Abstract
Pnn/DRS protein is associated with desmosomes and colocalizes with splicing factors in nuclear speckled domains. The potential interaction of Pnn with RNPS1, a pre-mRNA splicing factor and a component of the exon-exon junction complex, prompted us to examine whether Pnn is involved in nuclear mRNA processing. By immunoprecipitation, we found that Pnn associates preferentially with mRNAs produced by splicing in vitro. Oligonucleotide-directed RNase H digestion revealed that Pnn binds to the spliced mRNAs at a position immediately upstream of the splice junction and that 5′ splice site utilization determines the location of Pnn in alternatively spliced mRNAs. Immunoprecipitation further showed that Pnn binds to mRNAs produced from a transiently expressed reporter in vivo. Although associated with mRNPs, Pnn is a nuclear-restricted protein as revealed by the heterokaryon assay. Overexpression of an amino-terminal fragment of Pnn that directly interacts with RNPS1 leads to blockage of pre-mRNA splicing. However, although suppression of Pnn expression shows no significant effect on splicing, it leads to some extent to nuclear accumulation of bulk poly(A)+ RNA. Therefore, Pnn may participate, via its interaction with RNPS1, in mRNA metabolism in the nucleus, including mRNA splicing and export.
In eukaryotic cells, the step to remove the intron sequences from the pre-mRNA is carried out by the spliceosome, a macromolecular complex consisting of small nuclear ribonucleoproteins (snRNPs) and a number of protein factors (22). A family of Ser/Arg-dipeptide-rich proteins (SR proteins) play essential roles in constitutive splicing and/or can modulate alternative splice site selection (13). Atypical SR protein RNPS1 was previously characterized with a splicing activity that promotes utilization of distal alternative 3′ splice sites (32). However, recombinant RNPS1 instead synergizes with prototypical SR proteins to activate both constitutive and alternative pre-mRNA splicing, suggesting the role of RNPS1 as a general splicing activator (32). On the other hand, RNPS1 associates with SAP18 and acinus proteins to form the apoptosis and splicing-associated protein (ASAP) complex, which inhibits in vitro splicing and promotes apoptosis (42). It appears that RNPS1 functions to activate or suppress splicing by forming complexes with different regulatory proteins.
During the pre-mRNA splicing process, a multiprotein complex is deposited on the spliced mRNP (24, 25). This complex occupies a region 20 to 24 nucleotides (nt) upstream of the splice junctions of mature mRNA and is thus termed the exon-exon junction complex (EJC) (25). RNPS1 has been determined as a component of the EJC (25), which is consistent with the observation that RNPS1 specifically associates with spliced mRNAs in vitro (32). The EJC is thought to function as an adaptor platform that provides multiple postsplicing functions (26). This notion is apparently held true in the case of nonsense-mediated decay (NMD), which subjects aberrant mRNAs with premature termination codons to degradation (15). RNPS1 as well as another EJC component, Y14, have been shown to interact with Upf3, a component of the NMD machinery (12, 21, 29), providing a direct link between mRNA processing and the NMD.
Another putative function of the EJC is to promote mRNA export by recruiting an export adaptor, TAP (26). Although earlier observations suggested that splicing enhances mRNA export (5, 28), several recent lines of evidence make this attractive model disputable (11, 36). Specifically, suppression of RNPS1, Aly/REF, or SRm160 by the RNA interference technique has, at best, a minor effect on bulk mRNA export (11). However, the effect is enchanced when expression of more than one EJC component is suppressed (11). It is possible that down-regulation of a single EJC component does not completely abolish the ability of the EJC to recruit TAP, thus leading to only partial blockage of mRNA export. Moreover, multiple parallel pathways may exist for directing mRNA to export. A recent finding that SR protein 9G8 and SRp20 can recruit TAP directly to mRNAs certainly provides a support for the multiple adaptor model (17). Nevertheless, the possibility that the EJC plays some role in mRNP export cannot be completely excluded.
Following mRNA export, the EJC components may participate in the subsequent fate of mRNA in the cytoplasm, such as translation and localization. Earlier reports have already established that the Drosophila melanogaster Y14/Mago heterodimer is required for oskar mRNA transport to the posterior pole of the oocyte during oogenesis (14, 33, 34). Moreover, several reports showing that splicing enhances translation efficiency support the idea that the EJC also functions in modulating the translation activity of cytoplasmic mRNAs (27, 31, 35). Since Y14 is bound to mRNAs associated with ribosomes in the cytoplasm, it may be responsible for the translation control (8).
Pnn/DRS was originally identified as a protein closely associated with mature desmosomes of epithelial cells (37, 38). It was subsequently shown to exhibit dual localization at both sites of cell adhesion and the nucleus (3, 4, 39). Transiently expressed Pnn is located predominantly in the nucleus, with a prominent speckled pattern that coincides with that of splicing factors (4). Since nuclear Pnn shows different reactivity to monoclonal antibodies from that of Pnn expressed at cell surfaces, Pnn may assume distinct conformations at different locations (39). Increased expression of Pnn promotes cell-cell adhesion and negatively regulates cell migration and proliferation (43). Accordingly, the genes involved in cell cycle regulation and cell migration and invasion were altered at their expression levels by ectopically expressing Pnn, suggesting that Pnn has a direct impact on gene expression (44). Recently, a yeast two-hybrid screen has revealed RNPS1 to be a potential interacting partner of Pnn (P. Ouyang, unpublished data). We therefore attempted to characterize the role of Pnn in mRNA processing and export.
In this report, we show that Pnn is associated with mRNPs at a position immediately upstream of the exon-exon junctions of spliced mRNAs. Pnn interacts with RNPS1 and may help to coordinate multiple steps of mRNA maturation in the nucleus.
MATERIALS AND METHODS
Plasmids.
The plasmids for in vitro transcription of PIP85A and pSP64-5′D16X were kindly provided by B. J. Blencowe (Toronto, Canada) and A. Krainer (Cold Spring Harbor, N.Y.), respectively. pEGFP-hnRNP A1 and the pcDNA clones for expressing FLAG-tagged TAP, DEK, Aly/REF, Y14, hUpf3a, and SRm160 were kindly provided by J. Lykke-Andersen (Boulder, Colo.). The RNPS1 cDNA clone was obtained from E. M. Gardiner (Darlinghurst, Australia). The vector for in vitro translation of canine Pnn was described by Ouyang and Sugrue (38). The pBS-Δi PIP85A plasmid for in vitro transcription of spliced mRNA was constructed as described below. In vitro splicing using PIP85A as the substrate was carried out in HeLa cell nuclear extract to produce the spliced RNA. The cDNA of spliced PIP85A RNA (Δi RNA) was generated by reverse transcription-PCR and then inserted into pBluescript KS(+) (Stratagene). The RNase protection assay (RPA) probe for the intron-containing chloramphenicol acetyltransferase (CAT) reporter was described previously (23). The plasmid pCDNA3-FLAG-RNPS1 was constructed by placing the full-length RNPS1 open reading frame into a pcDNA-3-derived vector carrying the sequence coding for the FLAG epitope with appropriate restriction enzymes. The pcc-myc vector was constructed by inserting a DNA fragment coding for 6x c-myc tag into pcDNA3.1+ vector (Invitrogen). Subsequently, the DNA fragments encoding the coiled-coil domain (CCD; amino acids 1 to 284) and the PQLQ- and SR-rich region (PQLSR; amino acids 454 to 717) of Pnn were each cloned into pcc-myc to generate the vectors expressing 6x myc-fusion protein. The antisense RNA expression plasmids were constructed as follows. The cDNA segments of human Pnn (nt 1 to 434 of the open reading frame), TAP (nt 1 to 558), RNPS1 (nt 1 to 487), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH; nt 231 to 800) were each cloned into pcDNA3.1+ in the reverse direction to the cytomegalovirus promoter. To coexpress β-galactosidase (β-Gal), the neomycin resistance gene of the above vectors was replaced with the lacZ gene from pCH110 (Amersham Biosciences) to create pcDNA(lacZ)-antisense RNA vectors.
In vitro splicing, immunoprecipitation, and oligonucleotide-directed RNase H digestion.
Preparation of HeLa cell nuclear extract and splicing substrate was essentially as described previously (7, 46). Mixed SR proteins were isolated from frozen HeLa cell nuclei (Cell Culture Center) using a two-step salt precipitation method described by Zahler et al. (48). In vitro splicing reactions in 25-μl volumes containing 7.5 × 104 cpm of radiolabeled 5′D16X substrate were performed as described elsewhere (46). For immunoprecipitation, a 25-μl reaction mixture was incubated with 10 μl of antibody-bound protein A-Sepharose for 1 h at 4°C. Protein A-Sepharose was preabsorbed either with 15 μl of polyclonal anti-3a serum (anti-Pnn) (38) or with 20 μg of rabbit anti-mouse immunoglobulin G antibodies as control (Zymed). After incubation, the beads were washed extensively with NET-2 buffer containing 50 mM Tris-Cl (pH 7.4), 150 mM NaCl, and 0.05% NP-40. RNA was recovered from the immunoprecipitates for gel electrophoresis analysis. Oligonucleotide-directed RNase H digestion was performed after splicing by adding 25 μl of the mixture containing 0.5 μg of oligonucleotide, 0.5 U of RNase H (Roche), and 4 mM MgCl2 in buffer D (20 mM HEPES [pH 7.9], 50 mM KCl, 0.2 mM EDTA, 0.5 mM dithiothreitol, and 20% glycerol) to a 25-μl splicing reaction mixture. Incubation was continued for another 30 min prior to immunoprecipitation. Oligonucleotides used in RNase H digestion included H1 (5′-CACCGAGCACTTTCTTGC), H2 (5′-GGTAGACCACCAGCAGCC), H3 (5′-GGCAGACTTCTCCTCAGG), and H4 (5′-CTCTGTCTCCACATGCCC).
Transient transfection, immunoprecipitation, and RPA.
HeLa Tet-Off cells (Clontech) and HEK293 cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (Invitrogen). Transient transfection using Lipofectamine 2000 (Invitrogen) followed closely the protocol provided by the manufacturer.
To examine whether Pnn associates with mRNAs in vivo, HeLa Tet-Off cells were transiently transfected with pSV40-CAT(In1) (23) for 36 h. The cells were lysed in radioimmunoprecipitation assay buffer containing 10 mM sodium phosphate (pH 7.2), 150 mM NaCl, 2 mM EDTA, and 1% NP-40 for 30 min on ice. After centrifugation, cell debris was removed and the lysate was subjected to immunoprecipitation using anti-Pnn antibodies. Bound RNA was recovered and then treated with RNase-free DNase I (Promega) for 30 min at 37°C for the subsequent RPA. The RPA was performed according to the method of Lai et al. (23).
For coimmunoprecipitation of Pnn and EJC components, 5 × 105 HEK293 cells were transfected with a pcDNA-derived expression vector coding for FLAG-tagged EJC component. Cell lysate was prepared 30 h posttransfection according to the method described by Lykke-Andersen et al. (29), except that 0.5% NP-40 instead of Triton X-100 was used in the hypotonic buffer. Canine Pnn or human Pnn fragments (CCD or PQLSR) were in vitro translated in the TNT coupled reticulocyte lysate system (Promega) and subsequently mixed with cell lysate containing overexpressed EJC protein for 4 h at 4°C. The mixture was then incubated with 10 μl of anti-FLAG antibody agarose (Sigma) for 4 h at 4°C. Beads were extensively washed with NET-2 and then boiled in sodium dodecyl sulfate (SDS) sample buffer. Recovered proteins were fractionated in SDS-polyacrylamide gel electrophoresis (SDS-PAGE) gels and subjected to autoradiography.
Heterokaryon assay.
HeLa cells were cotransfected with the expression vector for myc-tagged canine Pnn and pEGFP-hnRNP A1 (gift of J. Lykke-Andersen) for 36 h. NIH 3T3 cells were fused with HeLa cells by polyethylene glycol 3350 (Sigma) followed by indirect immunofluorescence using monoclonal anti-myc antibody (clone 9E10, 1:1,000 dilution; BAbCO) as described previously (23). The cells were also counterstained with Hoechst dye (Sigma) to identify mouse nuclei.
Fluorescence in situ hybridization (FISH).
HeLa Tet-Off cells were transfected with antisense RNA- or Pnn subfragment-expressing construct, trypsinized, and seeded on glass coverslips 6 h after transfection. Cells were harvested 30 or 48 h posttransfection and subsequently fixed in 3% formaldehyde-phosphate-buffered saline (PBS) for 30 min, followed by permeabilization in 0.5% Triton X-100-PBS for 10 min and acetylation in 0.25% acetic acid-PBS for 10 min. For antisense experiments, positively transfected cells were located by immunostaining with monoclonal anti-β-Gal antibody (1:500 dilution; Promega) and secondary fluorescein isothiocyanate-conjugated anti-mouse antibody (1:250 dilution; Cappel). The cells were further fixed in 1% formaldehyde-PBS for 10 min and subsequently washed in hybridization buffer containing 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 30% formamide, 5% dextran sulfate, and 250 ng of fragmented salmon sperm DNA/μl for 30 min. Coverslips were inverted onto a drop of hybridization buffer containing 10 ng of denatured, digoxigenin (DIG)-labeled 25-mer oligo(dT)/μl. Hybridization was performed overnight at 37°C. Cells were washed in 2× SSC at room temperature twice and then at 37°C once; each wash was carried out for 10 min. DIG-oligo(dT) was stained with rhodamine-conjugated anti-DIG Fab fragment (1:200 dilution; Roche). The images of double-stained cells were generated with a Bio-Rad MRC-1000 confocal microscope. To count positively transfected (β-Gal-expressing) cells with a nuclear mRNA accumulation pattern, double-stained cells were examined under an Olympus BX60 fluorescence microscope.
RESULTS
Pnn is associated with spliced mRNA in vitro.
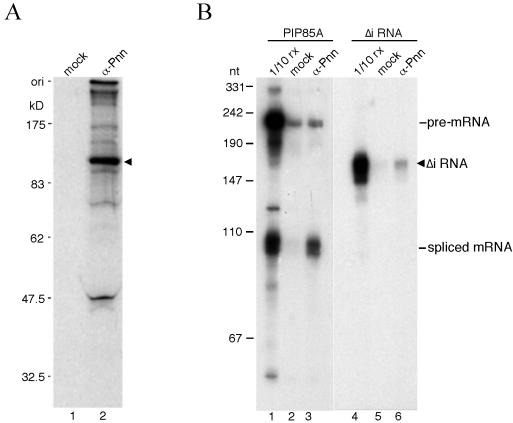
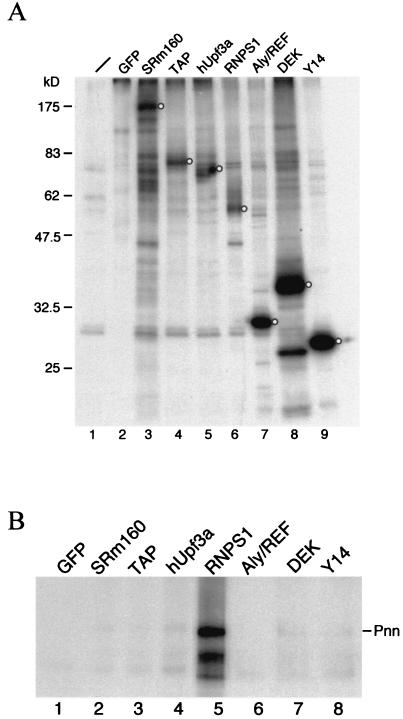
Nuclear Pnn is localized predominantly in speckles where splicing factors are concentrated, and it has been implicated in alternative splicing (3, 39, 47). Coincidentally, a yeast two-hybrid screen recently revealed splicing factor RNPS1 as a potential Pnn-interacting protein (P. Ouyang, unpublished data). To determine whether Pnn is involved in pre-mRNA splicing, immunoprecipitation of in vitro-assembled splicing complexes was examined using antiserum against the PQLQ-rich domain of canine Pnn (anti-3a, hereafter referred to as anti-Pnn) (38). The anti-Pnn serum specifically precipitated a band of ∼130 kDa from the lysate prepared from 35S-metabolically labeled HeLa cells (Fig. 1A, lane 2), which coincides with that detected by Western blotting (39). Precipitation of this 130-kDa protein was blocked by adding a recombinant protein bearing the Pnn PQLQ domain (data not shown), further confirming the specificity of the antiserum to a human homolog of Pnn.
FIG. 1.
Preferential binding of Pnn to spliced mRNA in vitro. (A) Immunoprecipitation was performed with 35S-labeled HeLa cell lysate using anti-mouse immunoglobulin G (lane 1, mock) or anti-Pnn antibodies (lane 2). Anti-Pnn antibodies specifically precipitated a protein of about 130 kDa (arrowhead). Protein size markers in kilodaltons are indicated at left. (B) In vitro splicing reaction was carried out at 30°C for 45 min with PIP85A (lanes 1 to 3) or its intron-lacking version (Δi RNA; lanes 4 to 6) as substrate. Immunoprecipitation was then performed with mock (lanes 2 and 5) or anti-Pnn (lanes 3 and 6) antibodies. Lanes 1 and 4 show 10% of the splicing reaction mixture used in immunoprecipitation. The Δi RNA (arrowhead) contains an extra 83-nt sequence derived from the vector at its 5′ end, thus being larger than the spliced mRNA. Size markers are radiolabeled MspI-digested pUC19 as indicated at left.
Next, the splicing reaction was performed in HeLa cell nuclear extract using PIP85A pre-mRNA as the substrate, followed by immunoprecipitation with anti-Pnn antiserum. As shown in Fig. 1B, only spliced mRNA was specifically precipitated with anti-Pnn antibodies (lane 3). In contrast, a monoclonal antibody against the snRNP Sm proteins quantitatively precipitated the precursor and splicing intermediates (data not shown). Similar analysis performed with β-globin and dsx(GAA)x6 pre-mRNAs consistently showed preferential association of Pnn with the spliced mRNAs (data not shown). This observation is similar to that reported with EJC components RNPS1, Y14, and Aly/REF (20, 32, 49).
To determine whether Pnn binding to mRNA is splicing dependent, the intron-lacking PIP85A RNA (Δi RNA) was incubated under the splicing conditions and subsequently subjected to immunoprecipitation with anti-Pnn antibodies. Figure 1B shows that Δi RNA was precipitated much less efficiently than PIP85A mRNA produced by splicing (lanes 3 and 6). Thus, the scenario in which Pnn becomes associated with the spliced mRNA during the splicing process is similar to that of the EJC components (25).
Pnn binds spliced mRNAs at a region immediately upstream of the exon-exon junction.
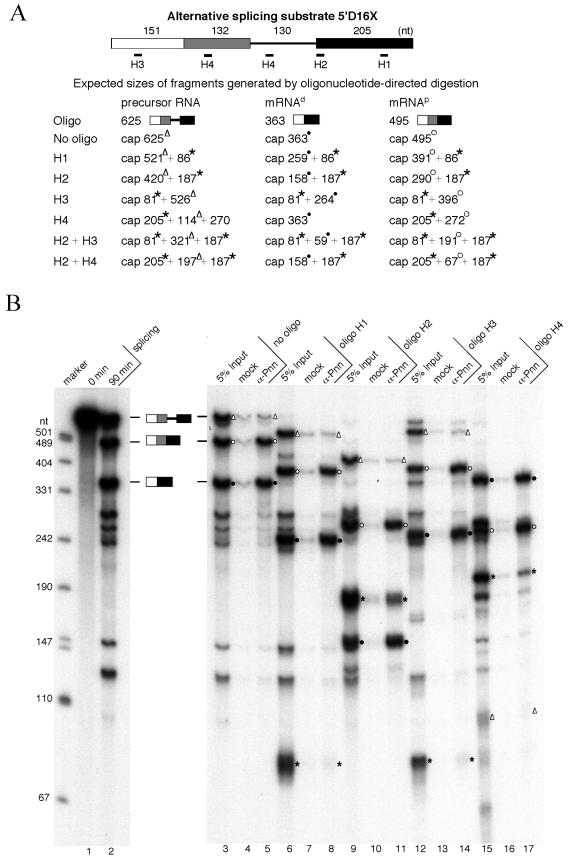
Since Pnn interacts with mRNAs produced by splicing, we next examined whether Pnn, like the EJC, locates upstream of the exon-exon junctions. The strategy adopted involves oligonucleotide-directed RNase H digestion followed by immunoprecipitation (25, 26). The modified β-globin pre-mRNA 5′D16X containing duplicated 5′ splice sites was used as the substrate in this experiment (Fig. 2A). SR proteins can modulate 5′ splice site utilization of 5′D16X pre-mRNA (9), producing two alternatively spliced mRNAs that differ from each other in the sequence upstream of the splice junction. Therefore, splicing of 5′D16X was carried out in HeLa cell nuclear extracts in the presence of exogenous SR proteins. By using this approach, we could test whether Pnn binding to mRNA is determined by a relative distance to the splice junction or by a specific sequence. Immunoprecipitation with anti-Pnn antibodies showed that both of the alternatively spliced RNA products were specifically precipitated (Fig. 2B, lanes 3 to 5) (hereafter, the mRNAs spliced at the distal and the proximal site are referred to as mRNAd and mRNAp, respectively), which was consistent with the above result that Pnn preferentially interacts with spliced mRNAs.
FIG. 2.
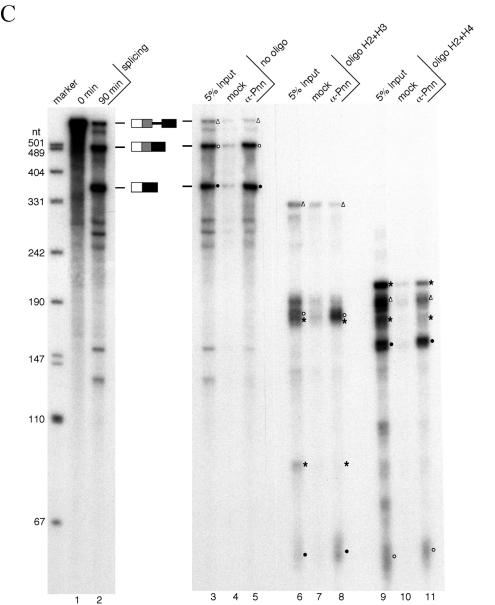
Pnn is associated with spliced mRNAs at the location immediately upstream of the exon-exon junction. (A) The 5′D16X pre-mRNA and the oligonucleotides used to direct RNase H cleavage are diagrammed. Note that oligonucleotide H4 anneals to a repeated sequence located in the intron and between the two 5′ splice sites (9). Expected sizes of the fragments generated by RNase H cleavage from the pre-mRNA and two alternatively spliced mRNAs (mRNAp and mRNAd) are listed. The mRNAp and mRNAd fragments carrying the region immediately upstream of the splice junction are labeled with open circles and solid circles, respectively. Open triangles indicate the fragments generated from the precursor RNA containing the site for EJC loading. Asterisks indicate the fragments that can be generated from both precursor and spliced mRNAs. (B) Single-oligonucleotide-directed RNase H digestion. The 5′D16X pre-mRNA was subjected to the splicing reaction at 30°C for 0 min (lane 1) or 90 min (lane 2). RNase H digestion was performed with 90-min splicing reactions in the absence of oligonucleotides (lanes 3 to 5) or in the presence of oligonucleotide H1 (lanes 6 to 8), H2 (lanes 9 to 11), H3 (lanes 12 to 14), or H4 (lanes 15 to 17). After digestion, immunoprecipitation was performed with mock (lanes 4, 7, 10, 13, and 16) or anti-Pnn (lanes 5, 8, 11, 14, and 17) antibodies. For comparison, 5% input of the reaction mixtures was loaded (lanes 3, 6, 9, 12, and 15). Size markers are as described in the legend for Fig. 1B. (C) RNase digestion with two oligonucleotides. Similar experiments were carried out as described for panel B except that oligonucleotides H2 and H3 (lanes 6 to 8) or H2 and H4 (lanes 9 to 11) were added simultaneously.
Next, RNase H digestion with oligonucleotides complementary to the exon sequences of 5′D16X was performed after splicing and followed by anti-Pnn immunoprecipitation. Figure 2A lists all the fragments produced from the pre-mRNA and two alternatively spliced mRNAs in the RNase H experiments. Oligonucleotides H1 and H2 directed cleavage in the 3′ exon at the position close to the 3′ end and immediately downstream of the 3′ splice site, respectively. When either of these two oligonucleotides was used, the 5′ RNA fragments released from the spliced mRNAs were preferentially precipitated by anti-Pnn antibodies (Fig. 2B, lanes 6 to 11). A low level of the 3′ exon fragment was precipitated (lane 11), probably because the integrity of the mRNP was not completely disrupted by RNase H digestion, as observed in an earlier report (26). In contrast, the 3′ mRNA fragment was preferentially precipitated by the use of oligonucleotide H3 that directs cleavage of the 5′ exon near its 5′ end (Fig. 2B, lanes 12 to 14). Furthermore, RNase H cleavage directed by oligonucleotide H4, which is complementary to a region between the two alternative 5′ splice sites, resulted in immunoprecipitation of the uncleaved mRNAd as well as the 3′ RNA fragment from mRNAp (lanes 15 to 17). The data indicated that Pnn is associated with the 5′ exon at a region upstream of the splice junction.
To further locate Pnn's binding site, RNase H digestion was performed simultaneously with two oligonucleotides. When both oligonucleotides H2 and H3 were added, two fragments of approximately 60 and 190 nt in length were enriched in anti-Pnn immunoprecipitates (Fig. 2C, lanes 6 to 8). It should be noted that cleavage of mRNAp with these two oligonucleotides produces two closely migrating bands at 191 and 187 nt. Although the identity of the precipitated ∼190-nt band (lane 8) was not further verified, its slightly slower migration than the nonprecipitated one on the gels suggested that it corresponds to the 5′ exon-containing 191-nt fragment. Digestion directed by oligonucleotides H2 and H4 resulted in precipitation of two fragments (∼60 and ∼160 nt) that likely correspond to the 67- and 158-nt fragments derived from mRNAp and mRNAd, respectively (lanes 9 to 11). Thus, Pnn associates with spliced mRNAs at a region between the −60 position and the splice junction, which overlaps with the binding site of the EJC (25). Indeed, oligonucleotides complementary to the −24 region were incapable of directing RNase H cleavage of the 5′D16X mRNPs (data not shown), indicating the occupancy of this region. Taken together, Pnn associates with a region of the spliced mRNAs where the EJC is expected to bind.
Pnn is associated with mRNA in vivo.
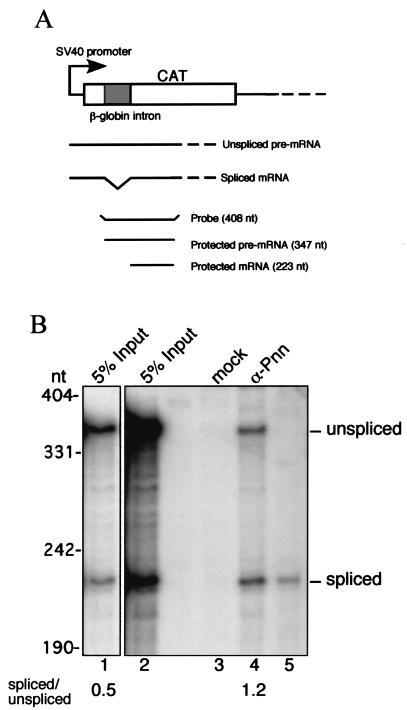
To assess the ability of Pnn to interact with mRNA in vivo, a reporter plasmid, pSV40-CAT(In1), was introduced into HeLa cells to produce a pre-mRNA containing the β-globin intron in the CAT coding region (23). Immunoprecipitation of cell lysates with anti-Pnn antibodies was performed and followed by RPA using a probe complementary to the 5′ part of the CAT pre-mRNA (Fig. 3A). Figure 3B shows that anti-Pnn, but not the control antibodies, could precipitate the CAT reporter RNAs (lanes 3 and 4). Although both spliced and unspliced mRNAs were precipitated, the results from independent experiments reproducibly showed that Pnn had a slightly higher preference to the spliced mRNA (Fig. 3B, compare lanes 1 and 4 for the ratios of spliced to unspliced mRNA). This correlates with the behavior of RNPS1, which associates with only spliced mRNA in vitro (32) but with both precursor and spliced mRNA in vivo (29). It is unclear why Pnn binds to pre-mRNAs in vivo, but it will be of great interest to explore whether Pnn plays additional roles in pre-mRNA processing or in nuclear export of unspliced mRNA (41) in intact cells. Nevertheless, Pnn is likely associated with spliced mRNAs as a component of mRNPs, further confirming the observation in vitro.
FIG. 3.
Pnn is associated with mRNPs in vivo. (A) A schematic diagram shows the intron-containing CAT reporter, pSV40-CAT(In1), and the experimental design for the RPA. Two RPA products of 347 and 223 nt correspond to unspliced and spliced mRNA, respectively. (B) Thirty-six hours after transfection with the reporter, cell lysates were prepared and then subjected to immunoprecipitation with mock (lane 3) or anti-Pnn (lane 4) antibodies. RPA with 5% total RNA prepared from transfected cells is shown in lane 2. Lane 1 is a lighter exposure of the autoradiogram of lane 2. Lane 5 shows RPA with in vitro-transcribed intron-lacking CAT RNA, indicating the size of protected spliced mRNA. The intensity of protected RNA fragments was measured using a PhosphorImager; the ratios of spliced to unspliced RNA in the input and anti-Pnn precipitate are shown, indicating preferential association of Pnn with spliced mRNPs.
Pnn interacts with the EJC component RNPS1.
Our data indicated that Pnn is associated with mRNPs at the position where the EJC locates. We therefore tested whether Pnn could directly interact with the EJC components. HEK293 cells were transiently transfected with vector expressing FLAG-tagged EJC protein. The anti-FLAG antibody successfully immunoprecipitated transiently expressed proteins from 35S-labeled HEK293 cell lysates (Fig. 4A), indicating their expression and accessibility to the antibody. We initially attempted to use Western blotting to examine if endogenous Pnn is detectable in the anti-FLAG immunoprecipitates from transfected cell lysates. However, anti-Pnn antibodies failed to reproducibly detect Pnn, probably due to low levels of Pnn in Nonidet P-40-solubilized extract. Disruption of nuclei by sonication indeed increased the solubility of Pnn (see the legend to Fig. 6, below) but could probably disrupt protein interactions or fragment mRNPs containing Pnn. Therefore, 35S-labeled canine Pnn was produced in reticulocyte lysates and subsequently mixed with transfected HEK293 cell lysates for immunoprecipitation. Figure 4B shows that Pnn was significantly coprecipitated only with RNPS1 but not with any others. Furthermore, the Pnn-RNPS1 coprecipitation was not disrupted by the presence of RNase (data not shown), indicating their RNA-independent interaction. These data suggest that Pnn forms a subcomplex with RNPS1 and probably thereby binds to the spliced mRNA through the EJC.
FIG. 4.
Pnn interacts with RNPS1, a component of the EJC. (A) HEK293 cells transfected with expression vector for FLAG-tagged EJC component were metabolically labeled with [35S]methionine. Cell lysates were subjected to immunoprecipitation with monoclonal anti-FLAG antibody. Precipitated proteins were analyzed by SDS-PAGE and autoradiography. Protein of correct size in each lane is indicated by the open circle. (B) Total lysates prepared from HEK293 cells transiently expressing FLAG-tagged EJC component were incubated with radiolabeled canine Pnn produced by in vitro translation, followed by immunoprecipitation with anti-FLAG antibody. Precipitates were analyzed by SDS-PAGE and autoradiography.
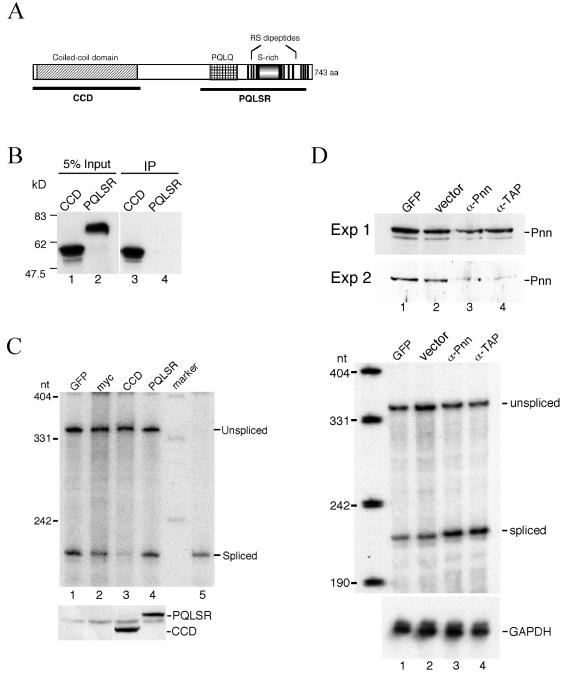
FIG. 6.
Effects of overexpression of Pnn protein subfragments or suppression of Pnn expression on pre-mRNA splicing. (A) Schematic representation of the domains of human Pnn protein. (B) In vitro-translated, 35S-labeled CCD or PQLSR fragment was incubated with total lysates prepared from HeLa cells transiently expressing FLAG-tagged RNPS1, followed by immunoprecipitation with anti-FLAG antibody (lanes 3 and 4). Precipitates were analyzed by SDS-PAGE and autoradiography. Lanes 1 and 2 show 5% input. Protein size markers are indicated at left. (C) HeLa cells were cotransfected with the pSV40-CAT(In1) reporter and vector for expressing GFP (lane 1), no fusion myc tag (lane 2), myc-CCD (lane 3), or myc-PQLSR domain (lane 4) for 30 h. Splicing of the CAT pre-mRNA was determined by RPA as described in the legend for Fig. 3B. Lane 5 is equivalent to lane 5 of Fig. 3B. Expression of CCD and PQLSR fragments was confirmed by Western blotting with anti-myc antibody (lower). (D) Vectors expressing anti-sense RNA against Pnn (α-Pnn) or TAP (α-TAP) were tested for their efficacy to suppress Pnn expression and their effect on splicing in vivo. Thirty hours after transfection, cell lysates were prepared as described in Materials and Methods, except that sonication was performed after cells were lysed, and then lysates were subjected to Western blotting with anti-Pnn antibodies (upper panel). Results of two independent experiments are shown. To examine the effect of Pnn down-regulation on pre-mRNA splicing, HeLa cells were cotransfected with pSV40-CAT(In1) and vector expressing GFP (lane 1), no antisense RNA (lane 2), anti-Pnn (lane 3), or anti-TAP (lane 4) RNA for 48 h. Total RNAs were analyzed by RPA to detect the levels of CAT and GAPDH mRNAs (lower panel).
Pnn is not a nucleocytoplasmic shuttle protein.
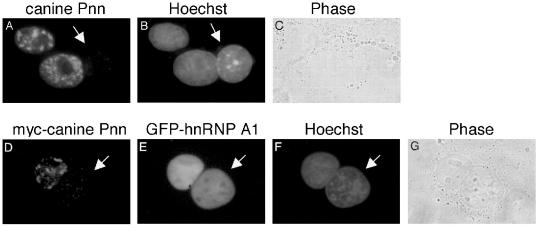
Since Pnn binds to spliced mRNAs, it was therefore of interest to test whether nuclear Pnn is able to export to the cytoplasm. We first examined whether endogenous Pnn shuttles in and out of the nucleus by an interspecies heterokaryon assay. Heterokaryons were formed by polyethylene glycol-induced fusion of canine MDCK cells and mouse NIH 3T3 cells, followed by immunostaining with anti-Pnn antibodies that had no detectable cross-reaction with the mouse Pnn homolog. Figure 5 shows that endogenous canine Pnn was restrained in the MDCK nuclei (panels A to C).
FIG. 5.
Pnn is not a nucleocytoplasmic shuttle protein. (A to C) Heterokaryons were formed between canine MDCK cells and mouse NIH 3T3 cells, followed by immunostaining with polyclonal anti-Pnn antibodies. (D to G) HeLa cells were cotransfected with vectors for myc-tagged canine Pnn and control protein GFP-hnRNPA1 and subsequently fused with NIH 3T3 cells after transfection for 36 h. Transiently expressed Pnn was immunofluorescently detected by monoclonal anti-myc antibody (D). The cells were stained with Hoechst 33258 to distinguish mouse (arrow) from canine (B) or human (F) nuclei. Panels C and G show cells visualized by phase-contrast microscopy.
Since anti-Pnn antibodies preferentially react with Pnn in the nucleus (39), a heterokaryon assay was also performed using myc-tagged Pnn to exclude the possibility that the exported Pnn fails in the detection by immunostaining with anti-Pnn antibodies. HeLa cells were cotransfected with vectors expressing myc-tagged canine Pnn and hnRNP A1 fused to the green fluorescent protein (GFP) as a positive control. Immunofluorescence showed that Pnn was detected in the transfected HeLa nuclei, but not in the mouse nuclei, of heterokaryons (Fig. 5D). In contrast, the shuttle protein hnRNP A1 appeared in both HeLa and 3T3 cell nuclei (Fig. 5E). Although Pnn could exhibit dual localization at sites of the cell surface and the nucleus (39), our observation suggests that the nuclear form of Pnn is incapable of exporting to the cytoplasm. This result is not completely unexpected, since Pnn was previously characterized as a component of the nuclear skeleton (3). Moreover, not all the EJC components are nucleocytoplasmic shuttle proteins (29). Given this result, it appears that Pnn is retained in the nucleus and dissociates from RNPS1 before or during the latter exports along with mRNPs to the cytoplasm.
Overexpression of the N-terminal domain of Pnn inhibits pre-mRNA splicing in vivo.
Since Pnn interacts with RNPS1 and is specifically associated with mRNPs, we thus speculated that Pnn plays some role in pre-mRNA splicing and/or mRNP export. To test the former possibility, we overexpressed Pnn protein fragments or suppressed Pnn expression in HeLa cells and subsequently examined the splicing of a transiently expressed reporter pre-mRNA by RPA. First, we tested if overexpression of Pnn fragments could have any effect on pre-mRNA splicing. The yeast two-hybrid assay revealed the N-terminal CCD (Fig. 6A) of Pnn interacting with RNPS1 (P. Ouyang, unpublished data). To confirm this, we performed coimmunoprecipitation experiments. The result showing that 35S-labeled CCD, but not the C-terminal PQLQ- and SR-rich domain (PQLSR), was coprecipitated with FLAG-RNPS1 confirmed the interaction of the CCD with RNPS1 (Fig. 6B). Next, vector expressing myc-tagged CCD or PQLSR was cotransfected with the pSV40-CAT(In1) reporter plasmid into HeLa cells. Transient expression of the CCD and PQLSR fragments was confirmed by Western blotting with the antibody against their fusion tag (Fig. 6C, lower panel). RPA showed that overexpression of the CCD, but not the PQLSR fragment, severely hampered the splicing of the CAT pre-mRNA (Fig. 6C, upper panel). Transiently expressed full-length Pnn was produced at a very low level, but it also showed a partial inhibitory effect on pre-mRNA splicing (data not shown). Thus, excess Pnn CCD (or full-length Pnn) could probably sequester RNPS1 or other essential splicing factors, thereby leading to inhibition of the splicing.
To further determine whether Pnn functions in splicing, we suppressed Pnn expression in cells by adopting the antisense RNA strategy. Introduction of the vector that produces the antisense RNA against Pnn nt 1 to 434 resulted in duplex formation with Pnn mRNA (data not shown) and reduced the level of Pnn protein (Fig. 6D, upper panel, lane 3). The residual Pnn protein observed (upper panel, Exp. 1) was probably from nontransfected cells, and between experiments its level varied upon the transfection efficiency. Nevertheless, reduction of Pnn (Fig. 6D, upper panel, lane 4) or lamin A/C proteins (data not shown) to different levels was also observed by expression of anti-TAP RNA. We reasoned that down-regulation of TAP that blocks nuclear export of mRNA and subsequent protein synthesis (11, 16) may result in depletion of a set of cellular proteins. The antisense clones were then tested for their effects on the splicing of the coexpressing CAT pre-mRNA. In contrast to the results obtained with overexpression of the Pnn CCD fragment, suppression of Pnn or TAP by antisense RNA showed no inhibitory effect on pre-mRNA splicing (Fig. 6D, lower panel). However, we cannot rule out the possibility that depletion of endogenous Pnn was not complete in transfected cells and, therefore, residual Pnn could still sustain splicing, leading to the observation that splicing was not affected. Taken together, pre-mRNA splicing appears to be blocked by overexpression of a Pnn domain that interacts with RNPS1, but probably not by depletion of Pnn.
Characterization of Pnn's role in mRNA export.
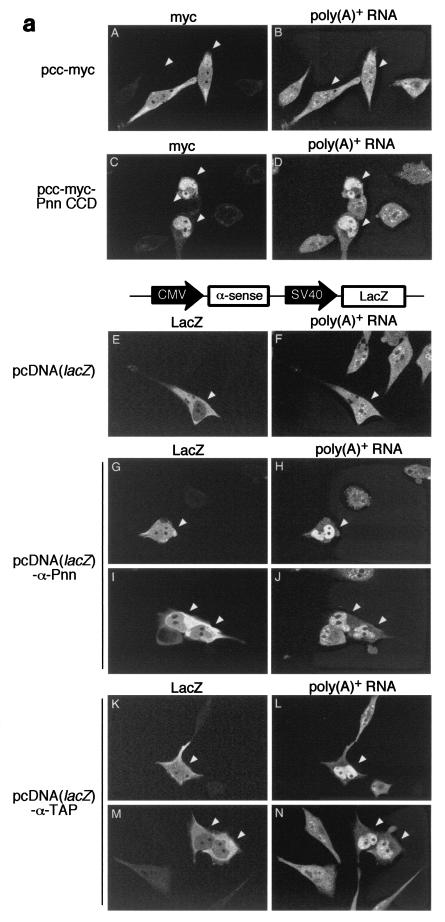
We next went on to investigate whether Pnn plays a role in the export of mRNA. The effect of overexpression of the Pnn CCD fragment on bulk mRNA accumulation in the nucleus was first evaluated by FISH. Immunofluorescence with anti-myc antibody was also conducted to indicate positively transfected cells and confirm the expression of myc-tagged proteins. In cells expressing a control protein composed of six consecutive myc epitopes (Fig. 7a, panels A and B) or myc-Pnn PQLSR fragment (data not shown), an even distribution of poly(A)+ RNA throughout the whole cell with additional intensively stained speckles inside the nucleus (10) was observed. However, overexpression of Pnn CCD resulted in considerable accumulation of poly(A)+ RNA in the nucleus (panel D). Since CCD overexpression hampered pre-mRNA splicing (Fig. 6C), the blockage of mRNA export was possibly a downstream effect of splicing inhibition (28).
FIG. 7.
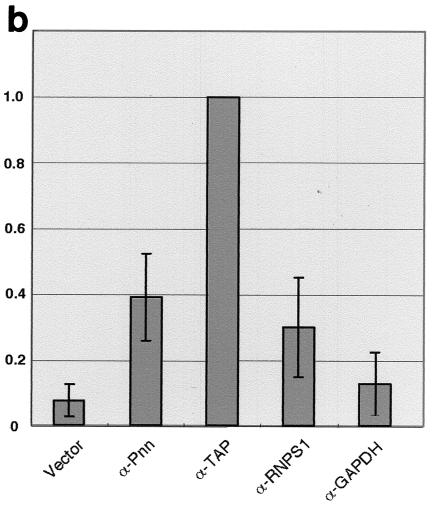
Effects of overexpression of Pnn protein subfragments or suppression of Pnn expression on mRNA export. (a) HeLa cells were transfected with vector as indicated at left. Thirty hours posttransfection, cells were immunostained with anti-myc antibody (A and C) or anti-β-Gal antibody (E, G, I, K, and M), followed by FISH with DIG-labeled oligo(dT) as probe (B, D, F, H, J, L, and N). Detection of oligo(dT) probe was made by staining with rhodamine-conjugated anti-DIG Fab fragment. Images were taken with a Bio-Rad MRC-1000 confocal microscope. Two samples are shown for cells expressing anti-Pnn RNA (G through J) and anti-TAP RNA (K through N). In all panels, positively transfected cells are indicated by an arrowhead. A diagram in the middle shows vector for coexpression of antisense RNA and β-Gal. (b) Similar FISH experiments were performed as in panel a, except that samples were collected 48 h posttransfection. The percentage of the cells with nuclear RNA accumulation over the positively transfected cells was measured. Note that ∼1,000 positively transfected cells were examined in each transfection and that ∼10% of anti-TAP RNA-expressing cells exhibited prominent nuclear accumulation of poly(A)+ RNA. The percentage obtained was then normalized against that of anti-TAP-expressing cells; averages and standard deviations were obtained from three to five independent experiments.
To examine the effect of Pnn depletion, a lacZ reporter was placed in the vectors that drive the production of antisense RNA in order to identify positively transfected cells by immunofluorescent detection of β-Gal. When the LacZ vector without expression of any antisense RNA was transfected, the distribution of poly(A)+ RNA appeared normal in the majority of the transfected cells (Fig. 7a, panel F), although few cells indeed showed nuclear poly(A)+ RNA accumulation (Fig. 7b). In contrast, expression of antisense RNA against Pnn or TAP led to nuclear accumulation of poly(A)+ RNA more significantly (panels H, J, L, and N).
In order to evaluate the effect of EJC factor depletion on mRNA export, we determined the percentage of the cells with nuclear RNA accumulation over the positively transfected cells. At 48 h after transfection, approximately 10% of anti-TAP RNA-expressing cells clearly showed nuclear poly(A)+ RNA accumulation. The percentage obtained with all other samples was thus normalized against that of the cells expressing anti-TAP RNA. Transfection with the empty vector or the vector expressing an antisense RNA against GAPDH was considered the negative control. Figure 7B shows that expression of either anti-Pnn or anti-RNPS1 RNA led to higher percentages of cells with nuclear poly(A)+ RNA accumulation than that of the negative controls but, nevertheless, lower than that of anti-TAP-expressing cells. Several independent experiments reproducibly revealed that the effect of depletion of Pnn on bulk mRNA export was minor but comparable to that of RNPS1, and thus similar to that reported with depletion of individual EJC proteins (11). The role of Pnn in facilitating mRNA export, hence, cannot be completely ruled out and is discussed below.
DISCUSSION
While Pnn was initially isolated as a desmosome-associated protein that promotes cell-cell adhesion when overexpressed (37, 38), later reports, however, showed that Pnn is also present in the nucleoplasm as a speckle-localized protein (3, 4, 39). Dual localization at both nucleus and cell surface has been reported for other cell junction proteins, such as ZO-2 and symplekin (18, 45). Nevertheless, transiently expressed Pnn is detected exclusively in the nucleus (Fig. 5) (4). This may be due to the lack of desmosomes in the cell lines used or localization of the majority of Pnn protein in the nucleus only by default. However, polyclonal anti-Pnn antibodies used in this study detected Pnn in nuclear speckles (Fig. 5) and specifically precipitated endogenous (Fig. 1) and transiently expressed (data not shown) Pnn from cell lysates. These observations eliminate the possibility that the antibodies are cross-reactive with other proteins and thus interfere with our interpretation of the data.
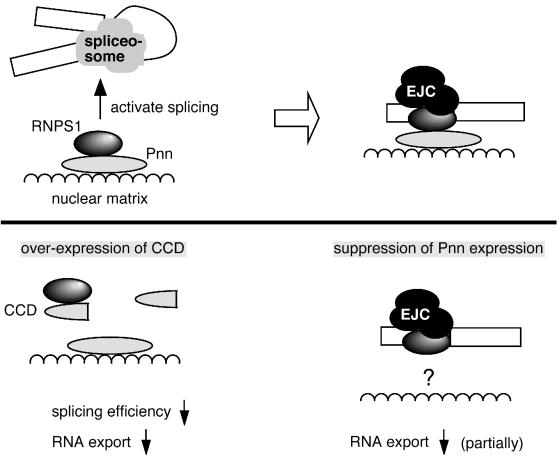
In this report, we demonstrate that Pnn binds preferentially to spliced mRNAs upon the splicing reaction (Fig. 1 and 3) and that the region of Pnn association with mRNAs is within ∼60 nt upstream of the 5′ splice sites (Fig. 2). Thus, Pnn behaves similarly to the EJC components in many aspects. A recent proteomic analysis of the spliceosome identified Pnn in catalytically active complex C (19), further supporting our result that Pnn can be an integral component of the mRNPs. However, the feature of Pnn interaction with mRNPs is not completely clear. Since there is no recognizable RNA binding motif in Pnn, it is less probable that Pnn binds mRNA directly. Our data showed that Pnn interacts with the EJC component RNPS1 (Fig. 4), and this interaction is neither RNA dependent nor splicing dependent (data not shown); thus, Pnn may form a subcomplex with RNPS1, which is in turn incorporated into the EJC. Moreover, we observed that nuclear Pnn was largely resistant to extraction under conditions (1) that solubilize vast cytoplasmic and nuclear proteins (data not shown), indicating association of Pnn with the nuclear matrix. We therefore suspected that Pnn functions to attach RNPS1 to the sites where RNPS1 activates splicing and joins the EJC (Fig. 8).
FIG. 8.
Model of a potential role of Pnn in mRNA processing and export. (Upper) Pnn probably functions to attach RNPS1 to nuclear components such as nuclear matrix, where RNPS1 activates splicing and joins the EJC. Pnn is associated with spliced mRNPs likely via its interaction with the EJC component RNPS1. (Lower) Pnn by itself is not crucial for splicing. However, overexpression of the Pnn CCD titrates RNPS1 and thereby results in splicing inhibition and subsequent nuclear export blockage. Depletion of Pnn protein shows a partial effect on bulk mRNA export. We suspect that Pnn only serves as a platform for mRNP assembly, and its role can be substituted by other matrix-associated proteins. Finally, Pnn is restricted in the nucleus and thus likely becomes detached from mRNPs prior to mRNA transport through the nuclear pore complex.
As to whether Pnn plays a role in pre-mRNA splicing, we initially attempted to use in vitro experiments to investigate the question. However, incomplete depletion of Pnn from extracts by antibodies yielded uninterpretable data (data not shown). In vivo suppression of Pnn did not show any significant effect on the splicing of the reporter CAT pre-mRNA (Fig. 6), suggesting that Pnn is dispensable for splicing. However, the possibility that residual amounts of Pnn could achieve its function in splicing still remains. On the other hand, we observed that overexpression of the Pnn CCD considerably interfered with the splicing of a reporter pre-mRNA (Fig. 6). The CCD is the only domain conserved from Drosophila to human Pnn homologs, indicating its functional importance. Our data suggest that overexpressed CCD acts as a dominant negative mutant that blocks mRNA splicing and perhaps its subsequent export, consistent with its ability to bind RNPS1. Notably, Pnn associates with pre-mRNAs at some extent in vivo (Fig. 3), as does RNPS1 (29). We suspect that Pnn serves a role, although not essential and partly via its interaction with RNPS1, in splicing in intact nuclei, such as coordinating splicing complex assembly or attaching the complexes to other nuclear components or matrix (Fig. 8); nevertheless, this hypothesis remains to be investigated further.
A fraction of cells transfected with the vector expressing anti-Pnn RNA shows clear nuclear accumulation of bulk poly(A)+ RNA (Fig. 7) that should not result from splicing blockage (Fig. 6). However, suppression of Pnn expression showed a less significant effect on nuclear mRNA accumulation than TAP down-regulation, consistent with the observation with depletion of any of the EJC components (11). Although the EJC was earlier proposed to play a significant role in recruiting TAP to spliced mRNA and subsequent export to the cytoplasm, recent reports argue against this hypothesis (11, 36). First, evidence suggesting that the EJC is not the signal, or not the only signal, for directing mRNA export comes from the observation that U1 RNA with a heterologous sequence insertion behaves as mRNA (36). Furthermore, while individual or multiple components of the EJC were depleted by RNA interference, only partial effects on bulk mRNA export in cultured Drosophila cells were observed (11). A recent report showing that SR protein 9G8 and SRp20 recruit TAP to mRNAs suggests that multiple independent pathways may exist for mRNA export (17). Nevertheless, whether Pnn facilitates mRNA export as a component of the EJC or whether it acts through other mechanisms, such as providing a docking site for export factors, remains to be answered by further investigations.
Like many EJC components (12, 14, 21, 27, 29, 30, 33-35), Pnn is possibly involved in multiple nuclear mRNA maturation events, although through its interaction with RNPS1. Particularly, Pnn is similar to SRm160 in several aspects. Both proteins are considered as nuclear matrix proteins and are associated with the mRNPs upstream of the exon-exon junction (this study and reference 25). SRm160 is an activator of pre-mRNA splicing and promotes 3′-end cleavage of nascent transcripts (2, 30). Furthermore, overexpression of SRm160 leads to leakage of unspliced RNA into the cytoplasm (30). Thus, SRm160, not only as a part of the EJC, may participate in multiple steps of mRNA processing. It is also of interest to compare Pnn to the tight junction protein symplekin, which has been suggested to function as a scaffold protein in the nucleus to help the assembly of the polyadenylation machinery or as a link between polyadenylation and other steps of mRNA metabolism (45). Recently, a yeast two-hybrid assay revealed the interaction of Pnn with mammalian PRP4 kinase that is also associated with the U5 snRNP and the N-CoR deacetylase complexes (6). Thus, Pnn may likewise serve as a platform to coordinate several steps of RNA metabolism in the nucleus, including transcription, pre-mRNA splicing, and mRNP export. Nevertheless, if Pnn functions to coordinate mRNP or export complex formation on the nuclear matrix, loss of Pnn would lead to retention of mRNA in the nucleus to some extent. However, the function of Pnn could be substituted by other matrix-associated proteins such as SRm160; therefore, depletion of Pnn has only minor effects on mRNA export. The simplified diagram in Fig. 8 depicts the current understanding of how Pnn functions in mRNA splicing and export. The more-detailed functions of Pnn await further characterization.
Acknowledgments
We are grateful to B. J. Blencowe, E. M. Gardiner, A. R. Krainer, and J. Lykke-Andersen for the generous gifts of plasmids and to J. Lykke-Andersen and S.-C. Cheng for critical reading of the manuscript. We acknowledge Chi-Hong Lou for his effort to initiate this study and C. V. Weaver for editing the manuscript.
This work was supported by the intramural fund of Academia Sinica.
REFERENCES
- 1.Aebi, U., J. Cohn, L. Buhle, and L. Gerace. 1986. The nuclear lamina is a meshwork of intermediate-type filaments. Nature 323:560-564. [DOI] [PubMed] [Google Scholar]
- 2.Blencowe, B. J., R. Issner, J. A. Nickerson, and S. A. Sharp. 1998. A coactivator of pre-mRNA splicing. Genes Dev. 12:996-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brandner, J. M., S. Reidenbach, and W. W. Franke. 1997. Evidence that “pnn,” reportedly a differentiation-specific desmosomal protein, is actually a widespread nuclear protein. Differentiation 62:119-127. [DOI] [PubMed] [Google Scholar]
- 4.Brandner, J. M., S. Reidenbach, C. Kuhn, and W. W. Franke. 1998. Identification and characterization of a novel kind of nuclear protein occurring free in the nucleoplasm and in ribonucleoprotein structures of the “speckle” type. Eur. J. Cell Biol. 75:295-308. [DOI] [PubMed] [Google Scholar]
- 5.Brodsky, A. S., and P. A. Silver. 2000. Pre-mRNA processing factors are required for nuclear export. RNA 6:1737-1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dellaire, G., E. M. Makarov, J. J. M. Cowger, D. Longman, H. G. E. Sutherland, R. Luhrmann, J. Torchia, and W. A. Bickmore. 2002. Mammalian PRP4 kinase copurifies and interacts with components of both the U5 snRNP and the N-CoR deacetylase complexes. Mol. Cell. Biol. 22:5141-5156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dignam, J. D., R. M. Lebovitz, and R. G. Roeder. 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11:1475-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dostie, J., and G. Dreyfuss. 2002. Translation is required to remove Y14 from mRNAs in the cytoplasm. Curr. Biol. 12:1060-1067. [DOI] [PubMed] [Google Scholar]
- 9.Fu, X., A. Mayeda, T. Maniatis, and A. R. Krainer. 1992. General splicing factors SF2 and SC35 have equivalent activities in vitro, and both affect alternative 5′ and 3′ splice site selection. Proc. Natl. Acad. Sci. USA 89:11224-11228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gallouzi, I. E., and J. A. Steitz. 2001. Delineation of mRNA export pathways by the use of cell-permeable peptides. Science 294:1895-1901. [DOI] [PubMed] [Google Scholar]
- 11.Gatfield, D., and E. Izaurralde. 2002. REF1/Aly and the additional exon junction complex proteins are dispensable for nuclear mRNA export. J. Cell Biol. 159:579-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gehring, N. H., G. Neu-Yilik, T. Schell, M. W. Hentze, and A. E. Kulozik. 2003. Y14 and hUpf3b form an NMD-activating complex. Mol. Cell 11:939-949. [DOI] [PubMed] [Google Scholar]
- 13.Graveley, B. R. 2000. Sorting out the complexity of SR protein functions. RNA 6:1197-1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hachet, O., and A. Ephrussi. 2001. Drosophila Y14 shuttles to the posterior of the oocyte and is required for oskar mRNA transport. Curr. Biol. 11:1666-1674. [DOI] [PubMed] [Google Scholar]
- 15.Hentze, M. W., and A. E. Kulozik. 1999. A perfect message: RNA surveillance and nonsense-mediated decay. Cell 96:307-310. [DOI] [PubMed] [Google Scholar]
- 16.Herold, A., T. Klymenko, and E. Izaurralde. 2001. NXF1/p15 heterodimers are essential for mRNA nuclear export in Drosophila. RNA 7:1768-1780. [PMC free article] [PubMed] [Google Scholar]
- 17.Huang, Y., R. Gattoni, J. Stevenin, and J. A. Steitz. 2003. SR splicing factors serve as adapter proteins for TAP-dependent mRNA export. Mol. Cell 11:837-843. [DOI] [PubMed] [Google Scholar]
- 18.Islas, S., J. Vega, L. Ponce, and L. Gonzalea-Mariscal. 2002. Nuclear localization of the tight junction protein ZO-2 in epithelial cells. Exp. Cell. Res. 274:138-148. [DOI] [PubMed] [Google Scholar]
- 19.Jurica, M. S., L. J. Licklider, S. R. Gygi, N. Grigorieff, and M. J. Moore. 2002. Purification and characterization of native spliceosomes suitable for three-dimensional structural analysis. RNA 8:426-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kataoka, N., J. Yong, V. N. Kim, F. Velazquez, R. A. Perkinson, F. Wang, and G. Dreyfuss. 2000. Pre-mRNA splicing imprints mRNA in the nucleus with a novel RNA-binding protein that persists in the cytoplasm. Mol. Cell 6:673-682. [DOI] [PubMed] [Google Scholar]
- 21.Kim, V. N., N. Kataoka, and G. Dreyfuss. 2001. Role of the nonsense-mediated decay factor hUpf3 in the splicing-dependent exon-exon junction complex. Science 293:1832-1836. [DOI] [PubMed] [Google Scholar]
- 22.Kramer, A. 1996. The structure and function of proteins involved in mammalian pre-mRNA splicing. Annu. Rev. Biochem. 65:367-409. [DOI] [PubMed] [Google Scholar]
- 23.Lai, M. C., B. H. Teh, and W. Y. Tarn. 1999. A human papillomavirus E2 transcriptional activator. The interactions with cellular splicing factors and potential function in pre-mRNA processing. J. Biol. Chem. 274:11832-11841. [DOI] [PubMed] [Google Scholar]
- 24.Le Hir, H., M. J. Moore, and L. E. Maquat. 2000. Pre-mRNA splicing alters mRNP composition: evidence for stable association of proteins at exon-exon junction. Genes Dev. 14:1098-1108. [PMC free article] [PubMed] [Google Scholar]
- 25.Le Hir, H., E. Izaurralde, L. E. Maquat, and M. J. Moore. 2000. The spliceosome deposits multiple proteins 20-24 nucleotides upstream of mRNA exon-exon junctions. EMBO J. 19:6860-6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Le Hir, H., D. Gatfield, E. Izaurralde, and M. J. Moore. 2001. The exon-exon junction complex provides a binding platform for factors involved in mRNA export and nonsense-mediated decay. EMBO J. 20:4987-4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu, S., and B. R. Cullen. 2003. Analysis of the stimulatory effect of splicing on mRNA production and utilization in mammalian cells. RNA 9:618-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luo, M., and R. Reed. 1999. Splicing is required for rapid and efficient mRNA export in metazoans. Proc. Natl. Acad. Sci. USA 96:14937-14942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lykke-Andersen, J., M. D. Shu, and J. A. Steitz. 2001. Communication of the position of exon-exon junctions to the mRNA surveillance machinery by the protein RNPS1. Science 293:1836-1839. [DOI] [PubMed] [Google Scholar]
- 30.McCracken, S., M. Lambermon, and B. J. Blencowe. 2002. SRm160 splicing coactivator promotes transcript 3′-end cleavage. Mol. Cell. Biol. 22:148-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matsumoto, K., K. M. Wassarman, and A. P. Wolffe. 1998. Nuclear history of a pre-mRNA determines the translational activity of cytoplasmic mRNA. EMBO J. 17:2107-2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mayeda, A., J. Badolato, R. Kobayashi, M. Q. Zhang, E. M., Gardiner, and A. R. Krainer. 1999. Purification and characterization of human RNPS1: a general activator of pre-mRNA splicing. EMBO J. 16:4560-4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Micklem, D. R., R. Dasgupta, H. Elliott, F. Gergely, C. Davidson, A. Brand, A. Gonzalez-Reyes, and D. St. Johnston. 1997. The mago nashi gene is required for the polarisation of the oocyte and the formation of perpendicular axes in Drosophila. Curr. Biol. 7:468-478. [DOI] [PubMed] [Google Scholar]
- 34.Mohr, S. E., S. T. Dillon, and R. Boswell. 2001. The RNA-binding protein Tsunagi interacts with Mago Nashi to establish polarity and localize oskar mRNA during Drosophila oogenesis. Genes Dev. 15:2886-2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nott, A., S. H. Meislin, and M. J. Moore. 2003. A quantitative analysis of intron effects on mammalian gene expression. RNA 9:607-617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ohno, M., A. Segref, S. Kuersten, and I. W. Mattaj. 2002. Identity elements used in export of mRNAs. Mol. Cell 9:659-671. [DOI] [PubMed] [Google Scholar]
- 37.Ouyang, P., and S. P. Sugrue. 1992. Identification of an epithelial protein related to the desmosome and intermediate filament network. J. Cell Biol. 118:1477-1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ouyang, P., and S. P. Sugrue. 1996. Characterization of Pinin, a novel protein associated with the desmosome-intermediate filament complex. J. Cell Biol. 135:1027-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ouyang, P. 1999. Antibodies differentiate desmosome-form and nucleus-form Pinin: evidence that Pinin is a moonlighting protein with dual location at the desmosome and within the nucleus. Biochem. Biophys. Res. Commun. 263:192-200. [DOI] [PubMed] [Google Scholar]
- 40.Reed, R., and E. Hurt. 2002. A conserved mRNA export machinery coupled to pre-mRNA splicing. Cell 108:523-531. [DOI] [PubMed] [Google Scholar]
- 41.Rodrigues, J. P., M. Rode, D. Gatfield, B. J. Blencowe, M. Carmo-Fonseca, and E. Izaurralde. 2001. REF proteins mediate the export of spliced and unspliced mRNAs from the nucleus. Proc. Natl. Acad. Sci. USA 98:1030-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schwerk, C., J. Prasad, K. Degenhardt, H. Erdjument-Bromage, E. White, P. Tempst, V. J. Kidd, J. L. Manley, J. M. Lahti, and D. Reinberg. 2003. ASAP, a novel protein complex involved in RNA processing and apoptosis. Mol. Cell. Biol. 23:2981-2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shi, Y., P. Ouyang, and S. P. Sugrue. 2000. Characterization of the gene encoding pnn/DRS/memA and evidence for its potential tumor suppressor function. Oncogene 19:289-297. [DOI] [PubMed] [Google Scholar]
- 44.Shi, Y., M. N. Simmons, T. Seki, S. P. Oh, and S. P. Sugrue. 2001. Change in gene expression subsequent to induction of Pnn/DRS/memA: increase in p21cip1/waf1. Oncogene 20:4007-4018. [DOI] [PubMed] [Google Scholar]
- 45.Takagaki, Y., and J. L. Manley. 2000. Complex protein interactions within the human polyadenylation machinery identify a novel component. Mol. Cell. Biol. 20:1515-1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tarn, W. Y., and J. A. Steitz. 1994. SR proteins can compensate for the loss of U1 snRNP functions in vitro. Genes Dev. 8:2704-2717. [DOI] [PubMed] [Google Scholar]
- 47.Wang, P., P. J. Lou, S. Leu, and P. Ouyang. 2002. Modulation of alternative pre-mRNA splicing in vivo by Pinin. Biochem. Biophy. Res. Commun. 294:448-455. [DOI] [PubMed] [Google Scholar]
- 48.Zahler, A. M., W. S. Lane, J. A. Stolk, and M. B. Roth. 1992. SR proteins: a conserved family of pre-mRNA splicing factors. Genes Dev. 6:837-847. [DOI] [PubMed] [Google Scholar]
- 49.Zhou, Z., M. J. Luo, K. Straesser, J. Katahira, E. Hurt, and R. Reed. 2000. The protein Aly links pre-messenger-RNA splicing to nuclear export in metazoans. Nature 407:401-405. [DOI] [PubMed] [Google Scholar]