Abstract
Cot, initially identified as an oncogene in a truncated form, is a mitogen-activated protein kinase kinase kinase implicated in cellular activation and proliferation. Here, we show that this truncation of Cot results in a 10-fold increase in its overall kinase activity through two different mechanisms. Truncated Cot protein exhibits a lower turnover rate (half-life, 95 min) than wild-type Cot (half-life, 35 min). The degradation of wild-type and truncated Cot can be specifically inhibited by proteasome inhibitors in situ. The 20S proteasome also degrades wild-type Cot more efficiently than the truncated protein. Furthermore, the amino acid 435 to 457 region within the wild-type Cot COOH-terminal domain confers instability when transferred to the yellow fluorescent protein and targets this fusion protein to degradation via the proteasome. On the other hand, the kinase specific activity of wild-type Cot is 3.8-fold lower than that of truncated Cot, and it appears that the last 43 amino acids of the wild-type Cot COOH-terminal domain are those responsible for this inhibition of kinase activity. In conclusion, these data demonstrate that the oncogenic activity of truncated Cot is the result of its prolonged half-life and its higher kinase specific activity with respect to wild-type Cot.
The Cot/tpl-2 gene encodes a mitogen-activated protein (MAP) kinase kinase kinase that is potentially capable of switching on several MAP kinase cascades, namely those leading to the activation of the MAP kinases ERK1/ERK2, JNK, p38γ, and ERK5 (13, 20, 25, 38, 47, 53). These signal transduction pathways link Cot activity with the up-regulation of several transcription factors, such as AP-1 (7, 13, 25), NFAT (7, 17, 62), NF-κB (8, 31, 38, 63), CREB (22), or E2F (65). As a result, Cot activity has been implicated in cellular activation as well as cell proliferation through the regulation of the cell cycle transitions G0/G1 and G1/S (6, 7, 20, 46, 54, 63, 65).
Cot has also been implicated in cellular transformation, as the genomic locus of the Cot gene is amplified in some human breast cancers (58) and malignant human Hodgkin/Reed-Sternberg cells exhibit abnormally high expression levels of Cot protein (21). The human Cot gene was originally identified in its truncated form, which is present in transformed SHOK cell foci obtained by the transfection of the genomic DNA from a human thyroid carcinoma cell line (42). This truncation unmasks the transforming capacity of this proto-oncogene, a property of the truncated Cot protein (trunc-Cot) in which the last 69 amino acids of the wild-type (wt) COOH-terminal (C-terminal) domain are replaced by an unrelated 18-amino-acid sequence (42). Nevertheless, it has since been shown that overexpression of the proto-oncogenic form is also capable of conferring a transformed phenotype to established cell lines (12, 13).
The rat homologue of the human Cot gene is the tpl-2 gene. Disruption of the last coding exon of tpl-2, due to the insertion of the Moloney leukemia provirus (MLV), leads to the expression of a truncated protein where the final 44 amino acids of the C-terminal domain of the normal cellular protein are replaced by a 10-amino-acid fragment of unrelated sequence. The truncation of the tpl-2 gene by the MLV insertion also unmasks its oncogenic potential (11, 40, 46). The C-terminal domains of both wt Cot and tpl-2 are almost identical. The only difference between the last 77 C-terminal amino acids of the human wt Cot and tpl-2 are the two conservative substitutions M437 for V and K439 for R. However, there is no similarity in the amino acids inserted in either trunc-Cot or trunc-tpl-2. Similarly, integration of the mouse mammary tumor provirus (MMTV) in the last intron of the mouse tpl-2 gene has also been implicated in epithelial cell transformation (23).
The tightly regulated expression of short-lived proteins implicated in specific signal transduction pathways is essential for the correct control of proliferation. Moreover, a deregulation of the turnover of these proteins can trigger transformation (34). Many such proteins are degraded by the proteasome (14, 27, 32, 34, 67), and an important aspect of their degradation is the identification of signals responsible for their efficient targeting to the proteasome machinery (68). Indeed, this kind of highly regulated degradation of specific proteins is often mediated by discrete regions of their primary sequences known as degrons (64). In most instances, a degron can function autonomously when inserted within the sequence of a heterologous protein, triggering to degradation the fusion protein by the same mechanisms that operate in the original substrate (24, 29, 33). The vast majority of proteins degraded by the proteasome are first modified by a set of enzymes that attach multiple copies of ubiquitin (for a review, see references 28 and 59). However, there are proteins, such as p21waf1 (60), ornithine decarboxylase (15, 66), casein (50), myelin basic protein (3, 39), and IκB (36), that can be degraded via the proteasome without prior ubiquitylation.
In this paper, we demonstrate that the C-terminal domain of wt Cot regulates its kinase activity by two different mechanisms. The amino acids 435 to 457 within the C-terminal domain of wt Cot contain a signal that targets this protein to proteasome degradation. Thus, wt Cot is expressed at steady-state levels that are 2.6-fold lower than those of trunc-Cot. On the other hand, wt Cot exhibits 3.8-fold lower specific kinase activity than trunc-Cot, indicating that the region encompassed by the amino acid 425 to 467 region within the C-terminal domain exercises an inhibitory control on its kinase activity. Consequently, transfected trunc-Cot cells exhibit a 10-fold-higher Cot activity per mg of total protein and a greater capacity to produce cell transformation than wt Cot transfected cells. These results provide an explanation of the molecular basis for the enhanced transformation capability of the trunc-Cot protein.
MATERIALS AND METHODS
Plasmids.
pcINEO HA-wt Cot (1 to 467), pcINEO HA-trunc-Cot, pcINEO HA plasmids (65), pEF-BOS wt Cot, pEF-BOS trunc-Cot (6, 7), and −73 pcol-Luc (7) have all been described previously. The GST-Cot390-467 construct containing the C-terminal domain of wt Cot fused to the C terminus of glutathione S-transferase (GST) was constructed by subcloning a PCR fragment of wt Cot into the pGEX4T3-GST plasmid (Amersham-Pharmacia-Biotech). The sequences of the primers used were as follows: F, 5′-GGCCCTGAAttCGCCCAGAG (where the lowercase letters indicate the mismatches to generate the EcoRI site for cloning); and R, 5′-CAAGACGACGcAgcTGGTTGTCCCCG (where the lowercase letters indicate the mismatches to generate the SalI site for cloning). The EcoRI- and SalI-digested PCR product was also inserted into the pEYFP-C1 plasmid (Clontech) to obtain EYFP-Cot390-467, in which the C terminal (390 to 467) of wt Cot is fused to the C terminus of EYFP. The different HA-Cot and EYFP-Cot constructs, with different stop codons (namely HA-Cot457 or EYFP-Cot390-457, HA-Cot446 or EYFP-Cot390-446, HA-Cot435 or EYFP-Cot390-435, and HA-Cot424 or EYFP-Cot390-424) were generated by in vitro-directed mutagenesis using the QuikChange directed mutagenesis kit and the following primers, respectively: forward (F) 5′-GGCTACTTCAATCTTtgaCGGGGACCACCAACGCTT-3′ and reverse (R) 5′-AAGCGTTGGTGGTCCCCGtcaAAAGATTGAAGTAGCC-3′; F 5′-CGCTCTCTC TACATCtAaCTCGGCGCTCTGGCTGGCTAC-3′ and R 5′-GTAGCCAGCCAGAGCGCCGAGtTaGATGTAGAGAGAGCG-3′; F 5′-GGAAGCACCGAGGAATCTtAGATGCTCAAGAGGCAA-3′ and R 5′-TTGCCTCTTGATCATCTaAGATTCCTCGGTGCTTCC-3′; and F 5′-GAGAACATTGCTtAaTCTTCGTGCACAGGAAGCACCG-3′ and R 5′-CGGTGCTTCCTGTGCACGAAGAtTaAGCAATGTTCTC-3′ (where the lowercase letters indicate the mismatches to generate the stop codon). EYFP-Cot445-457 was constructed by subcloning into the pEYFP-C1 plasmid the 5′-phosphorylated oligonucleotides F (5′gatcATCGACCTCGGCGCTCTGGCTGGCTACTTCAATCTTTAG, where the lowercase letters indicate the sequence for ligation to the BglII site) and R (5′tcgaCTAAAGATTGAAGTAGCCAGCCAGAGCGCCGAGGTCGATA, where the lowercase letters indicate the sequence for ligation to the SalI site), which contain the nucleotides corresponding to amino acids 445 to 457 of wt Cot. To obtain the three different constructs EYFP-Cot414-457, EYFP-Cot426-457, and EYFP-Cot435-457, the HA-Cot457 construct was used as a template to generate mutants with a BglII site in the nucleotides corresponding to amino acids 413, 425, and 434. The different mutants were obtained by in vitro mutagenesis using the following primers: to generate the 413 BglII site, primers F (5′AAGAGGCTGCTGAGTAGatctGAGCTGGAACTTCCT) and R (5′ AGGAAGTTCCAGctcaGATCTACTCAGCAGCCTCTT); to generate the 425 BglII site, primers F (5′CTTCCTGAGAACATTGCTagaTCTTCGTGCACAGGAAGC) and R (5′GCTTCCTGTGCACGAAGAtctAGCAATGTTCTCAGGAA); and to generate the BglII site at amino acid 434, primers F (5′ ACAGGAAGCACCGAGagATCTGAGATG CTCAAGAGG) and R (5′CCTCTTGAGCATCTCAGAtcTCTCGGTGCTTCCTGT) (where the lowercase letters indicate the mismatches to generate the BglII sites). These three different HA-Cot457 constructs with a new BglII site in the nucleotide corresponding to amino acid 413, 425, or 434 were subjected to digestion with BglII and SalI, and the three cDNA fragments were inserted in the pEYFP-C1 polylinker plasmid previously digested with BglII and SalI. All the constructs were verified by automatic DNA sequencing. The pCMV-His-ubiquitin construct was a generous gift of Dirk Bohmann and has been described previously (61). HA-PKB-DD and PKB-CaaX were generous gifts from Balduino Burgering (19). The −73 pcol-Luc plasmid has been described previously (reference 7 and references therein).
Cell transfection, labeling, and immunoprecipitation.
HEK293 cells (American Type Culture Collection) were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum at 37°C in a humidified atmosphere of 10% CO2-90% air. The cells were grown in 100-mm dishes and were transfected at 50% confluency. The transfection assays, unless otherwise indicated, were performed with 10 μg of DNA per 100-mm dish using the DEAE-dextran-chloroquine method. Inhibition of proteases was achieved by incubating the cells, 14.5 h after transfection, with different protease inhibitors for 7.5 h. The final concentrations of the protease inhibitors were as follows: 20 μM E64 (Boehringer Mannheim), 1 mM pefabloc AEBSF (Boehringer Mannheim), 20 μM phosporamidon (Calbiochem), 10 μM Z-AAF-CMK (Biomol), 20 μM MG132 or 10 μM lactacystin (Calbiochem), and 20 μM LLMet (Sigma-Aldrich). The phosphatidylinositol 3-kinase (PI3K) inhibitor LY 294002 (50 μM; Calbiochem) was added to the cells 2 h after transfection, and 8 h before lysis the cell medium was changed and a mixture containing 50 μM LY 294002, 100 μg of cycloheximide/ml with or without 10 μM lactacystin was added to the cells. For metabolic labeling, cells were cultured in methionine-, cysteine-, and glutamine-free DMEM for 20 min and then exposed to a 20-min pulse with 250 μCi of [35S]methionine and [35S]cysteine labeling mixture (Amersham-Pharmacia-Biotech)/ml. Subsequently, the cells were cultured in DMEM supplemented with 10% fetal bovine serum, 0.8 mM methionine, and 0.3 mM cysteine for the chase periods indicated in the figures. The proteasome inhibitors MG132 (20 μM) or lactacystin (10 μM) were added 2 h before the pulse and maintained during the chase periods as described previously (35). The radiolabeled cells were then washed twice with 10 ml of ice-cold phosphate-buffered saline (PBS) and lysed in 200 μl of ice-cold lysis buffer (50 mM HEPES [pH 7.4], 10% glycerol, 150 mM NaCl, 1% NP-40, 50 mM β-glycerophosphate, 1 mM sodium orthovanadate, 3 nM okadaic acid, 10 μg of pepstatin A/μl, 0.2 mM phenylmethylsulfonyl fluoride [PMSF] plus 1 Complete mini inhibitors tablet [Roche]/10 ml). The cell lysates were centrifuged at 24,100 × g for 10 min at 4°C, and the supernatants, containing the same amount of protein (400 to 900 μg), were subjected to immunoprecipitation with the antihemagglutinin (anti-HA) antibody covalently bound to protein A-agarose for 2 h at 4°C. The protein A-agarose was then washed once with ice-cold HNTG buffer (20 mM HEPES [pH 7.5], 10% glycerol [vol/vol], 0.1% Triton X-100) plus 200 mM NaCl and twice with the HNTG buffer plus 150 mM NaCl. The immunoprecipitated proteins were denatured with sodium dodecyl sulfate (SDS) loading buffer and separated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) on a 10% gel. After drying the gel, the radiolabeled proteins were visualized and quantified with an Instant Imager (Perkin-Elmer). To determine the amounts of immunoprecipitated proteins, immunoblotting was performed with anti-Cot antibodies using 50 μg of the distinct initial supernatants and the first supernatant after removal of the immunoprecipitated Cot, as described below. About 90% of the trunc-Cot and wt Cot was immunoprecipitated under the conditions described above.
Western blotting.
The transfected cells contained in one 100-mm dish were washed with ice-cold PBS and divided into two halves. One half of the cells was lysed in 200 μl of ice-cold lysis buffer and centrifuged at 24,100 × g for 10 min at 4°C. A total of 35 μg of protein from the supernatants of these cells was resolved by SDS-10% PAGE (soluble proteins). The pellets from the above extraction were directly solubilized in SDS loading buffer, and the volume corresponding to the same amount of soluble protein was also loaded onto an SDS-10% PAGE (insoluble proteins). The other half of the cells was directly resuspended in SDS loading buffer, and the volume corresponding to the same amount of soluble proteins was also subjected to electrophoresis in SDS-10% PAGE gels (total proteins). The separated proteins were transferred to polyvinylidene difluoride (PVDF) membranes and probed with polyclonal anti-Cot (6), monoclonal anti-Cot (Calbiochem), or monoclonal anti-ΗΑ (Babco) antibodies. As a control of protein loading in the SDS-PAGE gels, the PVDF membranes were also probed with a polyclonal anti-protein disulfide isomerase (PDI) antiserum as described previously (44). Antibodies against ubiquitin were purchased from Dako, and those against enhanced yellow fluorescent protein (EYFP) were from Clontech. Western blotting for phospho-Erk (P-Erk) and total Erk-2 was performed with specific anti-P-Erk (Santa Cruz) and anti-Erk-2 (Zymed) antibody, respectively. The anti-GST antibody was purchased from Sigma. The blots were developed using chemiluminiscence (ECL; Amersham-Pharmacia-Biotech), and the signals were quantified by optical densitometry using NIH Image 1.6 unless otherwise indicated.
RT-PCR.
Total RNA was isolated from cells transfected with pcINEO HA-wt Cot HA-trunc-Cot and pcINEO HA (20 h after transfection) with TRIzol (Gibco-BRL) according to the manufacturers' instructions. Two micrograms of total RNA was subjected to reverse transcription-PCR (RT-PCR) analysis as described previously (65), using β-actin primers as internal control.
AP-1-driven transcription assay.
Different concentrations of pEF-BOS wt Cot, pEF-BOS trunc-Cot (0.01, 0.03, 0.1, or 0.3 μg), normalized for the amount of DNA with empty pEF-BOS, or 0.15 μg of the different pcINEO Cot constructs were cotransfected together with 0.15 μg of −73 pcol-Luc into HEK293 cells. Twenty hours after transfection Luc activity was measured as described previously (7).
Cot kinase assay.
Cot kinase assays were performed as described previously (1, 53), but with some minor modifications. Cells transfected with pcINEO HA-Cot constructs and pcINEO HA for 20 h were lysed in 500 μl of ice-cold lysis buffer (50 mM Tris-HCl [pH 7.5], 1 mM EDTA, 1 mM EGTA, 1% [vol/vol] Triton X-100, 1 mM sodium orthovanadate [pH 10], 10 mM sodium glycerophosphate [pH 7.4], 5 mM sodium pyrophosphate, 0.27 mM sucrose, 0.1% [vol/vol] β-glycerophosphate, 0.1% β-mercaptoethanol, 1 mM microcystin-LR, 1 mM benzamidine, plus 1 Complete mini inhibitors tablet/10 ml [Roche]) and centrifuged at 24,100 × g for 10 min at 4°C. Ten micrograms of the different supernatants was incubated for 2 h with the anti-HA antibody covalently bound to protein A-agarose. The proteins immunoprecipitated were washed twice with ice-cold lysis buffer plus 0.5 M of NaCl, once with ice-cold lysis buffer, and finally with ice-cold 50 mM Tris HCl (pH 7.5), 0.1 mM EGTA, 0.03% Brij, 0.1% (vol/vol) β-mercaptoethanol. The agarose beads were then incubated in a final volume of 20 μl containing 12.5 mM Tris (pH 7.5), 0.0075% Brij, 0.025 mM EGTA, 0.0158 μg of MKK1/μl, 0.033 μg of MAPK/μl, 0.0025 mM microcystin-LR, 0.067 mM sodium orthovanadate, 0.025% (vol/vol) β-mercaptoethanol. The kinase reaction was then started by adding a mixture of ATP and MgCl2 to final concentrations of 330 μM and 6.5 mM, respectively, and was left to continue for 15 min at 30°C with agitation. During this period of time the kinase reaction was linear. Two microliters of this kinase reaction was then used for the second kinase assay, and the rest of the volume (18 μl) was subjected to Western blot analysis as described above. The second kinase reaction was carried out in a final volume of 50 μl: 2 μl of the first kinase reaction mixture, 37.5 mM Tris HCl (pH 7.5), 0.75 mM sodium orthovanadate (pH 10), 0.75 mM EGTA, 0.25 mg of myelin basic protein/ml, and 0.75% (vol/vol) β-mercaptoethanol. The reaction was started by adding a mixture of ATP and ΜgCl2 to a final concentration of 100 μM and 10 mΜ, respectively, and contained 1.66 μCi of [γ-32P]ATP. The reaction was incubated for 10 min at 30°C with shaking and stopped by spotting 40 μl of this mixture onto P81 Whatmann disk filters. The filters were dropped into a 75 mM phosphoric acid solution, washed extensively in the same solution, and dried after a final acetone wash. The 32P incorporated into myelin basic protein was quantified by Cerenkov counting. Under these reaction conditions, the incorporation of 32P into myelin basic protein was linear within the time span of the assay. We defined 1 U of Cot kinase activity as 1 nmol of phosphate incorporated into myelin basic protein in 1 min. The specific activity is expressed as milliunits of Cot activity divided by the amount of Cot protein.
Focus-forming transformation assays.
NIH 3T3 cells grown in 100-mm dishes were transfected with 10 μg of wt Cot, trunc-Cot, or the empty vector using the Lipofectin reagent according to the manufacturers' instructions (Life Technologies, Inc.). The day after transfection, the cells were kept in medium supplemented with 10% filtered calf serum. After 2 weeks, the cells were washed twice in PBS, pH 7.4, fixed for 10 min in 75% methanol-25% acetic acid, stained for 1 min with 0.4% crystal violet in 20% ethanol, and washed extensively with water until the excess stain was removed. All transfections were done in duplicate.
In vitro degradation assays.
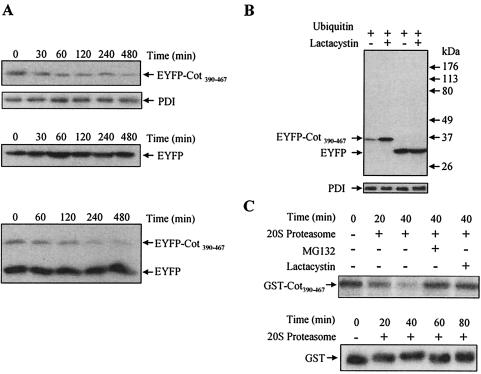
Transcription-translation assays were performed with the pcINEO HA-wt Cot and pcINEO HA-trunc-Cot constructs using the TNT coupled reticulocyte lysate system (Promega) and [35S]methionine and [35S]cysteine, according to the manufacturer's instructions. Degradation reactions were performed in a final volume of 20 μl containing 20 mM HEPES (pH 7.4), 5 mM EGTA, 5 mM EDTA, 1 μl of corresponding labeled kinase, and 0.2 to 0.4 μg of purified rat liver 20S proteasome (5). Control reaction mixtures contained either no proteasome or proteasome plus 20 μM MG132 or 10 μM lactacystin. Each reaction was terminated by adding SDS loading buffer, and the samples were resolved on SDS-10% PAGE gels and vacuum dried. The radiolabeled proteins were visualized and quantified by Instant Imager analysis (Perkin-Elmer). Degradation assays were also performed with 1.5 μg of recombinant GST-Cot390-466 or with 1.5 μg of recombinant GST. After the in vitro degradation reactions, GST-Cot390-466 and GST were visualized in SDS-10% PAGE gels stained either with Coomassie blue or immunoblotted with anti-Cot antibody or anti-GST-antibodies as described above.
RESULTS
trunc-Cot exhibits higher levels of steady-state expression than wt Cot.
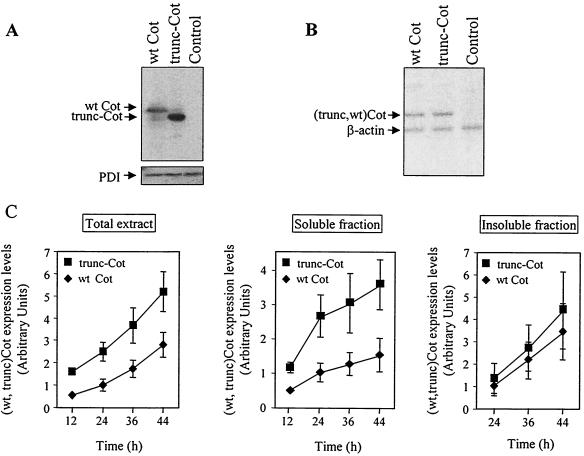
In order to determine the steady-state expression of wt Cot and trunc-Cot, we analyzed the amounts of these proteins in total extracts of HEK293 cells 24 h after transfection with 10 μg of pcINEO HA-wt Cot, pcINEO HA-trunc-Cot, or pcINEO HA. In Western blots, it was apparent that these cells expressed higher steady-state levels of trunc-Cot than wt Cot (Fig. 1A). However, RT-PCR analysis of the same transfected cells indicated that the mRNA levels of both wt Cot and trunc-Cot were very similar (Fig. 1B). The higher levels of protein expression of transfected trunc-Cot were observed with two different anti-Cot antibodies as well as with an anti-HA antibody, and the quantification of 10 independent experiments indicated that the expression of trunc-Cot was 2.6-fold ± 0.4 fold (mean ± standard deviation) higher than the steady-state levels of wt Cot.
FIG. 1.
Expression of wt Cot and trunc-Cot in HEK293 transfected cells. (A and B) Cells were transfected with pcINEO HA-wt Cot (10 μg), pcINEO HA-trunc-Cot (10 μg), or pcINEO HA (10 μg), and 24 h after transfection wt Cot and trunc-Cot protein and mRNA levels were analyzed. wt Cot and trunc-Cot protein levels were determined by Western blot analysis using the anti-Cot monoclonal antibody (Calbiochem) (A). The same PVDF membrane was probed with an anti-PDI antibody as a control of protein loading. The data shown correspond to 30 PCR cycles, and the amount of product amplified (62 to 373 nucleotides [nt] of HA-wt-Cot, 62 to 373 nt of HA-trunc-Cot, and 1,292 to 1,479 nt of β-actin) was proportional to the abundance of the starting material (B). (C) Quantification of the expression of wt Cot and trunc-Cot in total extracts at different times after transfection (12, 24, 36, and 44 h), as well as in soluble and insoluble 1% NP-40 fractions of HEK293 cells transfected with pcINEO HA-wt Cot (10 μg) or pcINEO HA-trunc-Cot (10 μg). wt Cot and trunc-Cot were detected by Western blot analysis with anti-Cot antibodies either generated in the laboratory (6) or commercially available (Calbiochem). As a control of protein loading, membranes were also probed with an anti-PDI antibody. The pixel intensities of the different wt Cot and trunc-Cot bands were quantified with GelDoc 200 (Bio-Rad) and corrected to the pixel intensity of PDI. The graphs shows the mean ± standard deviation of the intensities of the different wt Cot and trunc Cot bands from three different experiments, assuming a value of 1 for the intensity obtained for wt Cot 24 h after transfection. ♦, wt Cot; ▪, trunc-Cot.
The steady-state expression levels of wt Cot and trunc-Cot proteins were also examined at different times after transfection (12, 24, 36, and 44 h). Moreover, the Western blot analyses of the wt Cot and trunc-Cot proteins were performed with total cell extracts as well as with the 1% NP-40-soluble and -insoluble fractions from transfected cells. As expected, in total cell extracts the expression levels of both wt Cot and trunc-Cot proteins increased with time. However, at all times after transfection, more trunc-Cot accumulated in the cells than wt Cot (Fig. 1C). The higher expression levels of trunc-Cot were also observed after transfection of equal amounts of wt Cot and trunc-Cot plasmids in other cell lines (Jurkat, Cos-7, and HeLa) and appeared to be independent of the transfection method (DEAE-dextran, calcium-phosphate, Lipofectamine, or electroporation) or the type of vector (pcINEO or pEF-BOS). Nor was this effect influenced by the type of fusion protein transfected, as the difference in levels was also observed without tag or GST fused to wt Cot and to trunc-Cot (data not shown). The higher expression levels of trunc-Cot compared to wt Cot in the 1% NP-40-soluble fraction could be detected at different times after transfection (Fig. 1C). Furthermore, 24 h after transfection both wt Cot and trunc-Cot proteins appeared in the 1% NP-40-insoluble fraction (Fig. 1C), and the wt Cot and trunc-Cot from this fraction were only partially solubilized after treatment with 6 M guanidinium chloride (data not shown). Cytoplasmic aggregates of trunc-Cot and wt Cot could be detected by immunocytochemistry in HEK293 and Cos-7 cells 48 h after transfection with HA-trunc-Cot or HA-wt Cot, probably due to this insoluble protein fraction (data not shown). As a consequence, we performed all the subsequent experiments 14 to 20 h after cell transfection, when both wt Cot and trunc-Cot are completely soluble in 1% NP-40.
trunc-Cot is more slowly degraded than wt Cot.
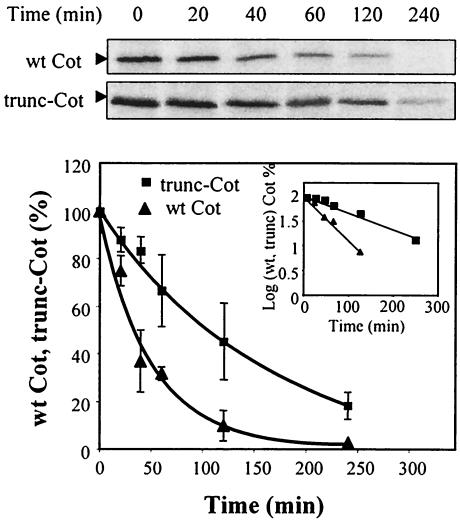
The higher protein expression levels of trunc-Cot compared to wt Cot could indicate that the truncated protein has a lower degradation rate. Thus, we carried out pulse-chase experiments in HEK293 cells 14 h after their transfection with pcINEO HA-wt Cot or pcINEO HA-trunc-Cot, and the t1/2 values for both proteins were determined. The radioactivity incorporated into wt Cot and trunc-Cot after a 20-min pulse was measured, as was its decay during the different periods of chase (0 to 240 min). As a result, it became clear that both wt Cot and trunc-Cot are short-lived proteins (Fig. 2), although the estimated t1/2 values calculated for wt Cot and trunc-Cot were consistently and significantly different. The t1/2 for wt Cot was 35 min while that for trunc-Cot was 95 min, indicating that wt Cot is more rapidly degraded than trunc-Cot.
FIG. 2.
Degradation of wt Cot and trunc-Cot in HEK293 transfected cells. Immunoprecipitation of 35S-labeled wt Cot and trunc-Cot from HEK293 cells transfected with 10 μg of pcINEO HA-wt Cot or 10 μg of pcINEO HA-trunc-Cot after a 20-min pulse-label and a chase for the times indicated (20, 40, 60, 120, and 240 min). The proteins immunoprecipitated were resolved by SDS-PAGE and visualized by autoradiography. The radioactivity incorporated was quantified using an Instant Imager. The figure shows one experiment, and the graph shows the mean ± standard deviation of the radioactivity incorporated in four experiments. The radioactivity incorporated into wt Cot or trunc-Cot after the 20-min pulse was considered 100%. The degradation of both proteins correlated with one-phase exponential equations fit to each data set, with R2 = 0.975 for wt Cot and R2 = 0.9736 for trunc-Cot. The graph also shows a representation of the decay as a log plot. ▴, wt Cot; ▪, trunc-Cot.
trunc-Cot and wt Cot are degraded by the proteasome in situ and in vitro.
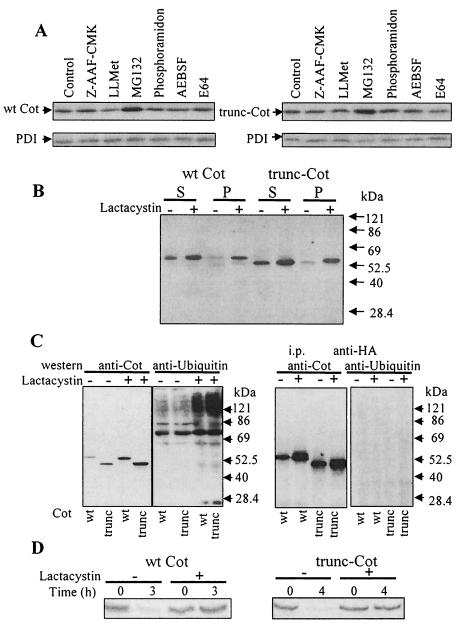
Having observed a difference in the degradation of both Cot proteins, we studied the pathways involved in their degradation. After transfection (14.5 h later) of HEK293 cells with pcINEO HA-wt Cot or pcINEO HA-trunc-Cot, the cells were treated with different protease inhibitors for 7.5 h. The protease inhibitors examined included cystein and a papain proteases inhibitor, such as E64; serine protease inhibitor, such as pefabloc AEBSF; metalloprotease inhibitors, such as phosporamidon; an inhibitor of tripeptidyl peptidase, such as Z-AAF-CMK; proteasome inhibitors, such as MG132 or lactacystin; and a calpain inhibitor, such as LLMet, as indicated in Materials and Methods. Total cell extracts were subjected to Western blot analysis, and the steady-state levels of both wt Cot and trunc-Cot proteins in the presence and absence of the inhibitors were determined. A significant increase in the amount of both wt Cot and trunc-Cot protein with respect to untreated control cells was only observed after treatment with the proteasome inhibitor 20 μM MG132 (Fig. 3A). Incubation of wt Cot- and trunc-Cot-transfected cells with 10 μM lactacystin (or 20 μM MG132 [data not shown]) resulted in the accumulation of both proteins not only in the 1% NP-40-soluble fraction but also in the insoluble fraction (Fig. 3B).
FIG. 3.
Levels of wt Cot and trunc-Cot expression after treatment of HEK293 transfected cells with different protease inhibitors. (A) Total extracts of HEK293 cells 14.5 h after transfection with 10 μg of pcINEO HA-wt Cot or 10 μg of pcINEO HA-trunc-Cot were further incubated for 7.5 h in the presence of different protease inhibitors (10 μM Z-AAF-CMK, a tripeptidyl peptidase inhibitor; 20 μM LLMet, a calpain II inhibitor; 20 μM MG132, a proteasome inhibitor; 20 μM phosporamidon, a metalloprotease inhibitor; 1 mM prefabloc AEBSF, a serine protease inhibitor; or 20 μM E64, a cysteine protease inhibitor) as indicated in Materials and Methods. Total extracts were analyzed by Western blotting using anti-Cot (Calbiochem) and anti-PDI antibodies. The figure shows representative results of one of the three experiments performed. (B) Cells transfected as indicated for panel A and incubated in the presence or absence of 10 μM lactacystin were lysed in lysis buffer, and both the 1% NP-40-soluble and -insoluble fractions were analyzed by Western blotting with an anti-Cot antibody (Calbiochem). (C) Total extracts of cells 14.5 h after transfection with 5 μg of pcDNA3.1 HA-wt Cot or 5 μg of pcDNA3.1 HA-trunc-Cot together with 5 μg of pCMV-His-ubiquitin, which were further incubated for 7.5 h in the presence or absence of 10 μM lactacystin, were analyzed by Western blotting with anti-Cot antibody (Calbiochem) or with antiubiquitin antibody. Both wt Cot and trunc-Cot were immunoprecipitated from the same transfected cells and analyzed by Western blotting with antiubiquitin antibody and anti-Cot antibody. The figure shows representative results of one of the three experiments performed. (D) Accumulation of newly synthesized wt Cot and trunc-Cot after lactacystin (10 μM) incubation. 35S-labeled wt Cot and trunc-Cot were immunoprecipitated from HEK293 cells transfected with 10 μg of pcINEO HA-wt Cot or 10 μg of pcINEO HA-trunc-Cot after a 20-min pulse and 3-h chase (wt Cot) or 4-h chase (trunc-Cot) in the presence or absence of 10 μM lactacystin. The proteins were resolved by SDS-PAGE and visualized by autoradiography. The figure shows representative results of one of the three experiments performed.
The accumulation of both proteins after proteasome inhibitor treatment did not result in the appearance of high-molecular-weight species of trunc-Cot or wt Cot, which would suggest the existence of polyubiquitylated forms of Cot. However, this was further examined by enhancing the possible formation of trunc-Cot or wt Cot polyubiquitylated forms. Thus, the cells were cotransfected with wt Cot or trunc-Cot plasmids together with pCMV-His-ubiquitin, and 14.5 h after transfection the cells were incubated for another 7.5 h in the presence or absence of proteasome inhibitors. Under these conditions, we were still unable to detect higher-molecular-weight forms of wt Cot or trunc-Cot in total cell extracts, even in the presence of 20 μM MG132 (data not shown) or 10 μM lactacystin (Fig. 3C). However, when an antiubiquitin antibody was used instead of the anti-Cot antibody, high-molecular-weight polyubiquitylated proteins did appear to accumulate in the same extracts in the presence of 10 μM lactacystin (Fig. 3C). In an attempt to enrich the fraction with wt Cot and trunc-Cot, both proteins were immunoprecipitated from 1 mg of protein extracted from the same cells transfected. However, in Western blotting with the antiubiquitin or anti-Cot antibody even in the presence of 10 μM lactacystin, no polyubiquitylated forms of wt Cot or trunc-Cot could be detected (Fig. 3C). On the other hand, neither wt Cot nor trunc-Cot could be detected by Western blot analysis with anti-Cot antibody in the enriched His-ubiquitin fraction obtained by affinity chromatography on Ni2+ columns (data not shown). Nevertheless, it does appear that the newly synthesized wt Cot and trunc-Cot are also catabolized via the proteasome, since in pulse-chase experiments of HEK293 cells transfected 12 h earlier with pcINEO HA-wt Cot or pcINEO HA-trunc-Cot and incubated or not with 10 μM lactacystin, the proteasome inhibitor clearly prevented the degradation of both newly synthesized trunc-Cot and wt Cot (Fig. 3D).
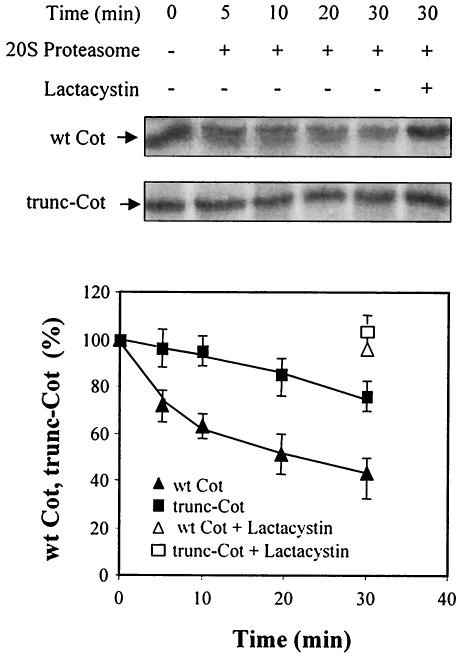
To further address the role of the proteasome in wt Cot and trunc-Cot stability, degradation experiments were conducted with in vitro-transcribed and -translated wt Cot and trunc-Cot together with 20S proteasome purified from rat liver (4, 5). In this assay, about 55% of wt Cot was degraded 30 min after incubating with the 20S proteasome, while only 20% of trunc-Cot was degraded in this time (Fig. 4A). No degradation was observed in the absence of the proteasome, and degradation was completely prevented by the addition of 10 μM lactacystin (Fig. 4A). All these data indicate that both wt Cot and trunc-Cot are targets for proteasome degradation, both in situ as well as in vitro, and that wt Cot is more sensitive to degradation than trunc-Cot.
FIG. 4.
Degradation of wt Cot and trunc-Cot by the 20S proteasome. Transcribed and translated wt Cot and trunc-Cot were incubated for the indicated times with 20S proteasome in the presence or absence of 10 μM lactacystin. The proteins were then resolved by SDS-PAGE and visualized by autoradiography, and the incorporated radioactivity was quantified by using an Instant Imager. The figure shows one representative experiment, and the graph shows the mean ± standard deviation of two experiments performed in duplicate.
The amino acid 435 to 457 region within the C terminal of wt Cot targets the protein to proteasome degradation.
The shorter half-life of wt Cot compared to trunc-Cot may be due to the main structural difference between the two proteins, which resides in the C terminal (12, 42). Thus, the C-terminal domain of wt Cot may contain a strong signal that targets the protein for degradation by the proteasome. To test this hypothesis, we transferred the wt Cot C terminal (amino acids 390 to 467) to the C terminus of EYFP, a fusion construct we named pEYFP-Cot390-467. Fluorescence-activated cell sorter analysis was performed 20 h after transfection of HEK293 cells with pEYFP or with pEYFP-Cot390-467. Both pools of transfected cells had a similar percentage of fluorescent cells (about 50%), but the mean fluorescence was systematically lower in cells expressing EYFP-Cot390-467 than in EYFP-transfected cells (data not shown).
To study the degradation of both proteins, HEK293 cells were transfected with either pEYFP, pEYFP-Cot390-467, or both plasmids together. After transfection (14.5 h later), cells were incubated with cycloheximide (100 μg/ml) for different time periods and the protein accumulated in the total cell extracts was examined using an anti-EYFP antibody in Western blotting. The expression of EYFP-Cot390-467 was lower than that of EYFP even before the addition of cycloheximide (Fig. 5A) and, in agreement with the t1/2 of >24 h previously reported for EYFP (16), the amount of EYFP did not change significantly over time in the presence of cycloheximide (Fig. 5A). However, in sharp contrast to the stability exhibited by EYFP, the amount of EYFP-Cot390-467 declined steadily after the addition of cycloheximide, indicating that the fusion of the C-terminal domain of wt Cot to EYFP conferred instability to the protein (Fig. 5A).
FIG. 5.
The C terminal of wt Cot triggers its degradation by the proteasome. (A) HEK293 cells 14.5 h after transfection with 10 μg of pEYFP-Cot390-467 or 10 μg of pEYFP or cotransfected with 5 μg of pEYFP-Cot390-467 together with 5 μg of pEYFP were further incubated with 100 μg of cycloheximide/ml for different times (0, 30, 60, 120, 240, and 480 min). The total extracts were Western blotted and probed with the anti-EYFP antibody. The figure shows one of two experiments performed in duplicate. (B) Total extracts of cells 14.5 h after transfection with 5 μg of pEYFP or 5 μg of pEYFP-Cot390-467 together with 5 μg of pCMV-His-ubiquitin and further incubated for 7.5 h in the presence or absence of 10 μM lactacystin. The extracts were Western blotted and probed with anti-EYFP. Similar results were obtained in two different experiments performed in duplicate. (C) Recombinant GST-Cot390-467 was subjected to in vitro degradation by the 20S proteasome for 20 and 40 min. GST-Cot390-467 was also incubated with 20S proteasome in the presence of 20 μM MG132 or 10 μM lactacystin. The different samples were resolved by SDS-PAGE and Western blotted to be probed with an anti-C-terminal Cot antibody. Similar results were obtained in three experiments performed in duplicate. Recombinant GST was subjected to in vitro degradation for 20, 40, 60, and 80 min by the 20S proteasome. The different samples were resolved by SDS-PAGE followed by Western blotting, which was probed with anti-GST antibody. Similar results were obtained in two experiments performed in duplicate.
Further experiments were performed on HEK293 cells 14.5 h after cotransfection of pEYFP or pEYFP-Cot390-467 together with pCMV-His-ubiquitin; the cells were incubated for a further 7.5 h in the presence or absence of 10 μM lactacystin. As a consequence of inhibiting the proteasome, EYFP-Cot390-467 but not EYFP accumulated in these cells (Fig. 5B). Thus, we examined whether the C terminal of wt Cot could also target a protein for degradation by the 20S proteasome in vitro. To this end, we fused the C terminal of wt Cot to the C terminus of GST, and the recombinant GST-Cot390-467 protein was analyzed in the in vitro degradation assay. The GST-Cot390-467 fusion protein was susceptible to degradation by the 20S proteasome and, as expected, 20 μM MG132 or 10 μM lactacystin prevented this degradation (Fig. 5C). In control experiments carried out with recombinant GST, the 20S proteasome was unable to degrade GST (Fig. 5C).
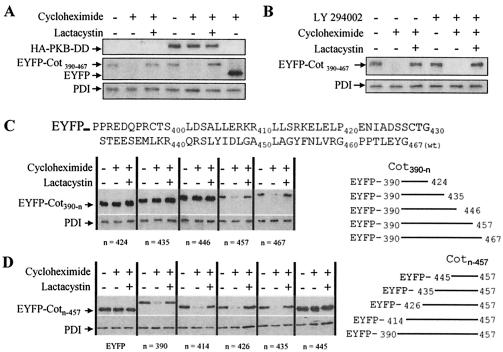
It has recently been reported that protein kinase B (PKB) regulates Cot-induced NF-κB activation by phosphorylating two residues in the C terminal of wt Cot (31). We therefore investigated whether PKB activity could modulate the sensitivity of EYFP-Cot390-467 to proteasome degradation in intact cells by cotransfecting HEK293 cells with pEYFP-Cot390-467 and a plasmid that expresses a constitutively active form of PKB, HA-PKB-DD (19). When the protein expression levels of EYFP-Cot390-467 were determined after treatment of the transfected cells with cycloheximide (100 μg/ml), in the presence or absence of lactacystin (10 μM), it was evident that the constitutively active form of PKB did not affect the steady-state expression of EYFP-Cot390-467 in intact cells or its susceptibility to proteasome degradation (Fig. 6A). To discard the possibility that the endogenous PI3K-PKB signaling pathway was already activated in the transfected cells, 2 h after transfection with EYFP-Cot390-467 the cells were incubated with an inhibitor of PI3K, LY 294002 (50 μM), and then for 8 h before the lysis with cycloheximide (100 μM) and LY 294002 (50 μM) in the presence or absence of lactacystin (10 μM). The presence of the PI3K inhibitor did not alter the steady-state expression levels of EYFP-Cot390-467 or its susceptibility to degradation via the proteasome (Fig. 6B). Similar results were obtained when EYFP-Cot390-467 was cotransfected with a construct that expresses a constitutive dominant-negative form of PKB (PKB-CaaX; data not shown).
FIG. 6.
The amino acid 435 to 457 region within the C terminal of wt Cot confers susceptibility to proteasome degradation and is independent of PKB activity. (A) HEK293 cells were cotransfected with 5 μg of EYFP-Cot390-467 together with 15 μg of HA-PKB-DD or empty vector, and 14.5 h after transfection the cells were further incubated with 100 μg of cycloheximide/ml in the presence or absence of 10 μM lactacystin as described in Materials and Methods. The figure is representative of the three experiments performed. (B) Two hours after the transfection of HEK293 cells with 10 μg of EYFP-Cot390-467, the cells were incubated with 50 μM LY 294002 and the medium was then supplemented with 100 μg of cycloheximide/ml and 50 μM LY 294002, with and without 10 μM lactacystin, for a further 8 h. The figure shows representative results of the three experiments performed. (C) HEK293 cells were transfected with 10 μg of pEYFP-Cot390-467, pEYFP-Cot390-457, pEYFP-Cot390-446, pEYFP-Cot390-435, or pEYFP-Cot390-424, and after 14.5 h cells were treated as described in the legend for Fig. 5B. Total extracts were Western blotted and probed with anti-EYFP and anti-PDI antibodies; the figure shows representative results of one of the three experiments. (D) HEK293 cells transfected for 14.5 h with pEYFP-Cot414-457, pEYFP-Cot426-457, pEYFP-Cot435-457, and pEYFP-Cot445-450 were treated as explained above for panel C. The figure shows one of the two experiments performed in duplicate.
To further define the region within the wt Cot C-terminal domain implicated in targeting the EYFP fusion protein for proteasome degradation, different pEYFP-Cot390-n constructs were analyzed, namely, pEYFP-Cot390-467, pEYFP-Cot390-457, pEYFP-Cot390-446, pEYFP-Cot390-435, and pEYFP-Cot390-424. When transfected into HEK293 cells and incubated 14.5 h after transfection for 8 h with 100 μg of cycloheximide/ml in the presence or absence of 10 μM lactacystin, EYFP-Cot390-457 presented similar levels of expression as EYFP-Cot390-467. Both proteins were hardly detectable in Western blotting after exposure for 8 h to cycloheximide (100 μg/ml); nevertheless, the presence of lactacystin (10 μM) inhibited their degradation (Fig. 6C). However, when a further 11 amino acids were deleted from the C terminal of EYFP-Cot390-457, the resulting protein, EYFP-Cot390-446, exhibited significantly higher steady-state levels of protein expression than those observed for EYFP-Cot390-457 or EYFP-Cot390-467. Moreover, treatment with cycloheximide (8 h) did not diminish its level of expression (Fig. 6C). Furthermore, EYFP-Cot390-435 and EYFP-Cot390-424 were expressed at steady-state protein levels similar to those of EYFP-Cot390-446 (Fig. 6C) and EYFP (data not shown).
Having defined what appears to be the C-terminal limit of the sequence in wt Cot that targets it for proteasome degradation, we set out to define the more-N terminal by using a similar strategy. The generated constructs pEYFP-Cot414-457, pEYFP-Cot426-457, pEYFP-Cot435-457, and pEYFP-Cot445-457 were transfected into HEK293 cells, and the transfected cells were treated as described above. The EYFP-Cot445-457 fusion protein was not susceptible to degradation via proteasome, indicating that although this 12-amino-acid region of the wt Cot C-terminal domain (amino acids 445 to 457) does contain part of the signal necessary for degradation (Fig. 6C), it is not sufficient to target the fusion protein to proteasome degradation (Fig. 6D). However, the fusion of the amino acid 435 to 457 region within the wt Cot C-terminal domain to EYFP did efficiently target the protein for degradation via proteasome (Fig. 6D).
trunc-Cot exhibits higher specific kinase activity than wt Cot.
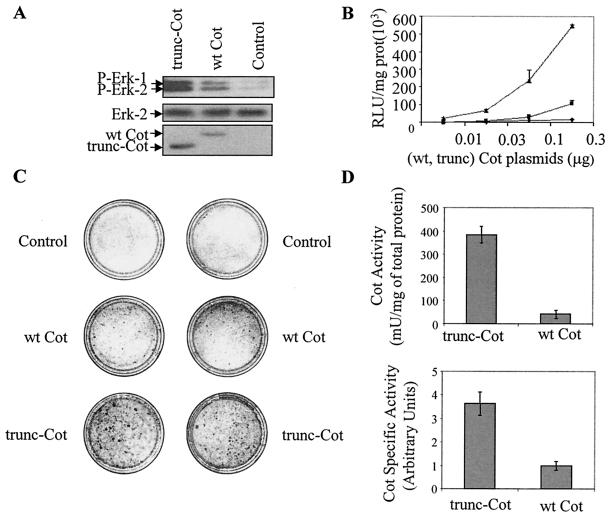
We also wanted to know whether the increased levels of protein expression of trunc-Cot were correlated with an increased activation of the different signal transduction pathways regulated by Cot. Cot up-regulates the classical MAP kinase pathway (20, 53). Therefore, we analyzed the phosphorylation state of endogenous Erk-1 and Erk-2 in total extracts of HEK293 cells 20 h after transfection with pcINEO HA-wt Cot and pcINEO HA-trunc-Cot. Both wt Cot and trunc-Cot activated endogenous Erk-1 and Erk-2, although a higher degree of activation was observed upon transfection of trunc-Cot (Fig. 7A).
FIG. 7.
trunc-Cot exhibits a higher transformation capacity and kinase activity than wt Cot. (A) Western blots of P-Erk in total extracts of HEK293 cells 20 h after transfecting with pcINEO HA-wt Cot (10 μg), pcINEO HA-trunc-Cot (10 μg), or pcINEO HA (10 μg). As a control of protein loaded, total Erk-2 levels were tested. The expression of wt Cot and trunc-Cot was also determined with the anti-Cot antibody (Calbiochem). Similar results were obtained in three different experiments. (B) Different amounts of pEF-BOS wt Cot, pEF-BOS trunc-Cot, or pEF-BOS (0.01, 0.03, 0.1, or 0.3 μg) together with 0.15 μg of −73 pcol-Luc were cotransfected in HEK293 cells, and Luc activity was measured 20 h after transfection. The graph shows the means ± standard deviations of three different experiments performed in duplicate. (C) NIH 3T3 cells were transfected with 10 μg of pcINEO HA-wt Cot, pcINEO HA-trunc-Cot, or pcINEO HA. Cells were cultured for 2 weeks with 10% filtered calf serum, fixed, and then stained as described in Materials and Methods. The figure shows a representative experiment of the three performed. (D) Cot activity per milligram of protein extract and Cot specific activity of wt Cot and trunc-Cot immunoprecipitated from HEK293 cells transfected as described for panel A. Cot activity indicates the milliunits of Cot kinase activity corresponding to 1 mg of transfected extract, with 1 U being 1 nmol of phosphate incorporated into myelin basic protein in 1 min. The specific activity indicates the milliunits of Cot kinase activity divided by the pixel intensity obtained from the scanned Western blots of the immunoprecipitated wt Cot or trunc-Cot, with a value of 1 being attributed to the specific activity of wt Cot divided by the pixel intensity of immunoprecipitated wt Cot. The graphs show the means ± standard deviations of three different experiments performed in triplicate.
It has previously been shown that Cot activity up-regulates the transcription factor AP-1 (7, 13). A reporter construct has been generated that contains the −73-bp collagenase promoter linked to the luciferase reporter gene (−73 pcol-Luc); this promoter region contains only the AP-1 response element and has been extensively used to study AP-1 activation (references 7 and 13 and references therein). Different amounts of pEF-BOS wt Cot, pEF-BOS trunc-Cot, and pEF-BOS were cotransfected together with the −73 pcol-Luc, and Luc activity was measured 20 h after transfection. Cells transfected with trunc-Cot up-regulated AP-1 transcription factor activity to a greater extent than cells expressing wt Cot (Fig. 7B). Interestingly, about 10-fold more wt Cot plasmid than trunc-Cot plasmid was required to reach similar activation levels of the AP-1 responsive element (Fig. 7B).
It has been proposed that the ability of Cot to induce cellular transformation (12, 13) depends on its ability to activate the AP-1 transcription factor (13). We therefore assessed whether the augmented activation of the AP-1 responsive element by trunc-Cot was correlated with a greater degree of cellular transformation. The transfection of pcINEO HA-wt Cot into NIH 3T3 cells induced the appearance of transformed foci after 2 weeks in culture as previously reported (Fig. 7C) (12, 13). However, the number of transformed foci was clearly higher in NIH 3T3 cells transfected with pcINEO HA-trunc-Cot (Fig. 7C). We then decided to measure the activity of Cot as MAP kinase kinase kinase 1 (MKKK1) in extracts prepared from cells transfected with wt Cot and trunc-Cot. From equal quantities (10 μg) of the 1% NP-40-soluble fractions of cells transfected with pcINEO HA-Cot wt, pcINEO HA-trunc-Cot (20 h after transfection), wt Cot, and trunc-Cot were immunoprecipitated and Cot activity was determined. The Cot activity (in milliunits per milligram of total protein extract) from trunc-Cot-transfected cells was 10-fold higher (±1) than that derived from wt Cot-transfected cells (Fig. 7D), These results are qualitatively not unexpected since, as mentioned above, trunc-Cot has higher protein expression levels than wt Cot. However, quantification of the amount of Cot protein that was immunoprecipitated showed an approximately 2.6-fold difference in the protein content in immunoprecipitates from trunc-Cot- and wt Cot-transfected extracts. This quantification allowed us to estimate the specific activity (in milliunits per amount of Cot protein) of both trunc-Cot and wt Cot. As a result, it appeared that trunc-Cot exhibited 3.8-fold (±0.4) higher kinase specific activity than wt Cot (Fig. 7D). A similar difference in the specific activity was also observed between trunc-Cot and wt Cot in autophosphorylation assays (data not shown).
Identification of the region within the C-terminal domain of Cot responsible for the inhibition of wt Cot activity.
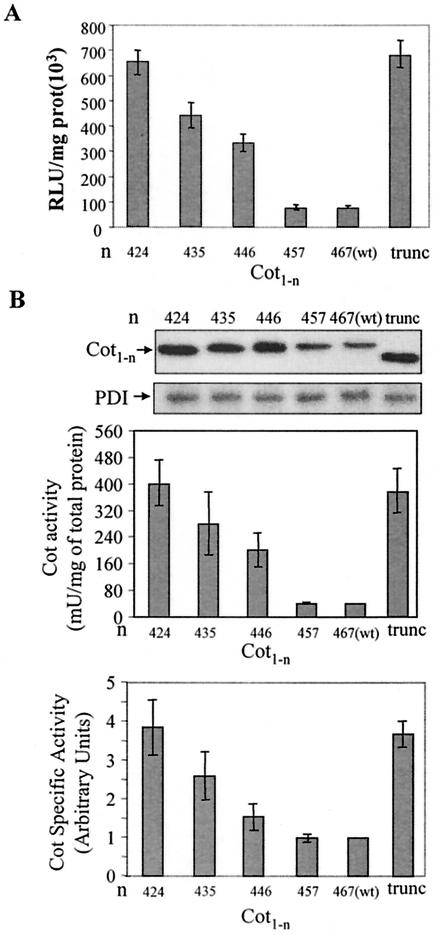
Finally, we set out to determine the region within the C-terminal domain of wt Cot that inhibits Cot specific activity, as well as to discard the possibility that the 18 amino acids inserted in trunc-Cot were responsible for its higher activity. For this purpose, we generated different wt Cot constructs with increasing portions of the C-terminal domain deleted, namely: pcINEO HA-Cot424, pcINEO HA-Cot435, pcINEO HA-Cot446, and pcINEO HA-Cot457. Each of these constructs, as well as pcINEO HA-wt Cot(467) and pcINEO HA-trunc-Cot, were cotransfected together with −73 pcol-Luc in HEK293 cells, and Luc activity was measured 20 h after transfection. While Cot424 activated the AP-1 response element to the same extent as trunc-Cot (Fig. 8A), the activation produced by the Cot435, Cot446, and Cot457 proteins decreased in a stepwise fashion, with Cot457 activating the reporter to the same extent as wt Cot(467). We next decided to determine the specific activity of the different Cot deletion mutants. The different Cot constructs were transfected in HEK293 cells, and Western immunoblot analysis of the steady-state expression levels as well as the Cot activity of the different Cot proteins were determined. In agreement with the data obtained above (Fig. 6C), trunc-Cot, Cot424, Cot435, and Cot446 exhibited expression levels approximately 2.6-fold higher than those of Cot457 and wt Cot(467) (Fig. 8B). The specific activity of the Cot457 protein was similar to that of wt Cot(467), and further deletions within the wt Cot C-terminal region produced a gradual increase in the specific activity of Cot mutants, with Cot424 demonstrating a similar specific activity as trunc-Cot (Fig. 8B). It is noteworthy that Cot446, with a transformation activity between that of wt Cot and trunc-Cot (data not shown), exhibited only 1.5-fold-higher Cot activity per mg of total protein extract than wt Cot; however its 2.6-fold-higher steady-state protein expression compared to wt Cot provided it about 4.6-fold-higher specific activity compared to wt Cot(467).
FIG. 8.
Identification of the region within the C-terminal domain of wt Cot that inhibits Cot specific activity. (A) HEK293 cells were cotransfected with 0.15 μg of pcINEO HA-wt Cot(467), pcINEO HA-Cot457, pcINEO HA-Cot446, pcINEO HA-Cot435, pcINEO HA-Cot424, or pcINEO HA-trunc-Cot together with 0.15 μg of −73 pcol-Luc, and Luc activity was measured 20 h after transfection. The graph shows the means ± standard deviations of two different experiments performed in duplicate. (B) HEK293 cells were transfected with 10 μg of pcINEO HA-wt Cot(467), pcINEO HA-Cot457, pcINEO HA-Cot446, pcINEO HA-Cot435, or pcINEO HA-Cot424. Western blotting of the total extracts of HEK293 cells 20 h after transfection with the different pcINEO Cot constructs was performed as described in the legend for Fig. 1A. The figure shows one representative result of the three experiments performed. As a control of protein loaded, total PDI levels were also assessed. Cot activity and Cot specific activity of the different Cot constructs [wt Cot(467), Cot 457, Cot446, Cot435, and Cot424] were determined as described in the legend for Fig. 7D. The graphs also show the means of three experiments ± standard deviations.
DISCUSSION
Cot/tpl-2 has been implicated in cellular activation and cellular transformation. Thus, Cot/tpl-2 (25, 53), together with c-Raf (18, 30, 37) and c-Mos (43, 51, 57), are three proto-oncogenes that code for MAP kinase kinase kinases that up-regulate the Erk-1/Erk-2 signal transduction pathway. However, different modifications convert these proto-oncogenes into oncogenes. The protein products of both the v-mil oncogene and the transforming variants of Raf differ from their proto-oncogenes in their N-terminal regions (10, 26, 48, 49). In contrast, the c-mos proto-oncogene and the v-mos oncogene differ as a consequence of a proviral insertion in the 5′ noncoding region that does not affect the sequence of the protein product (although some viral strains do also have point mutations within their coding sequence) (9, 69). The rearrangement of the 3′ region of the human Cot gene, as well as of its murine homologue tpl-2, unmasks their transformation activities, both oncogenes encoding C-terminal-truncated and -modified proteins (2, 11, 12, 42, 46). In this paper we demonstrate that the truncation of the C-terminal domain of wt Cot produces an increase in the total kinase activity of Cot through two different mechanisms: by an increase in the t1/2 of the protein and by augmenting the specific activity of the kinase. These two independent mechanisms potentiate one another to produce a 10-fold increase in the kinase activity of Cot. As a consequence, we observed a higher activation state of different MAP kinase transduction pathways in trunc-Cot-transfected cells, such as the Erk-1/Erk-2 and the Jnk-AP-1 pathways. Abnormal activation of these MAP kinase pathways has previously been associated with cellular transformation (41, 55), and indeed we have also shown that a higher number of transformed foci were generated in NIH 3T3 cells transfected with trunc-Cot than with wt Cot. In agreement with these data, it has been proposed that the ability of wt Cot to induce cellular transformation (12, 13) depends on its capacity to activate the AP-1 transcription factor (13).
The structural difference between wt Cot and trunc-Cot resides in the 69 amino acids of the C-terminal domain that in trunc-Cot are replaced by 18 amino acids of unrelated sequence. Here we show that the deletion of the last 43 C-terminal amino acids of wt Cot renders a protein with a similar activity to that of trunc-Cot. Thus, the domain that represses its kinase activity resides in amino acids 425 to 467 of wt Cot (Fig. 9), excluding the possibility that the 18-amino-acid insertion at the C terminal of trunc-Cot enhances its kinase specific activity. In this context, we also must bear in mind that the C terminals of wt tpl-2 and wt Cot are homologous, whereas the 10-amino-acid insert in trunc-tpl-2, which also behaves as an oncogene, does not share any homology with the 18 amino acids inserted into trunc-Cot (11, 46). Moreover, although the difference in the kinase specific activity between tpl-2 and trunc-tpl-2 is unknown, it has been shown that the C terminal of tpl-2 is capable of inhibiting the kinase activity of trunc-tpl-2 in vitro (11). These data, together with the fact that the Cot mutants with increasing C-terminal deletions exhibited increased specific activities, indicate that the C-terminal domain of wt Cot is an autoinhibitory domain. Therefore, by specifically up-regulating certain extracellular signals, it should be possible to abolish this C-terminal repression of kinase activity, perhaps by inducing posttranslational modifications in the Cot protein. In this context, it should be taken into account that the phosphorylation of the C-terminal domain of wt Cot by PKB (31) appeared to have no effect on the MKKK1 activity of wt Cot (M. Caivano, C. Rodriguez, P. Cohen, and S. Alemany, personal communication) or on AP-1 activation by wt Cot (31).
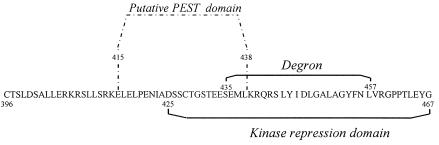
FIG. 9.
Representation of the different regions within C-terminal wt Cot. The analysis of the putative PEST sequences was performed with the pestfind program located at http://www.at.embnet.org/embnet/tools/bio/PESTfind/.
The C terminal of wt Cot also plays an important role in targeting the protein for degradation, as the amino acids 435 to 457 within the wt Cot C-terminal domain appear to be a region that controls the stability of wt Cot (Fig. 9). In this context, it is noteworthy that the region between amino acids 435 and 446, as well as the region between amino acids 445 and 457, are essential regions, but not sufficient, to target the EYFP C-terminal wt Cot fusion protein for degradation via the proteasome (Fig. 6C and D). This explains the similar levels of expression exhibited by Cot446 and trunc-Cot (Fig. 8A). The recognition-degradation signal contained in the region of amino acids 435 to 457 from wt Cot is capable of conferring instability to other unrelated proteins, an essential quality of the recognition-degradation sequences denominated degron, and we propose that this proteasome recognition sequence accounts for the lower t1/2 and the lower steady-state expression levels of wt Cot compared to trunc-Cot. To our knowledge, this region within wt Cot does not share similarities with other previously described degrons. Moreover, our data suggest a direct recognition of the Cot degron by the 20S proteasome. In this context, and considering that the C terminals of both wt Cot and wt tpl-2 are almost identical, it seems likely that the C terminal of tpl-2 also confers instability to the protein. Analysis of putative PEST domains (52) within the C terminal of wt Cot revealed that the region between amino acids 415 and 438 is a putative PEST sequence (Fig. 9). However, the degradation experiments performed with the different EYFP-Cot constructs revealed that this sequence does not play any role in the proteasome recognition-degradation of the C terminal of wt Cot.
It is accepted that short-lived proteins are mainly degraded via the proteasome (34, 67), a hypothesis supported by the results presented here for wt Cot and trunc-Cot. However, our data also suggest that both wt Cot and trunc-Cot are catabolized via the proteasome in an ubiquitylation-independent manner. In this context, it should be stressed that although the vast majority of proteins need to be polyubiquitylated prior to proteasome degradation, other proteins that do not need to be polyubiquitylated for proteasome degradation are starting to be identified (3, 15, 36, 39, 50, 60, 66). However, the possibility that an isopeptidase activity could hydrolyze the polyubiquitin chain cannot be excluded.
Interestingly, not only wt Cot but also trunc-Cot is degraded via the proteasome, indicating that wt Cot could contain more than one recognition site for proteasome degradation and that at least one of these is maintained in the trunc-Cot protein sequence. In this context, the region regulating stability has been identified in the N-terminal stability-regulating region of the MAP kinase kinase kinase c-Mos (45, 56). However, considering that the wt Cot sequence and the wt tpl-2 sequence do not contain the amino acids described as essential for the destabilization of c-mos at the specific positions (45, 56), it is unlikely that wt Cot or wt tpl-2 contain this stability-regulating region.
The insertion of MLV or MMTV provirus into the tpl-2 gene and the truncation of the Cot gene produce the deletion of the 3′ untranslated region of the tpl-2 or Cot gene that harbors three copies of the RNA-destabilizing sequence AUUUA. This results in higher expression levels of the corresponding mRNAs (23, 46). Thus, it has been proposed that this increase in the mRNA levels of the truncated Cot or tpl-2 gene may lead to higher levels of protein expression and therefore to cell transformation (23, 46, 63). However, we have shown here that trunc-Cot exhibits greater cellular transformation capacity and kinase activity than wt Cot and that transfection of both plasmids renders similar levels of mRNA expression. Thus, we conclude that, as a consequence of the Cot gene truncation, regulation is altered at three different levels: stabilization of the mRNA (23, 46), an increase in the Cot specific activity, and stabilization of the protein. Moreover, as shown here, the increase in the kinase specific activity and the stabilization of the truncated protein induce a 10-fold increase in Cot kinase activity and, consequently, a higher transformation capacity.
Acknowledgments
We thank Dirk Bohmann for providing the pCMV-His-ubiquitin construct, M. F. Balduino Burgering for the HA-PKB-DD and PKB-CaaX constructs, and Blanca Duarte and Nora Perez for their technical help.
This work was supported by CICYT grant SAF 2002-00566 to J.G.C. and BMC2002-00437, AICR, and Fundación La Caixa grants to S.A. M.L.G. was the recipient of a fellowship from the Fundación Marcelino Botin, and P.L. was the recipient of a fellowship from Fundación La Caixa.
REFERENCES
- 1.Alessi, D. R., P. Cohen, A. Ashworth, S. Cowley, S. J. Leever, and C. J. Marshall. 1995. Assay and expression of mitogen-activated protein kinase, Map kinase kinase, and RAF. Methods Enzymol. 255:279-290. [DOI] [PubMed] [Google Scholar]
- 2.Aoki, M., T. Akiyama, J. Miyoshi, and K. Toyoshima. 1991. Identification and characterization of protein products of the cot oncogene with serine kinase activity. Oncogene 6:1515-1519. [PubMed] [Google Scholar]
- 3.Arribas, J., P. Arizti, and J. G. Castano. 1994. Antibodies against the C2 COOH-terminal region discriminate the active and latent forms of the multicatalytic proteinase complex. J. Biol. Chem. 269:12858-12864. [PubMed] [Google Scholar]
- 4.Arribas, J., and J. G. Castano. 1993. A comparative study of the chymotrypsin-like activity of the rat liver multicatalytic proteinase and the ClpP from Escherichia coli. J. Biol. Chem. 268:21165-21171. [PubMed] [Google Scholar]
- 5.Arribas, J., and J. G. Castano. 1990. Kinetic studies of the differential effect of detergents on the peptidase activities of the multicatalytic proteinase from rat liver. J. Biol. Chem. 265:13969-13973. [PubMed] [Google Scholar]
- 6.Ballester, A., R. Tobena, C. Lisbona, V. Calvo, and S. Alemany. 1997. Cot kinase regulation of IL-2 production in Jurkat T cells. J. Immunol. 159:1613-1618. [PubMed] [Google Scholar]
- 7.Ballester, A., A. Velasco, R. Tobena, and S. Alemany. 1998. Cot kinase activates tumor necrosis factor-alpha gene expression in a cyclosporin A-resistant manner. J. Biol. Chem. 273:14099-14106. [DOI] [PubMed] [Google Scholar]
- 8.Belich, M. P., A. Salmeron, L. H. Johnston, and S. C. Ley. 1999. TPL-2 kinase regulates the proteolysis of the NF-κB-inhibitory protein NF-κB1 p105. Nature 397:363-368. [DOI] [PubMed] [Google Scholar]
- 9.Blair, D. G., M. K. Oskarsson, A. Seth, K. J. Dunn, M. Dean, M. Zweig, M. A. Tainsky, and G. F. Vande Woude. 1986. Analysis of the transforming potential of the human homolog of mos. Cell 46:785-794. [DOI] [PubMed] [Google Scholar]
- 10.Bonner, T. I., H. Oppermann, P. Seeburg, S. B. Kerby, M. A. Gunnell, A. C. Young, and U. R. Rapp. 1986. The complete coding sequence of the human raf oncogene and the corresponding structure of the c-raf-1 gene. Nucleic Acids Res. 14:1009-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ceci, J. D., C. P. Patriotis, C. Tsatsanis, A. M. Makris, R. Kovatch, D. A. Swing, N. A. Jenkins, P. N. Tsichlis, and N. G. Copeland. 1997. Tpl-2 is an oncogenic kinase that is activated by carboxy-terminal truncation. Genes Dev. 11:688-700. [DOI] [PubMed] [Google Scholar]
- 12.Chan, A. M., M. Chedid, E. S. McGovern, N. C. Popescu, T. Miki, and S. A. Aaronson. 1993. Expression cDNA cloning of a serine kinase transforming gene. Oncogene 8:1329-1333. [PubMed] [Google Scholar]
- 13.Chiariello, M., M. J. Marinissen, and J. S. Gutkind. 2000. Multiple mitogen-activated protein kinase signaling pathways connect the cot oncoprotein to the c-jun promoter and to cellular transformation. Mol. Cell. Biol. 20:1747-1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ciechanover, A., A. Orian, and A. L. Schwartz. 2000. Ubiquitin-mediated proteolysis: biological regulation via destruction. Bioessays 22:442-451. [DOI] [PubMed] [Google Scholar]
- 15.Coffino, P. 2001. Regulation of cellular polyamines by antizyme. Nat. Rev. Mol. Cell Biol. 2:188-194. [DOI] [PubMed] [Google Scholar]
- 16.Corish, P., and C. Tyler-Smith. 1999. Attenuation of green fluorescent protein half-life in mammalian cells. Protein Eng. 12:1035-1040. [DOI] [PubMed] [Google Scholar]
- 17.de Gregorio, R., M. A. Iniguez, M. Fresno, and S. Alemany. 2001. Cot kinase induces cyclooxygenase-2 expression in T cells through activation of the nuclear factor of activated T cells. J. Biol. Chem. 276:27003-27009. [DOI] [PubMed] [Google Scholar]
- 18.Dent, P., W. Haser, T. A. Haystead, L. A. Vincent, T. M. Roberts, and T. W. Sturgill. 1992. Activation of mitogen-activated protein kinase kinase by v-Raf in NIH 3T3 cells and in vitro. Science 257:1404-1407. [DOI] [PubMed] [Google Scholar]
- 19.Dufner, A., M. Andjelkovic, B. M. Burgering, B. A. Hemmings, and G. Thomas. 1999. Protein kinase B localization and activation differentially affect S6 kinase 1 activity and eukaryotic translation initiation factor 4E-binding protein 1 phosphorylation. Mol. Cell. Biol. 19:4525-4534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dumitru, C. D., J. D. Ceci, C. Tsatsanis, D. Kontoyiannis, K. Stamatakis, J. H. Lin, C. Patriotis, N. A. Jenkins, N. G. Copeland, G. Kollias, and P. N. Tsichlis. 2000. TNF-alpha induction by LPS is regulated posttranscriptionally via a Tpl2/ERK-dependent pathway. Cell 103:1071-1083. [DOI] [PubMed] [Google Scholar]
- 21.Eliopoulos, A. G., C. Davies, S. S. Blake, P. Murray, S. Najafipour, P. N. Tsichlis, and L. S. Young. 2002. The oncogenic protein kinase Tpl-2/Cot contributes to Epstein-Barr virus-encoded latent infection membrane protein 1-induced NF-κB signaling downstream of TRAF2. J. Virol. 76:4567-4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eliopoulos, A. G., C. D. Dumitru, C. C. Wang, J. Cho, and P. N. Tsichlis. 2002. Induction of COX-2 by LPS in macrophages is regulated by Tpl2-dependent CREB activation signals. EMBO J. 21:4831-4840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Erny, K. M., J. Peli, J. F. Lambert, V. Muller, and H. Diggelmann. 1996. Involvement of the Tpl-2/cot oncogene in MMTV tumorigenesis. Oncogene 13:2015-2020. [PubMed] [Google Scholar]
- 24.Glotzer, M., A. W. Murray, and M. W. Kirschner. 1991. Cyclin is degraded by the ubiquitin pathway. Nature 349:132-138. [DOI] [PubMed] [Google Scholar]
- 25.Hagemann, D., J. Troppmair, and U. R. Rapp. 1999. Cot proto-oncoprotein activates the dual specificity kinases MEK-1 and SEK-1 and induces differentiation of PC12 cells. Oncogene 18:1391-1400. [DOI] [PubMed] [Google Scholar]
- 26.Heidecker, G., M. Huleihel, J. L. Cleveland, W. Kolch, T. W. Beck, P. Lloyd, T. Pawson, and U. R. Rapp. 1990. Mutational activation of c-raf-1 and definition of the minimal transforming sequence. Mol. Cell. Biol. 10:2503-2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Henchoz, S., Y. Chi, B. Catarin, I. Herskowitz, R. J. Deshaies, and M. Peter. 1997. Phosphorylation- and ubiquitin-dependent degradation of the cyclin-dependent kinase inhibitor Far1p in budding yeast. Genes Dev. 11:3046-3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hershko, A., and A. Ciechanover. 1998. The ubiquitin system. Annu. Rev. Biochem. 67:425-479. [DOI] [PubMed] [Google Scholar]
- 29.Hochstrasser, M., and A. Varshavsky. 1990. In vivo degradation of a transcriptional regulator: the yeast alpha 2 repressor. Cell 61:697-708. [DOI] [PubMed] [Google Scholar]
- 30.Howe, L. R., S. J. Leevers, N. Gomez, S. Nakielny, P. Cohen, and C. J. Marshall. 1992. Activation of the MAP kinase pathway by the protein kinase raf. Cell 71:335-342. [DOI] [PubMed] [Google Scholar]
- 31.Kane, L. P., M. N. Mollenauer, Z. Xu, C. W. Turck, and A. Weiss. 2002. Akt-dependent phosphorylation specifically regulates Cot induction of NF-κB-dependent transcription. Mol. Cell. Biol. 22:5962-5974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.King, R. W., R. J. Deshaies, J. M. Peters, and M. W. Kirschner. 1996. How proteolysis drives the cell cycle. Science 274:1652-1659. [DOI] [PubMed] [Google Scholar]
- 33.King, R. W., M. Glotzer, and M. W. Kirschner. 1996. Mutagenic analysis of the destruction signal of mitotic cyclins and structural characterization of ubiquitinated intermediates. Mol. Biol. Cell 7:1343-1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kornitzer, D., and A. Ciechanover. 2000. Modes of regulation of ubiquitin-mediated protein degradation. J. Cell Physiol. 182:1-11. [DOI] [PubMed] [Google Scholar]
- 35.Krappmann, D., F. G. Wulczyn, and C. Scheidereit. 1996. Different mechanisms control signal-induced degradation and basal turnover of the NF-κB inhibitor IκBα in vivo. EMBO J. 15:6716-6726. [PMC free article] [PubMed] [Google Scholar]
- 36.Kroll, M., M. Conconi, M. J. Desterro, A. Marin, D. Thomas, B. Friguet, R. T. Hay, J. L. Virelizier, F. Arenzana-Seisdedos, and M. S. Rodriguez. 1997. The carboxy-terminus of IκBα determines susceptibility to degradation by the catalytic core of the proteasome. Oncogene 15:1841-1850. [DOI] [PubMed] [Google Scholar]
- 37.Kyriakis, J. M., H. App, X. F. Zhang, P. Banerjee, D. L. Brautigan, U. R. Rapp, and J. Avruch. 1992. Raf-1 activates MAP kinase-kinase. Nature 358:417-421. [DOI] [PubMed] [Google Scholar]
- 38.Lin, X., E. T. Cunningham, Jr., Y. Mu, R. Geleziunas, and W. C. Greene. 1999. The proto-oncogene Cot kinase participates in CD3/CD28 induction of NF-κB acting through the NF-κB-inducing kinase and IκB kinases. Immunity 10:271-280. [DOI] [PubMed] [Google Scholar]
- 39.Lucas, J., D. Lobo, E. Terry, E. L. Hogan, and N. L. Banik. 1992. Susceptibility of myelin proteins to a neutral endoproteinase: the degradation of myelin basic protein (MBP) and P2 protein by purified bovine brain multicatalytic proteinase complex (MPC). Neurochem. Res. 17:1261-1266. [DOI] [PubMed] [Google Scholar]
- 40.Makris, A., C. Patriotis, S. E. Bear, and P. N. Tsichlis. 1993. Genomic organization and expression of Tpl-2 in normal cells and Moloney murine leukemia virus-induced rat T-cell lymphomas: activation by provirus insertion. J. Virol. 67:4283-4289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mansour, S. J., W. T. Matten, A. S. Hermann, J. M. Candia, S. Rong, K. Fukasawa, G. F. Vande Woude, and N. G. Ahn. 1994. Transformation of mammalian cells by constitutively active MAP kinase kinase. Science 265:966-970. [DOI] [PubMed] [Google Scholar]
- 42.Miyoshi, J., T. Higashi, H. Mukai, T. Ohuchi, and T. Kakunaga. 1991. Structure and transforming potential of the human cot oncogene encoding a putative protein kinase. Mol. Cell. Biol. 11:4088-4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nebreda, A. R., C. Hill, N. Gomez, P. Cohen, and T. Hunt. 1993. The protein kinase mos activates MAP kinase kinase in vitro and stimulates the MAP kinase pathway in mammalian somatic cells in vivo. FEBS Lett. 333:183-187. [DOI] [PubMed] [Google Scholar]
- 44.Nieto, A., E. Mira, and J. G. Castano. 1990. Transcriptional regulation of rat liver protein disulphide-isomerase gene by insulin and in diabetes. Biochem. J. 267:317-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nishizawa, M., N. Furuno, K. Okazaki, H. Tanaka, Y. Ogawa, and N. Sagata. 1993. Degradation of Mos by the N-terminal proline (Pro2)-dependent ubiquitin pathway on fertilization of Xenopus eggs: possible significance of natural selection for Pro2 in Mos. EMBO J. 12:4021-4027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Patriotis, C., A. Makris, S. E. Bear, and P. N. Tsichlis. 1993. Tumor progression locus 2 (Tpl-2) encodes a protein kinase involved in the progression of rodent T-cell lymphomas and in T-cell activation. Proc. Natl. Acad. Sci. USA 90:2251-2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Patriotis, C., A. Makris, J. Chernoff, and P. N. Tsichlis. 1994. Tpl-2 acts in concert with Ras and Raf-1 to activate mitogen-activated protein kinase. Proc. Natl. Acad. Sci. USA 91:9755-9759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Patschinsky, T., and K. Bister. 1988. Structural analysis of normal and transforming mil(raf) proteins: effect of 5′-truncation on phosphorylation in vivo or in vitro. Oncogene 3:357-364. [PubMed] [Google Scholar]
- 49.Patschinsky, T., H. W. Jansen, H. Blocker, R. Frank, and K. Bister. 1986. Structure and transforming function of transduced mutant alleles of the chicken c-myc gene. J. Virol. 59:341-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pereira, M. E., T. Nguyen, B. J. Wagner, J. W. Margolis, B. Yu, and S. Wilk. 1992. 3,4-Dichloroisocoumarin-induced activation of the degradation of beta-casein by the bovine pituitary multicatalytic proteinase complex. J. Biol. Chem. 267:7949-7955. [PubMed] [Google Scholar]
- 51.Posada, J., N. Yew, N. G. Ahn, G. F. Vande Woude, and J. A. Cooper. 1993. Mos stimulates MAP kinase in Xenopus oocytes and activates a MAP kinase kinase in vitro. Mol. Cell. Biol. 13:2546-2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rechsteiner, M., and S. W. Rogers. 1996. PEST sequences and regulation by proteolysis. Trends Biochem. Sci. 21:267-271. [PubMed] [Google Scholar]
- 53.Salmeron, A., T. B. Ahmad, G. W. Carlile, D. Pappin, R. P. Narsimhan, and S. C. Ley. 1996. Activation of MEK-1 and SEK-1 by Tpl-2 proto-oncoprotein, a novel MAP kinase kinase kinase. EMBO J. 15:817-826. [PMC free article] [PubMed] [Google Scholar]
- 54.Sanchez-Gongora, E., C. Lisbona, R. de Gregorio, A. Ballester, V. Calvo, L. Perez-Jurado, and S. Alemany. 2000. COT kinase proto-oncogene expression in T cells: implication of the JNK/SAPK signal transduction pathway in COT promoter activation. J. Biol. Chem. 275:31379-31386. [DOI] [PubMed] [Google Scholar]
- 55.Schutte, J., J. Viallet, M. Nau, S. Segal, J. Fedorko, and J. Minna. 1989. jun-B inhibits and c-fos stimulates the transforming and trans-activating activities of c-jun. Cell 59:987-997. [DOI] [PubMed] [Google Scholar]
- 56.Sheng, J., A. Kumagai, W. G. Dunphy, and A. Varshavsky. 2002. Dissection of c-MOS degron. EMBO J. 21:6061-6071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shibuya, E. K., and J. V. Ruderman. 1993. Mos induces the in vitro activation of mitogen-activated protein kinases in lysates of frog oocytes and mammalian somatic cells. Mol. Biol. Cell 4:781-790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sourvinos, G., C. Tsatsanis, and D. A. Spandidos. 1999. Overexpression of the Tpl-2/Cot oncogene in human breast cancer. Oncogene 18:4968-4973. [DOI] [PubMed] [Google Scholar]
- 59.Stancovski, I., H. Gonen, A. Orian, A. L. Schwartz, and A. Ciechanover. 1995. Degradation of the proto-oncogene product c-Fos by the ubiquitin proteolytic system in vivo and in vitro: identification and characterization of the conjugating enzymes. Mol. Cell. Biol. 15:7106-7116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Touitou, R., J. Richardson, S. Bose, M. Nakanishi, J. Rivett, and M. J. Allday. 2001. A degradation signal located in the C-terminus of p21WAF1/CIP1 is a binding site for the C8 alpha-subunit of the 20S proteasome. EMBO J. 20:2367-2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Treier, M., L. M. Staszewski, and D. Bohmann. 1994. Ubiquitin-dependent c-Jun degradation in vivo is mediated by the delta domain. Cell 78:787-798. [DOI] [PubMed] [Google Scholar]
- 62.Tsatsanis, C., C. Patriotis, S. E. Bear, and P. N. Tsichlis. 1998. The Tpl-2 proto-oncoprotein activates the nuclear factor of activated T cells and induces interleukin 2 expression in T cell lines. Proc. Natl. Acad. Sci. USA 95:3827-3832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tsatsanis, C., C. Patriotis, and P. N. Tsichlis. 1998. Tpl-2 induces IL-2 expression in T-cell lines by triggering multiple signaling pathways that activate NFAT and NF-κB. Oncogene 17:2609-2618. [DOI] [PubMed] [Google Scholar]
- 64.Varshavsky, A. 1991. Naming a targeting signal. Cell 64:13-15. [DOI] [PubMed] [Google Scholar]
- 65.Velasco-Sampayo, A., and S. Alemany. 2001. p27kip protein levels and E2F activity are targets of Cot kinase during G1 phase progression in T cells. J. Immunol. 166:6084-6090. [DOI] [PubMed] [Google Scholar]
- 66.Verma, R., and R. J. Deshaies. 2000. A proteasome howdunit: the case of the missing signal. Cell 101:341-344. [DOI] [PubMed] [Google Scholar]
- 67.Verma, R., H. McDonald, J. R. Yates III, and R. J. Deshaies. 2001. Selective degradation of ubiquitinated Sic1 by purified 26S proteasome yields active S phase cyclin-Cdk. Mol. Cell 8:439-448. [DOI] [PubMed] [Google Scholar]
- 68.Voges, D., P. Zwickl, and W. Baumeister. 1999. The 26S proteasome: a molecular machine designed for controlled proteolysis. Annu. Rev. Biochem. 68:1015-1068. [DOI] [PubMed] [Google Scholar]
- 69.Wood, T. G., M. L. McGeady, B. M. Baroudy, D. G. Blair, and G. F. Vande Woude. 1984. Mouse c-mos oncogene activation is prevented by upstream sequences. Proc. Natl. Acad. Sci. USA 81:7817-7821. [DOI] [PMC free article] [PubMed] [Google Scholar]