar]
- 73.Virji, M., D. Evans, J. Griffith, D. Hill, L. Serino, A. Hadfield, and S. M. Watt. 2000. Carcinoembryonic antigens are targeted by diverse strains of typable and non-typable Haemophilus influenzae. Mol. Microbiol. 36:784-795. [DOI] [PubMed] [Google Scholar]
- 74.Virji, M., K. Makepeace, D. J. Ferguson, and S. M. Watt. 1996. Carcinoembryonic antigens (CD66) on epithelial cells and neutrophils are receptors for Opa proteins of pathogenic neisseriae. Mol. Microbiol. 22:941-950. [DOI] [PubMed] [Google Scholar]
- 75.Watanabe, N., P. Madaule, T. Reid, T. Ishizaki, G. Watanabe, A. Kakizuka, Y. Saito, K. Nakao, B. M. Jockusch, and S. Narumiya. 1997. p140mDia, a mammalian homolog of Drosophila diaphanous, is a target protein for Rho small GTPase and is a ligand for profilin. EMBO J. 16:3044-3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Watt, S. M., A. M. Teixeira, G. Q. Zhou, R. Doyonnas, Y. Zhang, F. Grunert, R. S. Blumberg, M. Kuroki, K. M. Skubitz, and P. A. Bates. 2001. Homophilic adhesion of human CEACAM1 involves N-terminal domain interactions: structural analysis of the binding site. Blood 98:1469-1479. [DOI] [PubMed] [Google Scholar]
- 77.Wikstrom, K., G. Kjellstrom, and B. Öbrink. 1996. Homophilic intercellular adhesion mediated by C-CAM is due to a domain 1-domain 1 reciprocal binding. Exp. Cell Res. 227:360-366. [DOI] [PubMed] [Google Scholar]
- 78.Yang, N., O. Higuchi, K. Ohashi, K. Nagata, A. Wada, K. Kangawa, E. Nishida, and K. Mizuno. 1998. Cofilin phosphorylation by LIM-kinase 1 and its role in Rac-mediated actin reorganization. Nature 393:809-812. [DOI] [PubMed] [Google Scholar]
- 79.Yap, A. S., and E. M. Novacs. 2003. Direct cadherin-activated cell signaling: a view from the plasma membrane. J. Cell Biol. 160:11-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhao, Z. S., E. Manser, X. Q. Chen, C. Chong, T. Leung, and L. Lim. 1998. A conserved negative regulatory region in alphaPAK: inhibition of PAK kinases reveals their morphological roles downstream of Cdc42 and Rac1. Mol. Cell. Biol. 18:2153-2163. [DOI] [PMC free article] [PubMed] [Google Scholar]