Abstract
A recent analysis of gene expression in renal cell carcinoma cells led to the identification of mRNAs whose translation was dependent on the presence of the von Hippel-Lindau (VHL) tumor suppressor gene product, pVHL. Here, we investigate the finding that pVHL-expressing RCC cells (VHL+) exhibited elevated levels of polysome-associated p53 mRNA and increased p53 protein levels compared with VHL-defective (VHL−) cells. Our findings indicate that p53 translation is specifically heightened in VHL+ cells, given that (i) p53 mRNA abundance in VHL+ and VHL− cells was comparable, (ii) p53 degradation did not significantly influence p53 expression, and (iii) p53 synthesis was markedly induced in VHL+ cells. Electrophoretic mobility shift and immunoprecipitation assays to detect endogenous and radiolabeled p53 transcripts revealed that the RNA-binding protein HuR, previously shown to regulate mRNA turnover and translation, was capable of binding to the 3′ untranslated region of the p53 mRNA in a VHL-dependent fashion. Interestingly, while whole-cell levels of HuR in VHL+ and VHL− cells were comparable, HuR was markedly more abundant in the cytoplasmic and polysome-associated fractions of VHL+ cells. In keeping with earlier reports, the elevated cytoplasmic HuR in VHL+ cells was likely due to the reduced AMP-activated kinase activity in these cells. Demonstration that HuR indeed contributed to the increased expression of p53 in VHL+ cells was obtained through use of RNA interference, which effectively reduced HuR expression and in turn caused marked decreases in p53 translation and p53 abundance. Taken together, our findings support a role for pVHL in elevating p53 expression, implicate HuR in enhancing VHL-mediated p53 translation, and suggest that VHL-mediated p53 upregulation may contribute to pVHL's tumor suppressive functions in renal cell carcinoma.
Von Hippel-Lindau (VHL) disease is a cancer syndrome that arises through inactivation of the VHL tumor suppressor gene and is inherited with an autosomal dominant pattern. Affected individuals develop numerous tumors in multiple organs, such as renal cell carcinomas (RCCs), retinal angiomas, pheochromocytomas, hemangioblastomas of the cerebellum and spine, endolymphatic sac tumors, and pancreatic adenomas. In addition, biallelic mutations in the VHL gene are also found in up to 80% of sporadic clear cell RCCs (15, 18, 52) and in hemangioblastoma (39).
pVHL, the product of the VHL gene (38), is expressed as two isoforms: a predominant 24- to 30-kDa isoform, and a less abundant 19-kDa isoform that is synthesized from an internal translation start site within the same transcript (4, 28, 50). Both isoforms appear to have similar functions and biochemical properties, so pVHL will be used to refer to both of them. pVHL is the substrate recognition component of a multisubunit complex (VCB-CUL2) that also contains elongin C, elongin B, cullin 2 (Cul2), and Rbx1/ROC1 (for review, see reference 29). The VCB-CUL2 complex functions as an E3 ubiquitin ligase that ubiquitinates certain proteins, targeting them for degradation by the proteasome.
The best-studied substrates of pVHL-mediated proteolysis are the α subunits of the hypoxia inducible factor (HIF). In normal cells, normal oxygen conditions signal the hydroxylation of HIFα at key proline residues, thereby rendering it a suitable target for pVHL-mediated ubiquitination and rapid proteasome-mediated degradation. However, under low-oxygen conditions, HIFα subunits accumulate and associate with HIF-1β/ARNT. The resulting heterodimer binds to specific hypoxia response elements (HREs) present in genes that control angiogenesis and erythropoiesis, such as those for vascular endothelial growth factor, Glut-1, platelet-derived growth factor alpha, and erythropoietin (6, 14, 51). Recently, pVHL was also shown to bind the large subunit (Rbp1) of RNA polymerase II when Rbp1 is phosphorylated and hydroxylated at proline residues. This interaction likewise targets Rbp1 for ubiquitin-mediated proteolysis (37).
However, pVHL appears to have biological functions unrelated to its influence on HIFα or Rbp1 levels. First, VHL alleles associated with type 2C VHL disease (in which affected individuals develop only pheochromocytoma) retain the ability to ubiquitinate HIFα. Second, analyses of gene expression profiles revealed that pVHL and HIFα regulate overlapping but not identical sets of genes (4, 59). Third, expression of HIF-1α variants that escape pVHL regulation does not recapitulate the formation of VHL-related tumors and cysts, although these effects remain to be tested in relevant target organs and with HIF-2α (11). Fourth, pVHL has been proposed to influence intracellular protein processing. Evidence that pVHL participates in such surveillance comes from studies showing that VHL-deficient cells are hypersensitive to the toxicity of agents that cause endoplasmic reticulum stress (22) and from reports suggesting pVHL's influence on the function of a cytosolic protein-folding complex called CCT (23). Furthermore, pVHL binds to fibronectin and affects its deposition in the extracellular matrix; while the precise mechanisms underlying this process remain poorly understood, they may be linked to pVHL's association with the endoplasmic reticulum and its potential involvement in the retrograde transport of misprocessed fibronectin (44). In addition, pVHL may influence extracellular matrix function by regulating integrin assembly (12) and the expression of enzymes (such as matrix metalloproteinases and tissue inhibitors of metalloproteinases) involved in remodeling the extracellular matrix (34).
Despite pVHL's widely recognized role as a regulator of protein stability, most studies to date have focused on the identification of pVHL's influence on the patterns of expressed mRNAs, both through single-gene and high-throughput gene expression analyses. For example, pVHL-defective cells have been found to express higher levels of mRNAs encoding carbonic anhydrases 9 and 12, plasminogen activator inhibitor 1, transforming growth factor alpha, vascular endothelial growth factor, platelet-derived growth factor, and the glucose transporter Glut-1 (7, 27, 48, 58, 60; reviewed in reference 29). pVHL has been postulated to contribute to such gene expression differences through its ability to influence (i) transcription initiation, through its effects on the half-life of transcription factors such as Jade-1 and HIFα (42, 45, 61); (ii) transcription elongation, through an inhibitory association with elongin/SIII-containing complexes, as illustrated for the tyrosine hydroxylase gene (10, 35), and through ubiquitination of Rbp1 (37); and (iii) mRNA turnover, as exemplified in vascular endothelial growth factor mRNA, the stability of which is markedly higher in VHL-deficient cells (19, 27, 42, 45).
While some controversy remains regarding the subcellular localization of pVHL, it has been reported to reside in both the nucleus and the cytoplasm and possibly shuttles between the two compartments (26, 40). Moreover, pVHL was shown to regulate the polysomal association of important RNA-binding proteins such as heteronuclear ribonucleoprotein A2 (whose abundance has been reported to be regulated by pVHL) and heteronuclear ribonucleoprotein L (46). Cytoplasmic HuR has been found in association with the endoplasmic reticulum and the polysomal fraction (46, 49). Based on these observations and the fact that other RNA-binding proteins that critically regulate mRNA stability and translation reside in the polysomal compartment of the cell, we recently investigated the influence of pVHL on the polysomal association of expressed transcripts (16). In that study, RCC cells that either lacked (VHL−) or expressed VHL through stable transfection (VHL+) were used to prepare RNA from cytosolic and polysome-bound fractions. Hybridization of cDNA arrays with RNA from each fraction revealed subsets of transcripts whose abundance in polysomes either increased or decreased in cells with restored VHL function. The tumor necrosis factor alpha (TNF-α) mRNA, identified as one of the transcripts that preferentially associated with polysomes in VHL− cells, was found to be one of the targets of translational repression by VHL (16).
Here, we investigated the role of VHL in enhancing translation of a different set of transcripts, among which was the mRNA encoding the p53 tumor suppressor protein. Our findings reported here support the notion that increased p53 mRNA translation in VHL+ cells contributes to the increased expression of p53. Moreover, the RNA-binding protein HuR, a member of the ELAV family of proteins involved in mRNA stability and translation (5, 36, 41, 43) was found to bind to the p53 mRNA in vitro and in vivo and to enhance p53 expression levels, in agreement with our previous findings (41). Interestingly, the cytoplasmic localization of HuR was significantly enhanced in VHL+ cells, an effect that was attributed to the reduced AMP-activated protein kinase (AMPK) activity observed in these cells, and HuR was specifically found in association with polysomes, in keeping with earlier reports (57).
In support of HuR's ability to regulate p53 expression, silent interfering (siRNA)-based approaches to reduce HuR levels effectively reduced p53 translation and p53 abundance. pVHL's influence on p53 expression likely represented a general phenomenon in this system, as restoration of pVHL expression in three different RCC lines (786-O, UMRC6, and UOK121) also led to elevations in p53 expression. Taken together, our findings support a role for pVHL in elevating p53 expression through enhanced p53 translation and implicate HuR in these regulatory events.
MATERIALS AND METHODS
Cell culture, treatment and transfection of plasmids and siRNA.
786-O (26) cells that either lacked VHL (VHL−) or expressed the wild-type VHL (VHL+) through stable transfection were a generous gift from W. G. Kaelin and O. Iliopoulos. UOK121 and UMRC6 cells (7), each lacking VHL function, were stably transfected with pCEP4-VHL (kindly obtained from B. Zbar and M. Lerman), and pooled populations selected in the presence of 400 μg of hygromycin B per ml (Sigma, St. Louis, Mo.). All cell lines were maintained in high-glucose Dulbecco's modified Eagle's medium (DMEM; Gibco-BRL) containing 10% fetal bovine serum (HyClone, Logan, Utah). HuR4 siRNA, targeting the coding region of HuR (GenBank BC003376), was AACACGCTGAACGGCTTGAGG; Ctrl siRNA (a control sequence not matching any known human gene) was AAGTGTAGTAGATCACCAGGC. siRNAs (Ambion, Inc., Austin, Tex.) were transfected at a concentration of 20 nM with Oligofectamine (Invitrogen, Carlsbad, Calif.). 5-Amino-imidazole-4-carboxamide riboside (AICAR) was from Sigma.
Pulse labeling of endogenous protein.
Populations of 786-O cells (200,000 cells per 6-cm dish) were cultured in complete DMEM (Gibco-BRL) supplemented with 10% fetal bovine serum for 24 h prior to a 30-min starvation period in methionine- and cysteine-free DMEM containing 5% dialyzed fetal bovine serum. Cells were then incubated for 20 min with 900 μCi of l-[35S]methionine and l-[35S]cysteine (NEG-072 Expre35S35S protein labeling mix; NEN/Perkin Elmer, Boston, Mass.) per well and immediately scraped into 500 μl of ice-cold phosphate-buffered saline. After centrifugation (1,800 rpm, 4°C, 1 min), pellets were shock-frozen in liquid nitrogen and resuspended in 100 μl of TSD lysis buffer (50 mM Tris [pH 7.5], 1% sodium dodecyl sulfate [SDS], and 5 mM dithiothreitol). After boiling and determining protein concentration, 100-μg protein aliquots were brought to a final volume of 1.2 ml in TNN buffer (50 mM Tris [pH 7.5], 250 mM NaCl, 5 mM EDTA, 0.5% NP-40, 1 mM phenylmethylsulfonyl fluoride, 2 ng of aprotinin per ml, and 2 ng of leupeptin per μl) and precleared with protein A/G (1:1)-Sepharose mix (Sigma) for 1 h at 4°C. Following a brief centrifugation, supernatants were incubated for 1 h at 4°C with 0.4 μg of either a monoclonal antibody that recognizes p53 (D0-1; Santa Cruz Biotechnology, Inc., Santa Cruz, Calif.) or immunoglobulin G1 (BD Biosciences Pharmingen, San Diego, Calif.) and precipitated with 50 μl of protein A/G (1:1) mix (Sigma) for 1 h at 4°C. Following washes in TNN buffer, immunoprecipitated material was resolved by electrophoresis in SDS-containing 12% polyacrylamide gels, transferred onto polyvinylidene difluoride filters, and visualized with a PhosphorImager (Molecular Dynamics).
Northern and Western blotting.
Total RNA was isolated from cells and Northern blot analysis was performed as previously described (20). The p53 mRNA as well as the 18S rRNA (used to monitor even loading and transfer of samples) were detected with specific oligonucleotides GTGAACCATTGTTCAATATCGTCCGGGGACAGCATCAAATCATCCATTGCTTGGG and ACGGTATCTGATCGTCTTCGAACC, respectively, that were end labeled with [α-32P]dATP and terminal transferase, as described previously (21).
For Western blot analysis, protein aliquots (typically 5 to 20 μg) were size-fractionated with Bio-Rad Tris-HCl gels and transferred onto polyvinylidene difluoride membranes. Hybridizations were carried out in the presence of monoclonal antibodies recognizing p53, p21, β-tubulin, HDAC1 (Santa Cruz Biotechnology, Inc., Santa Cruz, Calif.), pVHL (NeoMarkers, Fremont, Calif.), or β-actin (Abcam, Cambridge, United Kingdom). Following incubation with the appropriate secondary antibodies, Western signals were visualized with enhanced chemiluminescence (Amersham).
Polysome analysis.
Five million cells were used per sucrose gradient. Cells were incubated for 3 min with 0.1 mg of cycloheximide at 37°C per ml and then lysed in 1 ml of PEB lysis buffer (0.3 M NaCl, 15 mM MgCl2, 15 mM Tris-HCl [pH 7.6], 1% Triton X-100, 0.1 mg of cycloheximide per ml, 1 mg of heparin per ml). After a 10-min incubation on ice, nuclei were pelleted by centrifugation at 10,000 × g (4°C, 10 min), and the resulting supernatant was carefully layered onto a 10 to 50% linear sucrose gradient. Gradients were centrifuged at 35,000 × g for 3 h at 4°C, and fractions were collected at a rate of 1 ml per min with a set-up that comprised a syringe pump, needle-piercing system (Brandel, Gaithersburg, Md.), UV-6 detector, and fraction collector (ISCO, Lincoln, Neb.). For Western blotting, equal volumes (≈100 μl) of each fraction were added to 4× protein dye mix (400 mM dithiothreitol, 40% glycerol, 200 mM Tris-HCl [pH.6.8], 8% SDS, 0.01 mg of bromophenol blue per ml), boiled, and electrophoresed through SDS-12% polyacrylamide gels. Northern blotting was performed with RNA that was isolated from each fraction with STAT-60; equal volumes (≈15 μl) of purified RNA from each fraction were used for analysis.
cDNA array analysis.
RNA was prepared from polysome-bound or from unbound fractions, then reverse transcribed in the presence of [α-33P]dCTP, and used to hybridize cDNA arrays (focused array; 4,608 genes; described at http://www.grc.nia.nih.gov/branches/rrb/dna/array.htm) as previously described (13). All data were first normalized by Z score transformation (58). In brief, the log base 10 of each original spot intensity was adjusted to the mean and divided by the standard deviation of all the spot intensities. Changes in gene expression between different RNA groups were calculated by subtracting the average of triplicate measurements. This value, referred to as the Z difference (Zdiff, Z average in VHL− cells minus Z average in VHL+ cells), is tested for significance with a two-tailed Z test [Z ≤ 2.4] for the difference between two means. All values reported were significant at P ≤ 0.05. The data reflect three independent experiments. +, Zdiff between 0 and 0.25; ++, Zdiff between 0.25 and 0.5; +++, Zdiff between 0.5 and 0.75. Genes exhibiting no significant difference in their relative distribution (polysomal compared with nonpolysomal) in cells with different VHL statuses are described elsewhere (16).
In vitro transcription, REMSA supershift, and biotin pull-down assays.
Reverse-transcribed total RNA was used for the amplification of p53 cDNA segments to be used as templates for in vitro transcription of the p53 3′ untranslated region (3′UTR) and coding region. All 5′ oligonucleotides contain the T7 RNA polymerase promoter sequence CCAAGCTTCTAATACGACTCACTATAGGGAGA (T7) as previously described (56). Preparation of the template to synthesize the p53 coding region (positions 252 to 1439) was carried out with primers (T7)ATGGAGGAGCCGCAGTCAGATCCTAGC and AGAATGTCAGTCTGAGTCAGGC. Preparation of the p53 3′UTR (positions 1421 to 2629) was carried out with primers (T7)TGACTCAGACTGACATTCTCC and TGGCAGCAAAGTTTTATTGTAAAATAAGAGATCG. RNA transcripts were synthesized with T7 RNA polymerase in the presence of either [α-32P]UTP or biotinylated CTP and purified as described previously (55). Binding reactions in the presence of radiolabeled transcripts were followed by RNA electrophoretic mobility shift assays (REMSA) to visualize complexes (56); for supershift assays, 0.5 μg of either anti-HuR or anti-p38 antibodies (Santa Cruz Biotech, Santa Cruz, Calif.) was used. Proteins associating with biotinylated transcripts were pulled down with streptavidin-coated magnetic beads (55) and assessed by Western blot analysis with anti-p53 and anti-β-actin antibodies.
HuR immunoprecipitation for identification of target transcripts.
Immunoprecipitation of endogenous HuR-mRNA complexes, used to assess the association of endogenous HuR with endogenous p53 mRNA, was performed as previously described (53). Twenty million RCC cells were collected per sample, and lysates were used for immunoprecipitation for 4 h at room temperature in the presence of excess (30 μg) immunoprecipitation antibody [either mouse monoclonal anti-HuR antibody 3A2 (Santa Cruz) or IgG1 (BD Pharmingen)]. RNA in immunoprecipitation material was used in reverse transcription-PCRs to detect the presence of p53 mRNA; the p53 coding region was amplified with the primer pair described above and the following amplification conditions: 1 min at 94°C, 1 min at 55°C, and 1 min at 68°C for 35 cycles. PCR products were visualized by ethidium bromide staining of 1.5% agarose gels.
AMPK assay.
AMPK activity was assayed as described previously (25, 57). Briefly, AMPK was immunoprecipitated from 10 μg of cell lysate with 1 μg of anti-α1 and 1 μg of anti-α2 polyclonal antibodies (kindly provided by D. Carling and D. G. Hardie) in AMPK immunoprecipitation buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 50 mM NaF, 5 mM sodium pyrophosphate, 1 mM EDTA, 1 mM EGTA, 1 mM dithiothreitol, 0.1 mM benzamidine, 0.1 mM phenylmethylsulfonyl fluoride, and 5 μg of soybean trypsin inhibitor per ml) for 2 h at 4°C. Immunocomplexes were washed with immunoprecipitation buffer plus 1 M NaCl and then with a buffer containing 62.5 mM HEPES [pH 7.0], 62.5 mM NaCl, 62.5 mM NaF, 6.25 mM sodium pyrophosphate, 1.25 mM EDTA, 1.25 mM EGTA, 1 mM dithiothreitol, 1 mM benzamidine, 1 mM phenylmethylsulfonyl fluoride, and 5 μg of soybean trypsin inhibitor per ml. AMPK activity in immunocomplexes was determined by phosphorylation of the peptide HMRSAMSGLHLVKRR [SAMS (25)] in reaction buffer (50 mM HEPES [pH 7.4] 1 mM dithiothreitol, 0.02% Brij-35, 0.25 mM SAMS, 0.25 mM AMP, 5 mM MgCl2, 10 μCi of [α-32P]ATP) for 10 min at 30°C. Assay mixtures were spotted onto P81 filter paper and rinsed in 1% (vol/vol) phosphoric acid with gentle stirring to remove free ATP. Phosphorylated substrate was measured by scintillation counting.
Immunofluorescence.
Cells were seeded on coverslips, fixed for 15 min in phosphate-buffered saline containing 4% paraformaldehyde, and permeabilized for 15 min in phosphate-buffered saline containing 0.4% Triton X-100. After incubation for 16 h in blocking buffer (phosphate-buffered saline containing 2% bovine serum albumin and 0.1% Tween 20), coverslips were incubated for 1 h in a 1:500 dilution of mouse anti-HuR (Santa Cruz Biotech.) prepared in blocking buffer. Following washes with PBS containing 0.1% Tween 20, samples were incubated for 1 h with a mixture of horse anti-mouse immunoglobulin conjugated with Texas Red (1:200; Jackson Laboratories) and Hoechst 33342 (1:5,000; Molecular Probes). After washes with PBS containing 0.1% Tween 20, coverslips were mounted in Vectashield (Vector Laboratories) and visualized with an Axiovert 200 M microscope (Zeiss; 63× lens) with separate channels for the analysis of phase contrast images, red fluorescence, and blue fluorescence. Images were then processed with the AxioVision 3.0 program (Zeiss). Representative photographs from two independent experiments are shown.
RESULTS
p53 mRNA preferentially associates with the polysomal fraction of RCC cells expressing pVHL.
In a recent study, we assessed the influence of pVHL on the translation of specific mRNAs by an approach based on cDNA array analysis (16, 31, 62). Briefly, cytoplasmic lysates from both VHL-expressing (VHL+) and VHL-defective (VHL−) 786-O cells were centrifuged through sucrose gradients and then collected in fractions ranging from lightest (cytosolic samples devoid of ribosomes) to heaviest (largest polysomes). Untranslated RNA was prepared from fractions lacking any ribosomes or ribosome subunits (unbound mRNA). mRNA engaged in translation was prepared from fractions containing monosomes, low-molecular-weight polysomes, and high-molecular-weight polysomes (polysome-bound mRNA). Both sets of mRNAs (unbound and polysome-bound) from each cell line were used in reverse transcription reactions to prepare radiolabeled probes for hybridization of cDNA arrays [human focused arrays comprising 4,608 genes; http://www.grc.nia.nih.gov/branches/rrb//dna/array.htm (16)].
Pairwise comparisons of unbound (untranslated) and polysome-bound (translationally active) mRNAs were carried out between VHL− and VHL+ cells, and differences in the relative distribution of translationally active mRNAs were quantitated as Z averages (54). mRNAs that were found to be more abundant in polysomes of VHL+ cells, i.e., mRNAs that were potential targets of pVHL-mediated translational induction, were considered significant if they met the criteria outlined in the Materials and Methods section. Table 1 lists several genes on the array that met these selection criteria, out of a total of approximately 194 genes (4.2% induced genes of the 4,608 genes on the array). The degree to which each mRNA listed is preferentially associated with polysomes in VHL+ cells was assessed by comparing Z differences (Zdiff), calculated as the Z average in VHL+ cells compared with that in VHL− cells.
TABLE 1.
mRNAs preferentially associated with polysomes in VHL-expressing cellsa
| Unigene no. | Gene | Relative abundance |
|---|---|---|
| Hs.1030 | Ras inhibitor | ++ |
| Hs.75356 | Transcription factor 4 | + |
| Hs.75162 | MAPK-activated protein kinase 3 | ++ |
| Hs.118825 | MAPK kinase 6 | ++ |
| Hs.1846 | Tumor suppressor protein p53 | +++ |
| Hs.79362 | Retinoblastoma-like protein 2 | ++ |
For each gene on the cDNA arrays (described at http://www.grc.nia.nih.gov/branches/rrb/dna/dnapubs.htm), the relative presence of the encoded mRNA in polysome-bound and unbound fractions was compared between VHL-expressing (VHL+) and VHL− deficient (VHL−) 786-O cell lines. Cell fractionations and cDNA array analysis were performed in three independent experiments (16). The list includes genes potentially subject to translational upregulation by pVHL, since they are encoded by mRNAs that were significantly more abundant in polysome-bound fractions from VHL− cells than in those from VHL+ cells, as assessed through comparison of Z averages (described in reference 16). Briefly, for each gene on the array, mRNA abundance in each fraction (polysomal and nonpolysomal) was compared between VHL− and VHL+ cells. Data analysis was carried out as explained in the Materials and Methods section and in reference 16. Differences in Z averages (Zdiff) served to assess the degree to which a given transcript was preferentially associated with polysomes in VHL+ cells and was represented as follows: +, Zdiff between 0 and 0.25; ++, Zdiff between 0.25 and 0.50; +++, Zdiff between 0.50 and 0.75. Genes encoding mRNAs that exhibited no significant difference in their relative distribution (polysomal compared with nonpolysomal) in cells with different VHL statuses were described previously (16).
In the table, such differences are represented in the relative abundance column, and they ranged from moderate (+) to pronounced (+++) differences. A complete list is available athttp://www.grc.nia.nih.gov/branches/rrb/dna/dnapubs.htm. With a similar strategy, we recently demonstrated that the tumor necrosis factor alpha mRNA was preferentially associated with polysomes in VHL− cells and hence was subject to translational repression by VHL (16).
In VHL− RCC cells, ectopic expression of pVHL elevates p53 protein abundance and translation.
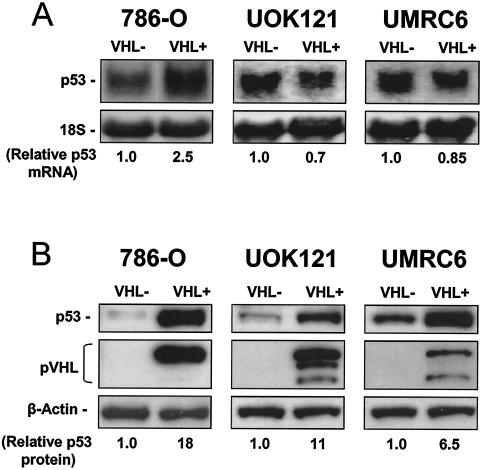
The finding that p53 mRNA was more abundant in the translating fraction of VHL+ cells compared with VHL− cells was of great potential interest, so we set out to further investigate these differences. First, we examined p53 mRNA and protein levels in 786-O cells. As shown in Fig. 1A, p53 mRNA signals were found to be either moderately upregulated (on the order of two- to threefold), as seen in VHL+ 786-O cells (26), or unchanged, as observed in two additional RCC lines lacking VHL function (UMRC6 and UOK121), in which wild-type pVHL had been reintroduced by transfection (Materials and Methods). Strikingly, however, 6- to 18-fold-higher p53 protein signals were seen when comparing VHL+ and VHL− cells (Fig. 1B).
FIG. 1.
Expression of p53 in RCC cells with different VHL statuses. (A) Representative Western blot analysis to monitor p53 and pVHL expression in 786-O, UOK121 and UMRC6 renal cell carcinoma (RCC) cells, in which pVHL expression was restored by transfection (described for 786-O cells in reference 2 and in Materials and Methods for UOK121 and UMRC6). β-Actin signals on the same filters illustrate even loading of samples. VHL-, parental populations (VHL-deficient); VHL+, VHL-restored populations (expressing wild-type VHL through stable transfection). (B) Representative Northern blot analysis of p53 mRNA abundance in RCC cells with different VHL statuses. 18S rRNA signals served to monitor loading differences among samples. Relative abundances of p53 mRNA and protein are the mean values obtained from three independent experiments.
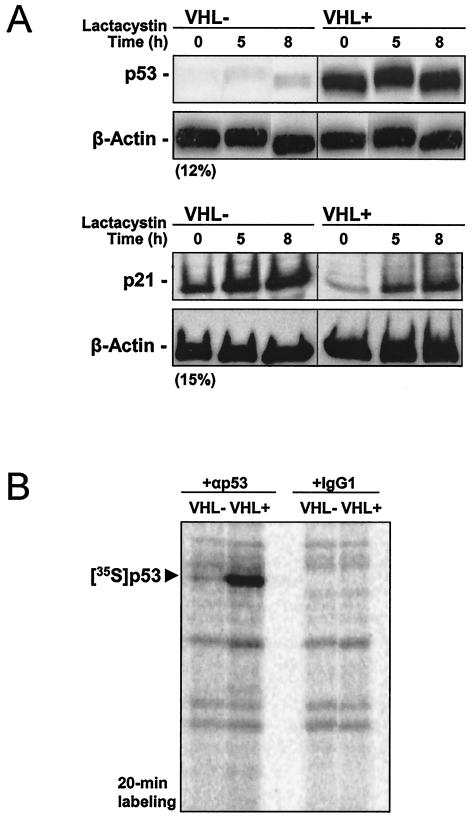
Given that the p53 protein is subject to rapid turnover (47), it was important to investigate whether the elevated steady-state p53 levels in VHL+ cells were due to decreased p53 degradation or increased p53 translation. First, we incubated VHL+ and VHL− 786-O cells in the presence of the proteasome inhibitor lactacystin to assess the contribution of ubiquitin-mediated proteolysis towards elevating p53 expression in this cell line. As shown in Fig. 2A, treatment of cells with lactacystin contributed little to upregulating p53 expression, even at relatively high concentrations (10 μM) of the proteasome inhibitor for extended time periods. Shorter incubations with lactacystin revealed comparable effects (not shown). Evidence that the cyclin-dependent kinase inhibitor p21, whose steady-state levels are modulated through proteasome-mediated proteolysis, was markedly more abundant following lactacystin treatment served to verify that the proteasome inhibitor was active at this concentration (Fig. 2A).
FIG. 2.
Analysis of p53 stability and translation in cells with different VHL statuses. (A) We prepared 20 μg of whole-cell protein extracts from 786-O cells that either lacked (VHL-) or expressed (VHL+) VHL after treatment with 10 μM lactacystin for the times indicated. Western blot analysis of expression of p53 (top) and p21 (bottom), as well as β-actin (loading control) after resolving samples by electrophoresis in SDS-containing gels with either 12% (top) or 15% (bottom) polyacrylamide. (B) Cells were incubated for 20 min in the presence of l-[35S]methionine and l-[35S]cysteine, whereupon nascent p53 was visualized by immunoprecipitation as described in the Materials and Methods section. Samples were resolved by electrophoresis in SDS-containing 12% polyacrylamide gels. Radiolabeled p53 signal is indicated.
Therefore, we turned our attention to examining the rate of p53 protein synthesis in VHL+ and VHL− cells. Cells were incubated in the presence of l-[35S]methionine and l-[35S]cysteine for 20 min, whereupon newly translated p53 was visualized through immunoprecipitation with an anti-p53 antibody (Materials and Methods). The brevity of the incubation period further ensured that we measured differences in p53 synthesis without having to account for potential effects of protein degradation. As shown in Fig. 2B, VHL+ cells expressed remarkably higher levels of newly synthesized p53 protein, lending strong support to the notion that VHL-expressing cells were capable of increased p53 translation.
Binding of the RNA-binding protein HuR to the p53 3′UTR is enhanced when lysates from VHL+ cells are used.
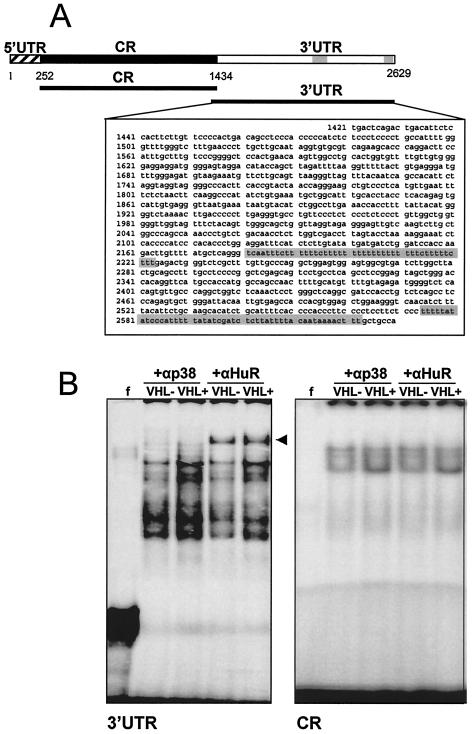
In order to investigate how pVHL influences p53 translation, we sought to focus on the association of specific regulatory proteins to the p53 mRNA. Given the reported ability of HuR and other ELAV proteins to influence translation as well as mRNA stability (reviewed in reference 33) and our previous finding that HuR was capable of elevating p53 translation following UV stress (41), we set out to directly investigate HuR's potential contribution to the VHL-dependent p53 upregulation in RCC cells. The p53 3′UTR is approximately 1.2 kb long and contains U- and AU-rich stretches, depicted as gray shading in the schematic of Fig. 3A. In order to investigate the association of p53 mRNA with proteins present in 786-O lysates, transcripts encompassing either the coding region or the 3′UTR of the p53 mRNA (Fig. 3A) were synthesized in vitro with biotinylated or radiolabeled nucleotides.
FIG. 3.
Association of HuR with the p53 3′UTR. (A) Schematic of p53 mRNA, depicting U- and AU-rich regions in the 3′UTR (shaded regions), as well as transcripts prepared for RNA-binding analysis. CR, coding region. (B) Radiolabeled transcripts encompassing either the p53 3′UTR or the p53 coding region were incubated with cytoplasmic lysates from 786-O cells (10 μg each) and subjected to REMSA supershift assays in the presence of either a control antibody recognizing mitogen-activated protein kinase p38 [a protein that lacks RNA-binding activity (+αp38 lanes)] or an antibody recognizing HuR (+αHuR lanes).
Evidence that the p53 mRNA indeed associated with HuR in RCC cells was obtained from experiments with radiolabeled p53 transcripts. Depicted in Fig. 3B are the results obtained with RNA electrophoretic mobility shift assays (REMSAs) to resolve complexes containing either the p53 3′UTR or the p53 coding region under native conditions. As shown, the abundance of complexes forming with the p53 3′UTR was greater with lysates from VHL+ cells compared with lysates from VHL− cells (Fig. 3B). The presence of HuR in such complexes was evidenced by use of an anti-HuR antibody, which specifically caused the appearance of a slower-migrating (supershifted) band, whereas control antibodies recognizing the mitogen-activated protein kinase (MAPK) p38, which does not bind RNA, or RNA-binding proteins such as AUF1, TTP, and TIAR (not shown) did not form supershifts. The enhanced association of p53 3′UTR with HuR from VHL+ cell lysates was further evidenced by the more intense supershift seen in REMSA analysis of VHL+ cells. By contrast, use of a transcript encompassing the coding region revealed faint complexes regardless of VHL status, and the complexes were not supershifted with anti-HuR antibodies (Fig. 3B), indicating the specificity of the association between HuR and the p53 3′UTR and its dependence on the VHL status of the cell.
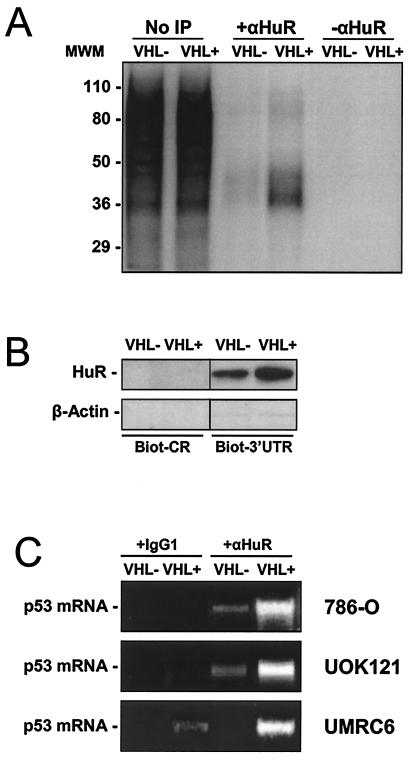
Additional evidence of VHL-dependent differences in the interaction between HuR and the p53 3′UTR was obtained through incubation of cytoplasmic lysates from either VHL+ or VHL− 786-O cells with radiolabeled p53 3′UTR, cross-linking the resulting complexes with UV light, and immunoprecipitating the resulting complexes in either the presence or absence of a specific anti-HuR antibody. Aliquots of UV-cross-linked material that was not immunoprecipitated revealed that the p53 mRNA associated with numerous cytoplasmic proteins (Fig. 4A, No IP lanes). However, radiolabeled p53 3′UTR transcript was more abundantly associated with HuR in lysates from VHL+ cells, as determined by use of a specific anti-HuR antibody in immunoprecipitation reactions. Immunoprecipitation of cross-linked material without a specific anti-HuR antibody produced no radiolabeled bands (Fig. 4A), in support of the specific association of HuR with the 3′UTR of p53.
FIG. 4.
Association of endogenous HuR with synthetic p53 transcripts and with endogenous p53 mRNA in RCC cells. (A) Radiolabeled p53 3′UTR transcript was incubated with cytoplasmic lysates from 786-O cells and then immunoprecipitated in either the presence or absence of anti-HuR antibody (αHuR). MWM, size markers (in kilodaltons). (B) Biotinylated p53 transcripts (1 μg each) encompassing either the coding region or the 3′UTR were tested for their ability to pull down HuR from cytoplasmic lysates of 786-O cells with different VHL statuses, as described in the Materials and Methods section. The presence of β-actin in pull-down material was monitored in order to assess the specificity of the assay. (C) Immunoprecipitation with either anti-HuR antibody or IgG1 under conditions that preserve the association of RNA-binding proteins with target mRNAs was followed by reverse transcription-PCR analysis to detect endogenous p53 mRNA in the three RCC cell lines shown. PCR products were resolved by electrophoresis in 1.5% agarose gels and visualized by staining with ethidium bromide.
Pull-down assays (described in the Materials and Methods section) were also carried out to assess the interaction between p53 3′UTR and HuR in RCC lines. Biotinylated transcripts encompassing either the p53 coding region or p53 3′UTR were incubated with proteins present in the cytoplasm of VHL+ and VHL− cells. As shown, the p53 3′UTR was capable of pulling down HuR present in cytoplasmic lysates from both VHL+ and VHL− cells, but HuR was more abundant in pull-down reactions with VHL+ lysates (Fig. 4B). The findings that a biotinylated p53 coding region transcript (devoid of AU-rich regions) did not pull down HuR and that neither p53 coding region nor p53 3′UTR transcripts could pull down β-actin (a protein lacking RNA-binding activity) served to verify the specificity of the assay (Fig. 4B).
Finally, evidence for the in vivo association of endogenous p53 mRNA and HuR in the three RCC lines was obtained through immunoprecipitation of HuR under conditions that preserved its association with target mRNAs, using a previously described method (53). Reverse transcription-PCR analysis revealed the presence of endogenous p53 mRNA in the material immunoprecipitated with anti-HuR antibodies, but no p53 mRNA (or much less p53 mRNA, as seen in UMRC6 cells) when nonspecific antibodies (IgG1) were employed (Fig. 4C). This association was much more prominent in VHL+ populations. Taken together, these data indicate that the p53 3′UTR forms complexes with proteins present in lysates from RCC cells, is a target of HuR, and forms more abundant complexes with HuR in VHL+ cells.
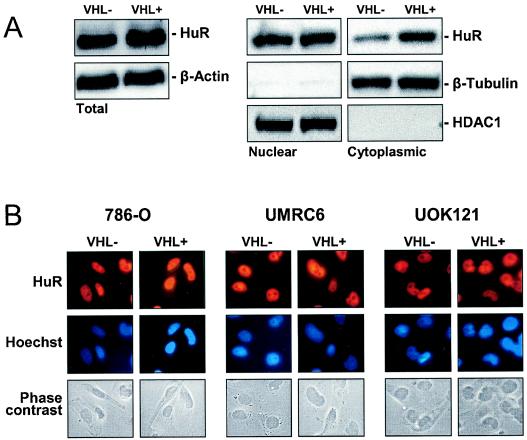
pVHL-expressing cells exhibit greater HuR presence in the cytoplasm.
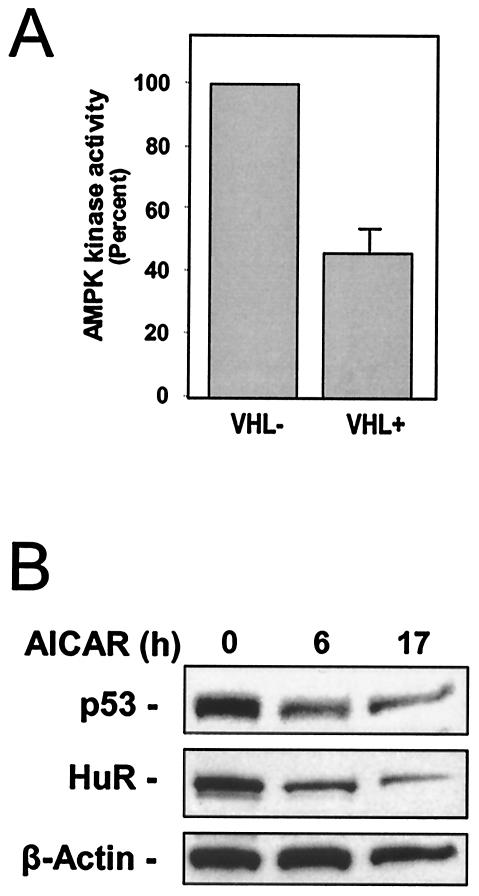
Interestingly, HuR abundance in whole-cell lysates was essentially unchanged when comparing VHL− and VHL+ cells. However, when comparing cytoplasmic levels of HuR, these were found to be markedly elevated in VHL+ compared with VHL− cells, as determined by Western blot analysis (Fig. 5A) and by immunofluorescence (Fig. 5B). The levels of HuR present in nuclear lysates (which typically comprise >90% of total cellular HuR) were unchanged, in keeping with earlier reports that nuclear HuR remains unchanged despite increased cytoplasmic HuR presence (55, 56). Elevated activity of the enzyme AMP-activated kinase (AMPK) was previously shown to promote the nuclear localization of HuR (57). Conversely, a reduction in AMPK activity caused increased cytoplasmic localization of HuR. As shown, basal AMPK activity in VHL+ cells was significantly lower than in VHL− cells (Fig. 6A). Reductions in AMPK activity of this magnitude, although modest, have been shown to significantly elevate the cytoplasmic levels of HuR (57). Accordingly, lowering of cytoplasmic HuR levels by treating cells with AICAR, an AMP analog widely used as a pharmacological activator of AMPK (57), caused a reduction in p53 expression (Fig. 6B).
FIG. 5.
Subcellular localization of HuR and AMPK activity in 786-O cells with different VHL statuses. (A) Whole-cell (20 μg), nuclear (10 μg), and cytoplasmic (40 μg) lysates were subjected to Western blot analysis to monitor the expression of HuR. Sequential hybridizations with antibodies against β-tubulin (a cytoplasmic protein) and HDAC1 (a nuclear protein) were carried out to assess the quality of the fractionation process and the uniformity in loading and transfer of samples. (B) Detection of HuR by immunofluorescence in RCC cells (786-O, UMRC6, and UOK121) with different VHL statuses. Top, HuR immunofluorescence; middle, Hoechst staining to visualize nuclei; bottom, phase contrast images. Representative photographs from two independent experiments are shown.
FIG. 6.
AMPK activity in 786-O cells with different VHL statuses and influence of AMPK activation on the cytoplasmic levels of HuR and p53. (A) Whole-cell lysates prepared from VHL+ and VHL− cells were used to measure AMPK activity. AMPK was immunoprecipitated with a polyclonal antibody that recognizes the AMPK α1 and α2 subunits; the in vitro kinase assay was performed with the synthetic peptide SAMS (described in the Materials and Methods section). (B) Western blot analysis of HuR and p53 expression in lysates (40 μg) prepared from 786-O cells treated for 6 h in the presence of AMPK activator AICAR (2 mM).
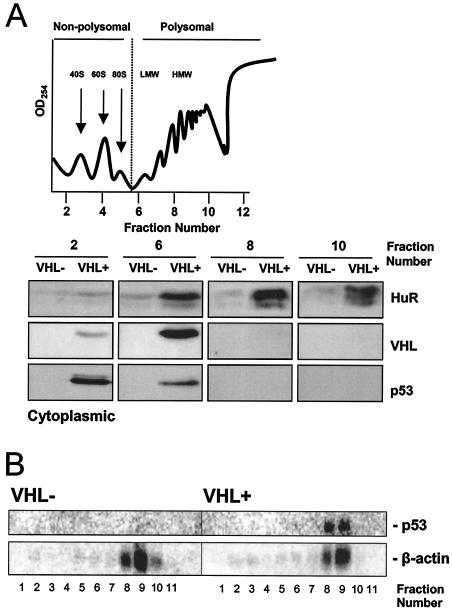
In order to gain further information on the subcellular localization of HuR in VHL+ cells, 786-O cytoplasmic lysates were fractionated by centrifugation through sucrose gradients, and HuR presence was assessed by Western blotting. As shown, VHL+ cells exhibited distinctly increased cytoplasmic HuR abundance in all fractions tested, with HuR localizing preferentially in the high-molecular-weight polysomal fractions, as previously observed in other cell types (17) (Fig. 7A). As shown, pVHL and p53 were also detected in both the nonpolysomal and low-molecular-weight polysomal fractions. The relative abundance of p53 mRNA in high-molecular-weight polysomes (Fig. 7B) was consistent with the presence of HuR in these fractions.
FIG. 7.
Cytoplasmic HuR and p53 mRNAs are associated with high-molecular-weight, actively translating polysomes in VHL+ cells. (A) Cytoplasmic lysates from 786-O cells were fractionated through sucrose gradients in order to separate unbound mRNA from mRNA bound to polysomes of increasing size. Top, representative profile. Bottom, representative Western blots depicting HuR signals in the unbound fraction 2, low-molecular-weight (LMW) polysome fraction 6, and high-molecular-weight (HMW) polysome fractions 8 and 10. Also shown are pVHL and p53 signals. (B) p53 mRNA is associated with high-molecular-weight, actively translating polysomes in VHL+ cells. Following fractionation of cytoplasmic lysates from 786-O cells, RNA was prepared from each of the 11 fractions and subjected to Northern blot analysis. Representative p53 mRNA signals are shown; β-actin mRNA signals were included to monitor differences in loading and transfer of samples.
Reduction of HuR expression decreases p53 translation and p53 steady-state levels.
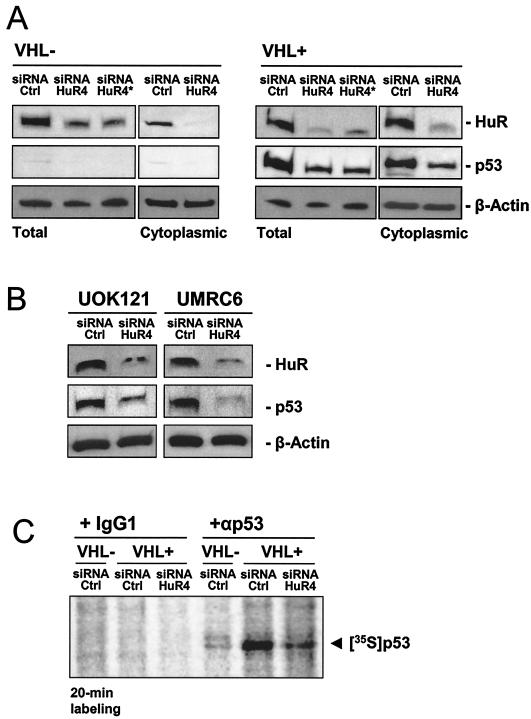
To directly investigate if HuR was regulating p53 expression in this system, we used an siRNA-based approach to diminish HuR expression in 786-O cells. Transfection of HuR-specific (HuR4) siRNA (Materials and Methods) specifically reduced HuR abundance in both VHL+ and VHL− 786-O cells compared with control transfections with a control siRNA (Fig. 8A). A sequential siRNA transfection protocol in which a second transfection was performed 24 h after the initial transfection achieved a reduction in HuR comparable to that resulting from a single transfection (Fig. 8A).
FIG. 8.
Small interfering RNA-mediated reduction of HuR expression causes a reduction in p53 levels in RCC cells. (A) 786-O cells were transfected with either control (Ctrl) or HuR-specific (HuR4) siRNA, and expression levels of HuR and p53 were assessed 2 days later in preparations of total protein or cytoplasmic protein (10 and 40 μg, respectively). *, second transfection with the same siRNA 24 h later; proteins were collected 2 days after the second transfection. (B) VHL+ RCC lines UOK121 and UMRC6 were transfected with either control (Ctrl) or HuR-specific (HuR4) siRNA, and expression levels of HuR and p53 were assessed 2 days later in whole-cell lysates (20 μg). Representative signals are shown. (C) Two days after transfection with either control siRNA or HuR-directed siRNA (HuR4), 786-O cells with different VHL statuses were incubated for 20 min in the presence of l-[35S]methionine and l-[35S]cysteine, whereupon nascent p53 was visualized by immunoprecipitation as described in the Materials and Methods section. Samples were resolved by electrophoresis in SDS-containing 12% polyacrylamide gels. Radiolabeled p53 signal is shown.
Reduced HuR levels caused a striking decrease in p53 abundance, an effect that was more evident in VHL+ cells (Fig. 8A). Similarly, siRNA-mediated reduction of HuR expression in UOK121 and UMRC6 cells also led to a decrease in p53 expression (Fig. 8B). To determine if this reduction was due to decreased p53 translation, siRNA-transfected 786-O cells were pulsed with l-[35S]methionine and l-[35S]cysteine, and cell lysates were subjected to immunoprecipitation with either an anti-p53 antibody or a control antibody. As shown (Fig. 8C), specific reduction of HuR expression caused a pronounced reduction in p53 translation compared with the control siRNA group. These findings strongly support a role for HuR as positive regulator of p53 translation in this system. In summary, restoration of pVHL expression in RCC cells increased HuR function, which in turn associated with the p53 mRNA and increased p53 translation.
DISCUSSION
In this investigation, we examined the positive influence of pVHL on p53 expression in RCC cells. Our findings reveal that pVHL elevates p53 expression by enhancing p53 translation and identify HuR as a central regulator of this process. A recent analysis of the influence of pVHL on the fraction of translating mRNAs with RCC cells led us to discover that translation of tumor necrosis factor alpha was inhibited in pVHL-expressing cells (16). Here, we focused on the subset of mRNAs whose translation is enhanced in a pVHL-dependent manner, prominent among them the p53 mRNA. With pairs of RCC cells in which tumor-derived, VHL-deficient cells were stably transfected to express pVHL, we observed that restoration of pVHL expression caused a marked increase in p53 expression in the absence of corresponding elevations in p53 mRNA levels (Fig. 1). The altered p53 abundance was not due to differential degradation of p53 in cells with different VHL statuses and instead appeared largely based on the increased recruitment of p53 mRNA to heavy polysomes and the consequent enhancement in p53 translation in VHL+ cells (Fig. 2 and 7).
Further studies revealed that the RNA-binding protein HuR bound the 3′UTR of the p53 mRNA, which bears U- and AU-rich sequences (Fig. 3 and 4). While the cellular abundance of HuR was not influenced by pVHL, cytoplasmic and polysome-bound HuRs were significantly more abundant in pVHL+ cells, linked to the elevated presence of p53 mRNA in polysomes of VHL+ cells (Fig. 7). Small interfering RNA-mediated reduction in HuR expression caused a potent inhibition in the translation and steady-state levels of p53 (Fig. 8). The finding that restoration of pVHL expression in three different RCC lines led to increased p53 expression provided strong support to the notion that pVHL widely regulates p53 expression in renal carcinoma cells.
In light of the well-established role of HuR in the regulation of mRNA turnover, we anticipated finding altered p53 mRNA stability in cells with different levels of cytoplasmic HuR. Our data, however, argued against this possibility, since p53 mRNA half-life was unchanged (data not shown) and HuR appeared to influence translation of p53 in this system, in keeping with our recent findings that HuR promoted p53 translation after UV irradiation (41). Two additional works have also linked HuR to the translational regulation of another protein, the cdk inhibitor p27, through binding of HuR to the p27 5′UTR. One demonstrates the inhibitory effects of HuR on p27 translation (36), while an earlier study in a different cell system favored a role for HuR in the enhanced translation of p27 (43). Our results support a model in which HuR positively influences p53 translation via a 3′UTR binding site (41), similar to the previously reported translational regulation of neurofilament M expression by Hel-N1 (a neuronal tissue-specific ELAV protein), which was also shown to involve the neurofilament M 3′UTR (2).
The precise mechanisms underlying the enhanced translation of p53 mRNA are unclear but may be linked to the joint presence of HuR on the transcript along with poly(A)-binding protein and possibly other hnRNPs (5, 41, 43). U-rich and AU-rich stretches are found in the p53 3′UTR (Fig. 3). Linking the entire p53 3′UTR to the luciferase coding region revealed no differences in the translation of the chimeric mRNA (unpublished observations), suggesting that the association of HuR to the 3′UTR was required but not sufficient for conferring enhanced p53 translation. It remains to be investigated whether sequences in the 5′UTR or coding region, possible targets of additional binding factors, cooperate in the translational upregulation by HuR in RCC cells.
Interestingly, heightened cytoplasmic HuR levels in VHL+ cells did not lead to elevated abundance of other known HuR target mRNAs, as might have been expected. On the contrary, HuR target mRNAs encoding vascular endothelial growth factor, p21, and TNF-α, to cite a few examples, were substantially more abundant in VHL− cells (16, 17, 27; unpublished observations). Expression of such HuR target mRNAs likely relies on additional regulatory events governed by pVHL, particularly in light of the aforementioned effects of pVHL on factors that regulate transcription and mRNA turnover. For example, vascular endothelial growth factor mRNA levels may be elevated in VHL− cells due to increased HIF-1α expression and HIF-1α-driven transcription. In addition, these transcripts are also targets of other RNA-binding proteins (3, 7), so potential mRNA-stabilizing effects by HuR in this context may be overridden by juxtaposed gene regulatory mechanisms (transcription, transport, etc). Additional experiments involving immunoprecipitation of HuR along with the collection of HuR-associated mRNAs in RCC cells are under way. This approach (described in reference 53) will allow the identification and comparison of subsets of mRNAs associated with HuR in VHL+ cells with those in VHL− cells, as well as the composition of mRNA subsets in polysome-bound HuR versus unbound HuR in each cell type.
Elevated function of the AMP-activated kinase (AMPK) was previously shown to inhibit the cytoplasmic localization of HuR, while AMPK inhibition effectively elevated the cytoplasmic presence of HuR (57). Accordingly, the increased abundance of cytoplasmic HuR in VHL+ cells is likely due to the reduced AMPK activity observed in 786-O cells (Fig. 6) and UMRC6 cells (unpublished results). In this regard, treatment of VHL+ cells with the AMPK activator AICAR caused a marked decrease in cytoplasmic HuR levels as well as a reduction in p53 expression levels, both in whole-cell lysates (not shown) and in cytoplasmic lysates (Fig. 6B). Unexpectedly, treatment of VHL− RCC cells with AICAR caused an increase in cytoplasmic HuR (not shown).
Given the physiological importance of gene products encoded by HuR-target mRNAs, it will be of great interest to further investigate the differences in HuR subcellular localization under pVHL- and AMPK-mediated control. AMPK has also been termed the fuel gauge of the cell, as its activation is tightly dependent on elevated AMP/ATP ratios, which reflect low cellular energy levels. In support of the notion that VHL− cells may have reduced energy levels is the fact that their growth is critically dependent on glucose availability (22). However, the precise reasons underlying the elevated AMPK activity in VHL− cells remain unknown. Ongoing studies are aimed at studying AMP/ATP ratios as well as phosphorylation by the upstream kinase AMPK kinase, two AMPK regulatory mechanisms (25), in RCC cells with different VHL statuses.
Increased p53 expression has been linked to elevated HIF-1α function through mechanisms that include interaction between HIF-1α and Mdm-2, resulting in a p53 protein with greater stability (1, 9). In turn, elevated p53 contributes to maintaining reduced levels of HIF-1α in its unhydroxylated form (24). In the RCC model system described in these studies, however, it has been widely reported that cells lacking VHL function exhibit elevated HIFα activity (30, 42). However, as shown here, p53 levels are instead markedly reduced in VHL-deficient cells. This unexpected finding suggests that additional mechanisms, possibly involving pVHL, may be required for HIF-1α-mediated p53 stabilization. The finding that restoring pVHL expression in three independent RCC lines increased p53 levels lends support to the exciting possibility that p53 may be an important component of the VHL tumor-suppressive program in RCC.
Acknowledgments
We are grateful to J. Gnarra for providing the UOK121 cells, B. Zbar and M. Lerman for the UMRC6 cells, and O. Iliopoulos and W. G. Kaelin for the 786-O cells. We thank K. Becker and C. Cheadle (DNA Array Unit, NIA-IRP) for helpful discussions and for providing cDNA arrays for analysis.
S.G. was sponsored by a German Academic Exchange Service (DAAD) grant through the Johannes Gutenberg University in Mainz, Germany.
REFERENCES
- 1.An, W. G., M. Kanekal, M. C. Simon, E. Maltepe, M. V. Blagosklonny, and L. M. Neckers. 1998. Stabilization of wild-type p53 by hypoxia-inducible factor 1. Nature 392:405-408. [DOI] [PubMed] [Google Scholar]
- 2.Antic, D., N. Lu, and J. D. Keene. 1999. ELAV tumor antigen, Hel-N1, increases translation of neurofilament M mRNA and induces formation of neurites in human teratocarcinoma cells. Genes Dev. 13:449-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhattacharya, S., T. Giordano, G. Brewer, and J. S. Malter. 1999. Identification of AUF-1 ligands reveals vast diversity of early response gene mRNAs. Nucleic Acids Res. 27:1464-1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blankenship, C., J. Naglich, J. Whaley, B. Seizinger, and N. Kley. 1999. Alternate choice of initiation codon produces a biologically active product of the von Hippel-Lindau gene with tumor suppressor activity. Oncogene 18:1529-1535. [DOI] [PubMed] [Google Scholar]
- 5.Brennan, C. M., and J. A. Steitz. 2001. HuR and mRNA stability. Cell. Mol. Life Sci. 58:266-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bunn, H. F., J. Gu, L. E. Huang, J. W. Park, and H. Zhu. 1998. Erythropoietin: a model system for studying oxygen-dependent gene regulation. J. Exp. Biol. 201:1197-1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caldwell, M. C., C. Hough, S. Fürer, W. M. Linehan, S., P. J. Morin, and M. Gorospe. 2002. Serial analysis of gene expression in renal carcinoma cells reveals VHL-dependent sensitivity to TNF-α cytotoxicity. Oncogene 21:929-936. [DOI] [PubMed] [Google Scholar]
- 8.Carballo, E., W. S. Lai, and P. J. Blackshear. 1998. Feedback inhibition of macrophage tumor necrosis factor-alpha production by tristetraprolin. Science 281:1001-1005. [DOI] [PubMed] [Google Scholar]
- 9.Chen, D., M. Li, J. Luo, and W. Gu. 2003. Direct interactions between HIF-1 alpha and Mdm2 modulate p53 function. J. Biol. Chem. 278:13595-13598. [DOI] [PubMed] [Google Scholar]
- 10.Duan, D. R., A. Pause, W. Burgess, T. Aso, D. Y. T. Chen, K. P. Garrett, R. C. Conaway, J. W. Conaway, W. M. Linehan, R. D. Klausner, D. R. Duan, et al.1995. Inhibition of transcriptional elongation by the VHL tumor suppressor protein. Science. 269:1402-1406. [DOI] [PubMed] [Google Scholar]
- 11.Elson, D. A., G. Thurston, L. E. Huang, D. G. Ginzinger, D. M. McDonald, R. S. Johnson, and J. M. Arbeit. 2001. Induction of hypervascularity without leakage or inflammation in transgenic mice overexpressing hypoxia-inducible factor-1alpha. Genes Dev. 15:2520-2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Esteban-Barragan, M. A., P. Avila, M. Alvarez-Tejado, M. D. Gutierrez, A. Garcia-Pardo, F. Sanchez-Madrid, and M. O. Landazuri. 2002. Role of the von Hippel-Lindau tumor suppressor in the formation of β1-integrin fibrillar adhesions. Cancer Res. 62:2929-2936. [PubMed] [Google Scholar]
- 13.Fan, J., X. Yang, W. Wang, W. H. Wood 3rd, K. G. Becker, and M. Gorospe. 2002. Global analysis of stress-regulated mRNA turnover by with cDNA arrays. Proc. Natl. Acad. Sci. USA 99:10611-10616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Forsythe, J. A., B. H. Jiang, N. V. Iyer, F. Agani, S. W. Leung, R. D. Koos, and G. L. Semenza. 1996. Activation of vascular endothelial growth factor gene transcription by hypoxia-inducible factor 1. Mol. Cell. Biol. 16:4604-4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Foster, K., A. Prowse, A. van den Berg, S. Fleming, M. M. Hulsbeek, P. A. Crossey, F. M. Richards, P. Cairns, N. A. Affara, M. A. Ferguson-Smith, et al. 1994. Somatic mutations of the von Hippel-Lindau disease tumour suppressor gene in non-familial clear cell renal carcinoma. Hum. Mol. Genet. 3:2169-2173. [DOI] [PubMed] [Google Scholar]
- 16.Galbán, S., J. Fan, J. L. Martindale, C. Cheadle, B. Hoffman, M. P. Woods, G. Temeles, J. Brieger, J. Decker, and M. Gorospe. 2003. VHL-mediated repression of TNFα translation revealed through use of cDNA arrays. Mol. Cell. Biol. 23:2316-2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gallouzi, I. E., C. M. Brennan, M. G. Stenberg, M. S. Swanson, A. Eversole, N. Maizels, and J. A. Steitz. 2000. HuR binding to cytoplasmic mRNA is perturbed by heat shock. Proc. Natl. Acad. Sci. USA 97:3073-3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gnarra, J. R., K. Tory, Y. Weng, L. Schmidt, M. H. Wei, H. Li, F. Latif, S. Liu, F. Chen, F. M. Duh, et al. 1994. Mutations of the VHL tumour suppressor gene in renal carcinoma. Nat. Genet. 7:85-90. [DOI] [PubMed] [Google Scholar]
- 19.Gnarra, J. R., S. Zhou, M. J. Merrill, J. R. Wagner, A. Krumm, E. Papavassiliou, E. H. Oldfield, R. D. Klausner, and W. M. Linehan. 1996. Post-transcriptional regulation of vascular endothelial growth factor mRNA by the product of the VHL tumor suppressor gene. Proc. Natl. Acad. Sci. USA 93:10589-10594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gorospe, M., and N. J. Holbrook. 1996. Role of p21 in prostaglandin A2-mediated cellular arrest and death. Cancer Res. 55:475-479. [PubMed] [Google Scholar]
- 21.Gorospe, M., Y. Liu, Q. Xu, F. J. Chrest, and N. J. Holbrook. 1996. Inhibition of G1 cyclin-dependent kinase activity during growth arrest of human breast carcinoma cells by prostaglandin A2. Mol. Cell. Biol. 16:762-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gorospe, M., J. M. Egan, B. Zbar, M. Lerman, L. Geil, I. Kuzmin, and N. J. Holbrook. 1999. Protective function of von Hippel-Lindau protein against impaired protein processing in renal carcinoma cells. Mol. Cell. Biol. 19:1289-1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hansen, W. J., M. Ohh, J. Moslehi, K. Kondo, W. G. Kaelin, and W. J. Welch. 2002. Diverse effects of mutations in exon II of the von Hippel-Lindau (VHL) tumor suppressor gene on the interaction of pBHL with the cytosolic chaperonin and pVHL-dependent ubiquitin ligase activity. Mol. Cell. Biol. 22:1947-1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hansson, L. O., A. Friedler, S. Freund, S. Rudiger, and A. R. Fersht. 2002. Two sequence motifs from HIF-1alpha bind to the DNA-binding site of p53. Proc. Natl. Acad. Sci. USA 99:10305-10309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hardie, D. G., I. P. Salt, and S. P. Davies. 2000. Analysis of the role of the AMP-activated protein kinase in the response to cellular stress. Methods Mol. Biol. 99:63-74. [DOI] [PubMed] [Google Scholar]
- 26.Iliopoulos, O., A. Kibel, S. Gray, and W. G. Kaelin. 1995. Tumor suppression by the human von Hippel-Lindau gene product. Nat. Med. 1:822-866. [DOI] [PubMed] [Google Scholar]
- 27.Iliopoulos, O., A. P. Levy, C. Jiang, W. C. Kaelin, Jr., and M. A. Goldberg. 1996. Negative regulation of hypoxia-inducible genes by the von Hippel-Lindau protein. Proc. Natl. Acad. Sci. USA 93:10595-10599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Iliopoulos, O., M. Ohh, and W. G. Kaelin. 1998. pVHL19 is a biologically active product of the von Hippel-Lindau gene arising from internal translation initiation. Proc. Natl. Acad. Sci. USA 95:11661-11666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ivan, M., and W. G. Kaelin. 2001. The von Hippel-Lindau tumor suppressor protein. Curr. Opin. Genet. Dev. 11:27-34. [DOI] [PubMed] [Google Scholar]
- 30.Ivan, M., K. Kondo, H. Yang, W. Kim, J. Valiando, M. Ohh, A. Salic, J. M. Asara, W. S. Lane, and W. G. Kaelin, Jr. 2001. HIFα targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science 292:464-468. [DOI] [PubMed] [Google Scholar]
- 31.Johannes, G., M. S. Carter, M. B. Eisen, P. O. Brown, and P. Sarnow. 1999. Identification of eukaryotic mRNAs that are translated at reduced cap binding complex eIF4F concentrations with a cDNA microarray. Proc. Natl. Acad. Sci. USA 96:13118-13123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaelin, W. G. 2002. Molecular basis of the VHL hereditary cancer syndrome. Nat. Rev. Cancer 2:673-682. [DOI] [PubMed] [Google Scholar]
- 33.Keene, J. D. 2001. Ribonucleoprotein infrastructure regulating the flow of genetic information between the genome and the proteome. Proc. Natl. Acad. Sci. USA 98:7018-7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koochekpour, S., M. Jeffers, P. H. Wang, C. Gong, G. A. Taylor, L. M. Roessler, R. Stearman, J. R. Vasselli, W. G. Stetler-Stevenson, W. G. Kaelin, Jr., W. M. Linehan, R. D. Klausner, J. R. Gnarra, and G. F. Vande Woude. 1999. The von Hippel-Lindau tumor suppressor gene inhibits hepatocyte growth factor/scatter factor-induced invasion and branching morphogenesis in renal carcinoma cells. Mol. Cell. Biol. 19:5902-5912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kroll, S. L., W. R. Paulding, P. O. Schnell, M. C. Barton, J. W. Conaway, R. C. Conaway, and M. F. Czyzyk-Krzeska. 1999. von Hippel-Lindau protein induces hypoxia-regulated arrest of tyrosine hydroxylase transcript elongation in pheochromocytoma cells. J. Biol. Chem. 274:30109-30114. [DOI] [PubMed] [Google Scholar]
- 36.Kullmann, M., U. Gopfert, B. Siewe, L. Hengst. 2002. ELAV/Hu proteins inhibit p27 translation via an IRES element in the p27 5′UTR. Genes Dev. 16:3087-3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kuznetsova, A. V., J. Meller, P. O. Schnell, J. A. Nash, M. L. Ignacak, Y. Sanchez, J. W. Conaway, R. C. Conaway, and M. F. Czyzyk-Krzeska. 2003. von Hippel-Lindau protein binds hyperphosphorylated large subunit of RNA polymerase II through a proline hydroxylation motif and targets it for ubiquitination. Proc. Natl. Acad. Sci. USA 100:2706-2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Latif, F. et al. 1993. Identification of the von Hippel-Lindau disease tumor suppressor gene. Science 260:1317-1320. [DOI] [PubMed] [Google Scholar]
- 39.Lee, J. Y., S. M. Dong, W. S. Park, N. J. Yoo, C. S. Kim, J. J. Jang, J. G. Chi, B. Zbar, I. A. Lubensky, W. M. Linehan, A. O. Vortmeyer, and Z. Zhuang. 1998. Loss of heterozygosity and somatic mutations of the VHL tumor suppressor gene in sporadic cerebellar hemangioblastomas. Cancer Res. 58:504-508. [PubMed] [Google Scholar]
- 40.Los, M., G. H. Jansen, W. G. Kaelin, C. J. Lips, G. H. Blijham, and E. E. Voest. 1996. Expression pattern of the von Hippel-Lindau protein in human tissues. Lab. Investig. 75:231-238. [PubMed] [Google Scholar]
- 41.Mazan-Mamczarz, K., S. Galbán, I. López de Silanes, J. L. Martindale, U. Atasoy, J. D. Keene, and M. Gorospe. 2003. RNA-binding HuR enhances p53 translation in response to ultraviolet light irradiation. Proc. Natl. Acad. Sci. USA 100:8354-8359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maxwell, P. H., M. S. Wiesener, G.-W. Chang, S. C. Clifford, E. C. Vaux, M. E. Cockman, C. C. Wykoff, C. W. Pugh, E. R. Maher, and P. J. Ratcliffe. 1999. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature 399:271-275. [DOI] [PubMed] [Google Scholar]
- 43.Millard, S. S., A. Vidal, M. Markus, and A. Koff. 2000. A U-rich element in the 5′ untranslated region is necessary for the translation of p27 mRNA. Mol. Cell. Biol. 20:5947-5959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ohh, M., R. L. Yauch, K. M. Lonergan, J. M. Whaley, A. O. Stemmer-Rachamimov, D. N. Louis, B. J. Gavin, N. Kley, W. G. Kaelin, Jr., and O. Iliopoulos. 1998. The von Hippel-Lindau tumor suppressor protein is required for proper assembly of an extracellular fibronectin matrix. Mol. Cell 1:959-968. [DOI] [PubMed] [Google Scholar]
- 45.Ohh, M., C. W. Park, M. Ivan, M. A. Hoffman, L. E. Huang, V. Chau, and W. G. Kaelin. 2000. Ubiquitination of hypoxia-inducible factor requires direct binding to the beta-domain of the von Hippel-Lindau protein. J. Biol. Chem. 275:25733-25741. [DOI] [PubMed] [Google Scholar]
- 46.Pioli, P. A., and W. F. C. Rigby. 2001. The von Hippel-Lindau protein interacts with heteronuclear ribonucleoprotein A2 and regulates its expression. J. Biol. Chem. 276:40346-40352. [DOI] [PubMed] [Google Scholar]
- 47.Prives, C., and P. A. Hall. 1999. The p53 pathway. J. Pathol. 187:112-126. [DOI] [PubMed] [Google Scholar]
- 48.Sato, K., K. Terada, T. Sugiyama, S. Takahashi, M. Saito, M. Moriyama, H. Kakinuma, Y. Suzuki, M. Kato, and T. Kato. 1994. Frequent overexpression of vascular endothelial growth factor gene in human renal cell carcinoma. J. Tohoku Exp. Med. 173:355-360. [DOI] [PubMed] [Google Scholar]
- 49.Schoenfeld, A. R., E. J. Davidowitz, and R. D. Burk. 2001. Endoplasmic reticulum/cytosolic localization of von Hippel-Lindau gene products is mediated by a 64-amino acid region. Int. J. Cancer 91:457-467. [DOI] [PubMed] [Google Scholar]
- 50.Schoenfeld, A., E. Davidowitz, and R. A. Burk. 1998. A second major native von Hippel-Lindau gene product, initiated from an internal translation start site, functions as a tumor suppressor. Proc. Natl. Acad. Sci. USA 95:8817-8822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Semenza, G. L., and G. L. Wang. 1992. A nuclear factor induced by hypoxia via de novo protein synthesis binds to the human erythropoietin gene enhancer at a site required for transcriptional activation. Mol. Cell. Biol. 12:5447-5454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shuin, T., K. Kondo, S. Ashida, H. Okuda, M. Yoshida, H. Kanno, and M. Yao. 1999. Germline and somatic mutations in von Hippel-Lindau disease gene and its significance in the development of kidney cancer. Contrib. Nephrol. 128:1-10. [DOI] [PubMed] [Google Scholar]
- 53.Tenenbaum, S. A., P. J. Lager, C. C. Carson, and J. D. Keene. 2002. Ribonomics: identifying mRNA subsets in mRNP complexes with antibodies to RNA-binding proteins and genomic arrays. Methods 26:191-198. [DOI] [PubMed] [Google Scholar]
- 54.Vawter, M. P., T. Barrett, C. Cheadle, B. P. Sokolov, W. H. Wood, D. Donovan, M. Webster, W. J. Freed, and K. G. Becker. 2001. Application of cDNA microarrays to examine gene expression differences in schizophrenia. Brain Res. Bull. 55:641-650. [DOI] [PubMed] [Google Scholar]
- 55.Wang, W., S. Lin, C. M. Caldwell, H. Furneaux, and M. Gorospe. 2000. HuR regulates cyclin A and cyclin B1 mRNA stability during the cell division cycle. EMBO J. 19:2340-2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang, W., H. Furneaux, H. Cheng, M. C. Caldwell, D. Hutter, Y. Liu, N. J. Holbrook, and M. Gorospe. 2000. HuR regulates p21 mRNA stabilization by UV light. Mol. Cell. Biol. 20:760-769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang, W., J. Fan, X. Yang, S. Fürer, I. Lopez de Silanes, C. von Kobbe, J. Guo, S. N. Georas, F. Foufelle, D. G. Hardie, D. Carling, and M. Gorospe. 2002. AMP-activated kinase regulates cytoplasmic HuR. Mol. Cell. Biol. 22:3425-3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wizigmann-Voos, S., G. Breier, W. Risau, and K. Plate. 1995. Up-regulation of vascular endothelial growth factor and its receptors in von Hippel-Lindau disease-associated and sporadic hemangioblastomas. Cancer Res. 55:1358-1364. [PubMed] [Google Scholar]
- 59.Wykoff, C., C. Pugh, P. Maxwell, A. Harris, and P. Ratcliffe. 2000. Identification of novel hypoxia-dependent and -independent target genes of the von Hippel-Lindau (VHL) tumor suppressor by mRNA differential expression profiling. Oncogene 19:6297-6305. [DOI] [PubMed] [Google Scholar]
- 60.Zatyka, M., N. F. da Silva, S. C. Clifford, M. R. Morris, M. S. Wiesener, K. U. Eckardt, R. S. Houlston, F. M. Richards, F. Latif, and E. R. Maher. 2002. Identification of cyclin D1 and other novel targets for the von Hippel-Lindau tumor suppressor gene by expression array analysis and investigation of cyclin D1 genotype as a modifier in von Hippel-Lindau disease. Cancer Res. 62:3803-3811. [PubMed] [Google Scholar]
- 61.Zhou, M. I., H. Wang, J. J. Ross, I. Kuzmin, C. Xu, and H. T. Cohen. 2002. The von Hippel-Lindau (VHL) tumor suppressor stabilizes novel PHD protein Jade-1. J. Biol. Chem. 277:39887-39898. [DOI] [PubMed] [Google Scholar]
- 62.Zong, Q., M. Schummer, L. Hood, and D. R. Morris. 1999. Messenger RNA translation state: the second dimension of high-throughput expression screening. Proc. Natl. Acad. Sci. USA 96:10632-10636. [DOI] [PMC free article] [PubMed] [Google Scholar]