Abstract
The last unidentified gene encoding an enzyme involved in ergosterol biosynthesis in Saccharomyces cerevisiae has been cloned. This gene, designated ERG27, encodes the 3-keto sterol reductase, which, in concert with the C-4 sterol methyloxidase (ERG25) and the C-3 sterol dehydrogenase (ERG26), catalyzes the sequential removal of the two methyl groups at the sterol C-4 position. We developed a strategy to isolate a mutant deficient in converting 3-keto to 3-hydroxy-sterols. An ergosterol auxotroph unable to synthesize sterol or grow without sterol supplementation was mutagenized. Colonies were then selected that were nystatin-resistant in the presence of 3-ketoergostadiene and cholesterol. A new ergosterol auxotroph unable to grow on 3-ketosterols without the addition of cholesterol was isolated. The gene (YLR100w) was identified by complementation. Segregants containing the YLR100w disruption failed to grow on various types of 3-keto sterol substrates. Surprisingly, when erg27 was grown on cholesterol- or ergosterol-supplemented media, the endogenous compounds that accumulated were noncyclic sterol intermediates (squalene, squalene epoxide, and squalene dioxide), and there was little or no accumulation of lanosterol or 3-ketosterols. Feeding experiments in which erg27 strains were supplemented with lanosterol (an upstream intermediate of the C-4 demethylation process) and cholesterol (an end-product sterol) demonstrated accumulation of four types of 3-keto sterols identified by GC/MS and chromatographic properties: 4-methyl-zymosterone, zymosterone, 4-methyl-fecosterone, and ergosta-7,24 (28)-dien-3-one. In addition, a fifth intermediate was isolated and identified by 1H NMR as a 4-methyl-24,25-epoxy-cholesta-7-en-3-one. Implications of these results are discussed.
Keywords: fungi, sterol biosynthesis
The yeast Saccharomyces cerevisiae is used extensively as a model system for studying lipid biosynthesis (1). Yeast accumulates ergosterol, the sterol end-product equivalent of cholesterol in animals (2) and sitosterol in plants (3). Squalene, the first dedicated sterol precursor in the ergosterol pathway, is converted to the end-product sterol through a sequence of 15 enzymatic reactions. Only one gene in the ergosterol pathway, the gene encoding the 3-ketoreductase required in C-4 demethylation, has neither been cloned nor has had its gene product characterized. C-4 demethylation can be separated into three reactions: (i) a C-4 methyloxidase reaction in which the 4α-methyl group is converted to an alcohol, then an aldehyde, and finally to a carboxylic acid; (ii) a C-3 sterol dehydrogenation, which removes the 3α-hydrogen leading to the decarboxylation of a 3-ketocarboxylic acid sterol intermediate, and (iii) a 3-keto reduction, which converts the 3-keto to the β-hydroxy sterol.
Two consecutive rounds of C-4 demethylation in yeast and animals cells are required to demethylate 4,4-dimethylzymosterol. In higher plants, the sequence of C-4 demethylation is interrupted by the removal of the C-14 methyl group, as suggested by differences in substrate specificity of the plant demethylating enzymes. Thus, demethylation occurs in the sequence: C-4, C-14, C-4 in plants (4, 5) and C-14, C-4, C-4 in animals and yeast. Neither C-4 methyl nor 4,4-dimethyl sterols can substitute for end-product sterols in yeast (6, 7).
Previously, we cloned both the ERG25 (C-4 methyloxidase) and the ERG26 (C-3 sterol dehydrogenase, C-4 decarboxylase) genes. The ERG25 gene encodes a protein containing three histidine clusters and a KKXX golgi-to-endoplasmic reticulum retrieval signal (7). Histidine clusters are common among non-heme diiron enzymes such as hydroxylases and fatty acid desaturases (8). An ERG25 auxotroph was obtained by mutagenizing an erg1 strain (CP3, capable of growth on lanosterol) and isolating mutants unable to grow on lanosterol or other C-4 dimethylsterols. The ERG26 gene encoding the C-3 sterol dehydrogenase was cloned and disrupted based on its similarity to a cholesterol dehydrogenase from Nocardia spp. capable of converting cholesterol to its 3-keto derivative (9). Here, we describe the isolation of a 3-ketoreductase mutant and the subsequent cloning and characterization of the gene and gene products. To clone the 3-keto reductase gene, we mutagenized a sterol auxotroph and selected for mutants unable to grow on 3-ketosterol-supplemented media.
Several groups have demonstrated the accumulation of 3-keto sterol intermediates. Pascal et al. (4) have demonstrated 3-keto reductase activity in plant microsomes and have established that sterone reduction was exclusively dependent on NADPH and insensitive to azoles, and Bilheimer et al. (10) demonstrated that the rat liver microsomal enzyme could be solubilized and partially purified, resulting in a 24-fold increase in activity. Additionally, a Chinese hamster ovary cell line selected for cholesterol auxotrophy was found to accumulate 3-keto sterols (11). Treatment of Cryptococcus neoformans with azoles showed accumulation of 3-keto sterols (12).
Although loss of the 3-ketoreductase enzyme activity, like other enzymes involved in C-4 demethylation, results in sterol auxotrophy, many of the genes encoding sterol biosynthetic enzymes in yeast are not essential for viability, such as those encoding enzymes that convert zymosterol to ergosterol. Genes responsible for converting lanosterol to zymosterol appear to be essential. However, we were able to demonstrate that the erg11erg25slu1 (or slu2) triple mutants (slu mutants are leaky heme mutants) grow poorly and accumulate only lanosterol (13).
The cloning of the 3-keto sterol reductase gene completes the elucidation of the biosynthetic reactions leading to ergosterol biosynthesis in yeast. We demonstrate that 3-keto sterols do not accumulate in erg27 mutants unless lanosterol is added to the growth media. The marked reduction in lanosterol formation in erg27 strains suggests that the Erg27p may have an architectural role in other reactions in sterol biosynthesis, such as the conversion of squalene epoxide to lanosterol.
Materials and Methods
Strains and Plasmids.
The genotypes of strains used in this study are listed in Table 1. The ERG27 allele in strains SDG105, SDG110, and SDG115 was disrupted by using one-step PCR deletion–disruption (14). The position of all primers used within the ERG27 gene are shown in Fig. 1. Briefly, the primers DGK1(GGAAAGTAGCTATCGTAACGGGTACTAATAGTAATCTTGGTCTGAACATTGTGTTCCGtggcgggtgtcggggctggc) and DGK2 (CGGAGAAGGAATGGCTTGTAAATATGCCCGGTTGAACTACATACTGATTTATGCCCAGCTttgccgatttcggcctattg) were used to generate a 1.5-kilobase (kb) erg27 recombinogenic DNA fragment containing HIS3 or URA3 as a selectable marker and using pRS303 or pRS306 (15) vector as template, respectively. Lowercase bases correspond to conserved regions bordering the HIS3 and URA3 genes in the pRS303 and pRS306 vectors, respectively, and uppercase bases refer to the ERG27 DNA sequence. The PCR fragment used to disrupt ERG27 contains a 0.6-kb deletion of the ERG27-coding region (200 amino acid residues). A third primer, DGK3 (GCTGCAACAGTTGAACAAGC), 0.32 kb upstream of the deletion–substitution, was used in combination with DGK2 to verify each disruption. ERG27 cells transformed to erg27 auxotrophs were initially screened as colonies able to grow on ergosterol but not on lanosterol-supplemented media.
Table 1.
Genotypes of strains used in this study
| Strain | Genotype |
|---|---|
| W303-2A | MAT a his3 ura3-1 leu2-112 trp1 |
| CP3 | MAT α upc2 ade2 his3 ura3-52 erg1∷URA3 |
| CP3/26 | MAT α upc2 ade2 his3 ura3-52 erg1∷URA3 erg27-1 |
| SDG100 | MAT α upc2 ade2 his− ura3-52 erg27-1 |
| SDG105 | MAT α upc2 ade2 his3 ura3-52 erg1∷URA3 erg27Δ:HIS3 |
| SDG110 | MAT a upc2 ade2 his3 ura3-52 erg27Δ∷HIS3 |
| SDG127 | MAT a upc2 ade2 his3 erg27Δ∷H1S3 ura3-52∷pDG27(ERG27) |
| SDG116 | MAT a upc2 ade2 his3 erg27Δ∷HIS3 ura3-52/pDG7(ERG7) |
| WA1/6 | MAT α/a ade5/ade5 his7/his7 leu2-3,112/leu2-3,112 ura3-52/ura3-52 |
| SDG115 | MAT a ade5 his7 leu2-3,112 ura3-52 erg27Δ∷URA3 |
Figure 1.
Nucleotide sequence of the ERG27 gene and the deduced amino acid sequence. Double-underlined sequences are the amplifying primer sequences used for disruptions, and the single underlined sequence indicates the primer sequence used to confirm the disruption.
Strain SDG127 (an erg27 strain carrying an integrated version of ERG27) was made by integrating a 2.1-kb SacI–SalI ERG27 DNA fragment into the multiple-cloning site of pRS306 (to generate the plasmid pDG27) linearizing with StuI within the URA3 gene and transforming SDG110 (erg27). A multicopy plasmid containing the ERG7 gene was constructed by cleaving pZS11 (obtained from J. Griffin, Stanford University, Stanford, CA) with XhoI and ligating the 3.4-kb fragment into the high-copy-number vector YEp24 at the SalI site. All plasmids were maintained in Escherichia coli DH5α cultured in LB medium supplemented with ampicillin at 50 μg/ml. PCR were performed on a Thermocycler apparatus by using the Promega Taq polymerase kit.
Media and Chemicals.
Yeast sterol auxotrophs that were upc2 were grown aerobically or anaerobically at 30°C on YPD medium (1% yeast extract/2% Bacto peptone/2% glucose) supplemented with sterol (0.002% wt/vol) dissolved in ethanol/Tween 80 (1:1, vol/vol). Otherwise, yeast sterol auxotrophs were grown anaerobically (7). Transformants were grown on complete synthetic (CSM)-dropout media containing 0.67% yeast nitrogen base, 2% glucose, and amino acid and nitrogenous base supplements at 0.8% (Bio 101).
Ergosta-7,22-dien-3-one and 4,4-dimethyl-cholesta-8,14-dien-3β-ol were gifts from L. Frye (Rennsalear Polytechnic Institute, Troy, NY). Cholesta-7-en-3-one and pure lanosterol were gifts from D. B. Sprinson (Columbia University, New York). All other chemicals were purchased from Sigma unless specified otherwise. Methoxylamine⋅HCl, used to derivatize sterones, was dissolved in pyridine at a concentration of 10 mg/ml. All solvents were GC grade except acetonitrile, which was HPLC grade, and were purchased from Fisher Scientific. Chromerge was purchased from Matheson Coleman & Bell.
Complementation and Sequencing.
Yeast transformations were performed as described in the Bio 101 kit. A genomic library of S. cerevisiae fragments inserted into the vector YCp50 (16) was used to complement the erg27 strain (SDG100). The complementing plasmid was isolated using standard methods (17). DNA sequencing was performed by the Biochemistry Biotechnology Facility at the Indiana University School of Medicine by using the Applied Biosystems model 373 automated DNA sequencer.
Sterol Extraction and Separations.
Total lipids were extracted from freshly grown cells in the presence of washed glass beads according to the method of Bligh and Dyer (18). The sterol fraction was isolated, and phospholipids were precipitated at −20°C in the presence of acetone/chloroform (10:1, vol/vol), followed by centrifugation at 12,300 × g as described in Pleminitas et al. (19). Alternatively, the sterol fraction was saponified overnight at room temperature in 6% (wt/vol) KOH in methanol. 3-Ketosterols (sterones) were purified by TLC and were derivatized by treatment with methoxylamine in pyridine (using excess reagent overnight at room temperature). O-Methoxylimine and acetyl derivatives were resolved on 60 TLC F254 precoated silica gel TLC plates (Merck) with methylene chloride as the solvent system. Organic compounds were detected by spraying with a 0.1% ethanolic berberin sulfate solution after exposure to short-wavelength UV light. Sterols and sterones were eluted from scraped silica by using methylene chloride. HPLC (Shimadzu, LC10AD) purification of the epoxy-sterone was carried out with a Lichrosphere 100 RP18, 5-μm reversed-phase column (250 × 4.6 mm) by elution with acetonitrile (flow rate at 1 ml/min and monitoring at 210 nm). Oxidation of 4-methyl-fecosterol to its sterone derivative was accomplished by reacting sterol with a mixture of Chromerge (CrO3)/10% H2SO4 (1:20 vol/vol) in acetone for 30 min at 0°C.
Sterol Analyses.
GC Analysis of sterols and sterones was conducted on an HP5890 series II GC, by using a DB-5 capillary column (15 m × 0.25 mm i.d., 0.2-μm film thickness) with nitrogen as carrier gas (30 cm/sec) and was programmed from 195°C to 300°C (195°C for 3 min, 5.5°C/min to 300°C, then held for 10 min). GC/MS analyses were performed with an HP5890 GC coupled to a HP5972 mass selective detector. Electron impact GC/MS (70 eV, scanning from 40 to 700 or 650 amu, at 1-sec intervals) was performed by using the following conditions: (i) DB-5MS column (20 m × 0.18 mm i.d., 0.18-μm film thickness), He as carrier gas (30 cm/sec), detector temperature 180°C, column temperature 100–300°C (100°C for 1 min, 10°C/min to 300°C, then held for 15 min); and (ii) DB-5 (10 m × 0.25 mm i.d., 0.25-μm film thickness), He as carrier gas (50 cm/sec), column temperature 40–300°C (40°C for 1 min, 30°C/min to 300°C, then held for 4 min). All injections were run in splitless mode.
NMR Analyses.
1H NMR measurements (24°C) were made at 500 MHz on a Varian INOVA-500 NMR spectrometer equipped with a high-stability variable-temperature controller and triple-resonance, z-axis pulsed-field gradient probe. Samples consisted of 1–2 mM sterol in CDCl3 and were referenced to the residual protonated chloroform resonance at 7.24 ppm. Correlation spectroscopy spectra were acquired as 256 increments of 8 transients, with 3,800-Hz spectral width in each dimension, a 2-ms trim pulse, and 60-ms spin lock at one-fourth the power level of the standard 90° observe pulse. Correlation spectroscopy spectra were processed by double Fourier transformation with a 0.062-Hz Gaussian filter on the directly detected dimension and a 0.019 Hz Gaussian filter on the zero-filled indirectly detected dimension.
Results and Discussion
Isolation and Identification of a 3-Keto Sterol Reductase Mutant.
CP3, a sterol epoxidase auxotroph, erg1∷URA3, was previously used to isolate a C-4 sterol methyl oxidase mutant by screening mutagenized cells for the ability to grow on ergosterol-supplemented medium but unable to grow on lanosterol-supplemented medium. CP3 is able to efficiently take up and convert sterol intermediates to ergosterol. In this investigation, we mutagenized CP3 cells with UV light (97% kill) and screened for colonies that grew slowly on a YPD medium supplemented with ergosta-7,22-dien-3-one/cholesterol (9:1) and 20 μg/ml nystatin. Our reasoning was that efficient conversion of the ergosta-3-one to ergosterol would render the cells sensitive to nystatin, and inefficient conversion to ergosterol would render cells somewhat resistant to nystatin, which targets ergosterol in the cell membrane. Cholesterol was added to ensure that some growth would occur if little or no conversion of ergosta-7,22-dien-3-one (3-ketoergostadiene) occurred. A similar protocol was implemented by Karst and Lacroute (20), who obtained some of the first temperature-sensitive ergosterol mutants. Eight resistant colonies were obtained, and GC analysis indicated decreased ability to convert the 3-ketoergostadiene to ergosterol. CP3/26 grew very poorly on 3-ketoergostadiene and converted only 18% of this sterone to ergosterol, relative to the parent CP3 strain. CP3/26 was crossed to a wild-type strain, WA6, to isolate the erg27 mutation free of erg1. An erg27 segregant (SDG100) was then used to clone the ERG27 gene by complementation.
Cloning and Disruption of the ERG27 3-Keto Reductase Gene.
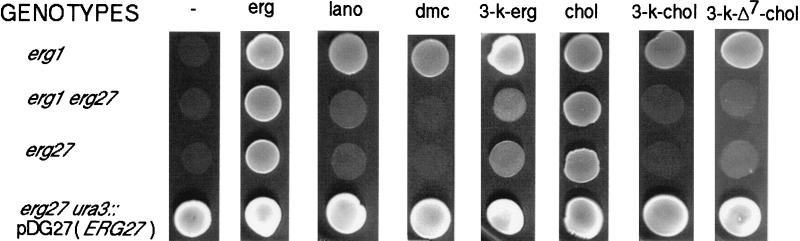
A centromeric genomic libary, RB239, was transformed into SDG100, and ≈11,800 transformants were obtained on CSM−ura+ergosterol medium incubated anaerobically. Five complementing clones, able to grow on CSM−ura, were investigated further, and four contained the same 7-kb insert (pDG100). DNA sequencing of a 1.2-kb HindIII fragment inserted into centromeric vector, pRS316, indicated that the complementing DNA was from chromosome X. The entire insert contained several ORFs, among which was YLR100w, which encodes polypeptides similar to mammalian steroid dehydrogenases. Indeed, the complementing clone was YLR100w, and a 2.1-kb SacI–SalI fragment from pDG100 was subcloned into pRS316 to generate pDG101. YLR100w overlaps a questionable ORF, YLR101c, which does not complement erg27 strains (data not shown). Fig. 1 shows the YLR100w DNA and derived protein sequence (39.7 kDa). Disruption of the ERG27 gene was accomplished and verified as described in Methods. Fig. 2 shows the growth responses of erg1 (CP3–2), erg1erg27 (SDG110), and erg27 (SDG115) strains grown on YPD + various sterols and sterones. All strains grew on ergosterol and cholesterol, but only erg1 grew on the two C-4 dimethylsterols: lanosterol and 4,4-dimethyl-cholesta-8,14-dien-3β-ol. The erg1 mutant also grew on the three 3-keto sterones: ergosta-7,22-dien-3-one, cholesta-3-one, and cholesta-7-en-3-one. The erg27-disrupted strains grew on ergosterol and cholesterol but showed no growth or marginal growth on C-4 dimethylsterols. We also observed either no growth or very marginal growth on 3-ketosterols, suggesting an inability to demethylate at C-4 and, in particular, an inability to reduce the 3-keto double bond at C-3. None of the erg27 strains grew on YPD medium without sterol supplementation. The integration of the ERG27 wild-type allele (strain SDG127) restored growth on all media, including sterol-unsupplemented YPD media.
Figure 2.
Growth responses on YPD media supplemented with various end-product sterols (ergosterol or cholesterol), 3-ketosterols (ergosta-7,22-dien-3-one, cholesta-3-one, cholesta-7-en-3-one), and 4,4-dimethylsterols (4,4-dimethyl-cholesta-8,24-dien-3β-ol and lanosterol). Cells were pregrown on YPD+ergosterol medium anaerobically and serially transferred two to three times onto the indicated sterol- or sterone-supplemented media. Erg, ergosterol; lano, lanosterol; dmc, 4,4-dimethyl-cholesta-8,24-dien-3β-ol; 3-k-erg, ergosta-7,22-dien-3-one; chol, cholesterol; 3-k-chol, cholesta-3-one; 3-k-Δ7-chol, cholesta-7-en-3-one.
Characterization of Sterol Intermediates in erg27 Mutants.
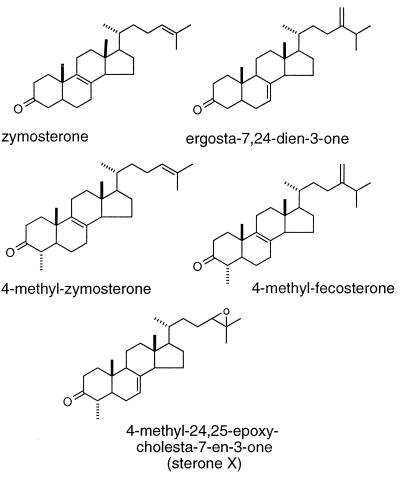
Surprisingly, the sterol profile of erg27 strains did not indicate accumulation of 3-ketosterols. In erg1erg27upc2 (SDG105), erg27upc2 (SDG110), and erg27 (SDG115) grown with cholesterol supplementation, only cholesterol and very small amounts of lanosterol accumulated (Table 2). The upc2 mutation allows sterol uptake to occur aerobically even when cells are heme-competent (21). The accumulation of sterols in these strains is similar to the sterol profile seen in erg1upc2, which is blocked in the conversion of squalene to its epoxide. Rather, SDG110 and SDG115 appear to accumulate squalene and squalene oxides (the epoxide and dioxide). To observe accumulation of 3-ketosterols (sterones), we added to erg27 strains SDG105, SDG110, and SDG115 a mixture of lanosterol and cholesterol (10:0.5 μg/ml) and grew all strains anaerobically. In the presence of excess lanosterol, we observed the accumulation of five different sterones in SDG110 and SDG115 and four in SDG105, albeit in small amounts. GC/MS analysis indicated that 3-hydroxysterols showed loss of 33 amu consistent with a loss of water and a methyl radical, but four of these 3-sterones failed to show loss of 33 amu. However, derivatization of the purified compounds with methoxylamine to give the methyloxime derivatives supported the premise that all five were 3-sterones. Finally, none of these compounds could be acetylated, again suggesting absence of a free hydroxyl group. Table 3 lists the MS parent and fragment ion masses of the free sterones and the methyloxime derivatives, as well as the TLC Rf values and GC relative retention times of each of the purified compounds. The proposed structures of the accumulated sterones are indicated in Fig. 3. One of the accumulated sterones, called sterone X, has a very low Rf value of 0.08 relative to the other four sterones, as well as a longer GC retention time. The mass spectrum of sterone X showed a molecular ion at m/z 412, and both this compound and its corresponding methyloxime derivative had fragmentation patterns indicative of a steroidal structure. The presence of m/z 243, which is likely due to loss of the side chain and D ring, also suggested unsaturation within the steroid nucleus, although its exact location remained ambiguous. Based on the structural assignments made for the other four sterones isolated from SDG127, we initially identified sterone X as 4,24-dimethylcholesta-8-en-3-one. However, neither the compound’s Rf nor its MS (22) or 1H NMR spectra (see below) was entirely consistent with this structural assignment.
Table 2.
Sterols and sterones accumulated in various erg27 genetic backgrounds
| Nonsaponifiable accumulation products, %† | Strains and genotypes
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CP3 erg1upc2
|
SDG105 erg1erg27 upc2
|
SDG110 erg27 upc2
|
SDG127 erg27 upc2 pDG27 (ERG27)
|
SDG116 erg27 upc2/pDG7 (ERG7)*
|
SDG115 erg27
|
|||||||
| C | L/C | C | L/C | C | L/C | C | L/C | C | L/C | C | L/C | |
| Cholesterol | 3 | 1 | 1 | 2.5 | 63 | 3 | 29 | — | 51 | — | 18 | 1.5 |
| Ergosterol | 0 | 9 | 0 | 0 | 0 | 0 | 69 | — | 0 | — | 0 | 0 |
| Lanosterol | 0 | tr | 0 | 0.8 | 0 | 1 | 0 | — | 7 | — | 1 | 1.5 |
| Squalene | 97 | 90 | 99 | 90 | 0 | 0 | 0 | — | 8 | — | 46 | 30 |
| Squalene epoxide | 0 | 0 | 0 | 0 | 4 | 16 | 0 | — | 23 | — | 34 | 45 |
| Squalene diepoxide | 0 | 0 | 0 | 0 | 33 | 62 | 0 | — | 7 | — | 1 | 13 |
| Zymosterone | 0 | 0 | 0 | 0.4 | 0 | 1 | 0 | — | 0 | — | tr | 1 |
| Ergosta-7,24(28)-dien-3-one | 0 | 0 | 0 | 1.2 | 0 | 4 | 0 | — | 0 | — | tr | 1 |
| 4α-methyl-zymosterone | 0 | 0 | 0 | 2.5 | 0 | 3.5 | 0 | — | 4 | — | tr | 4.5 |
| 4α-methyl-fecosterone | 0 | 0 | 0 | 1.2 | 0 | 4 | 0 | — | 0 | — | tr | 2 |
| 4α-methyl-24,25-epoxy-Cholesta-7-en-3-one (sterone X) | 0 | 0 | 0 | 0 | 0 | 5.5 | 0 | — | 1 | — | 0 | 0.5 |
All strains with the exception of SDG116 were grown onto YPD media anaerobically for 48 hours with cholesterol at 10 μg/ml (C) or lanosterol/cholesterol at 10:0.5 μg/ml (L/C). SDG116 was grown on CSM−ura medium anaerobically to ensure plasmid maintenance.
pDG27 is an integrated plasmid whereas pDG7 is a high-copy-number autonomous plasmid.
Sterols were extracted by using the glass beads method followed by overnight precipitation with acetone or saponification at room temperature.
Table 3.
Chromatographic properties of 3-keto compounds accumulated in SDG 110 (erg27 upc2) growing on YPD medium supplemented with lanosterol and cholesterol
| Sterol compound | Nonderivatized
|
Methyloxime derivatives
|
||||
|---|---|---|---|---|---|---|
| TLC, Rf | GC, RRT | MS (m/z mass fragment) | TLC, Rf | GC, RRT | MS (m/z mass fragment) | |
| Zymosterone | 0.28 | 1.06 | 382(M+), 367(-Me), | 0.32 | 1.092* | 411(M+), 396(-Me), 381(-O=CH2), |
| 271(-SC), 269(-SC-2H), | 380(-OMe), 300(-SC), 298(-SC-2H), | |||||
| 229(-SC-42) | 1.10† | 258(-SC-42), 228(-SC-42-O=CH2) | ||||
| Ergosta-7, 24(28)-diene-3-one | 0.28 | 1.12 | 396(M+), 381(-Me), | 0.32 | 1.153* | 425(M+), 410(-Me), 395(-O=CH2), |
| 312(-C6H12), 271(-SC) | 394(-OMe), 314(-C6H12), 326(- | |||||
| 269(-SC-2H), 229(-SC-42) | C6H12-Me), 300(-SC), 298(-SC-2H), | |||||
| 1.159† | 268(-SC-O=CH2-2H), 258(-SC-42) | |||||
| 4-Methyl- | 0.35 | 1.09 | 396(M+), 381(-Me), | 0.56 | 1.127 | 425(M+), 410(-Me), 395(-O=CH2), |
| zymosterone | 285(-SC), 283(-SC-2H), | 394(-OMe), 314(-SC), 312(-SC-2H), | ||||
| 243(-SC-42) | 272(-SC-42) | |||||
| 4-Methyl-fecosterone | 0.35 | 1.13 | 410(M+), 395(-Me), | 0.56 | 1.162 | 439(M+), 424(-Me), 409(-O=CH2), |
| 326(-C6H12), 311 (-C6H12-Me), 285(-SC), 283(-SC-2H), 243(-SC-42) | 408(-OMe), 341 (-C7H14), 314(-SC), 312(-SC-2H), 272(-SC-42) | |||||
| 4-methyl-24,25-epoxy-cholesta-7-en-3-one (sterone X) | 0.08 | 1.19 | 412(M+), 379(-33), 285(-SC), 283(-SC-2H), 243(-SC-42) | 0.16 | 1.22 | 441(M+), 426(-Me), 411(-O=CH2), 410(-OMe), 314(-SC), 312(-SC-2H), 272(-SC-42) |
TLC plates were developed in methylene chloride. RRT, retention time relative to cholesterol on a DB5 capillary column; SC, side chain.
syn isomer.
anti isomer.
Figure 3.
Chemical structures of the five sterone intermediates that accumulate in erg27 strains.
NMR Analysis of Sterone X.
To further elucidate the structure of sterone X, its 500-MHz 1H NMR spectrum was obtained and compared with that of 4α-methyl-fecosterone, which was prepared by oxidation of the corresponding alcohol by using a CrO3/H2SO4 solution. From two-dimensional NMR analysis, the C-2 and C-4 hydrogen and the C-4 methyl chemical shifts could be assigned, and because they were coincident in the two spectra, we confirmed the stereochemistry of sterone X to be 4α- methyl and not 4β. Although the 1H NMR spectra of sterone X and 4α-methyl-fecosterone were similar and provided strong evidence that the former was a 4α-methyl-3-keto-steroid, there were also several differences (Table 4). As might be expected for a 24-methyl-cholestane side chain, resonances absent in the NMR spectrum of sterone X included those of the C-24 exo-methylene and the H-25 methine. However, also absent was the predicted resonance at 0.77 ppm for the C-24 methyl group (23). Instead, additional shifts at 5.32 (broad s), 2.67 (t), and 2.33 (t) were present, and the C-26 and C-27 methyl signals were shifted to 1.2 and 1.3 ppm. Thus, it appeared that the side chain of sterone X was not the same as that of 4-methyl-ergosta-8-en-3-one (fully saturated in the side chain) and that unsaturation was Δ7,8 and not Δ8,9. The fact that both the C-26 and C-27 methyl resonances had been shifted downfield and that there was a triplet at 2.38 ppm was also consistent with the presence of a 24,25-epoxy-cholestane side chain (24). Hence, sterone X was assigned the structure of 4α-methyl-24,25-epoxy-cholesta-7-en-3-one. Because large quantities of squalene 2,3;22,23-dioxide were found to accumulate in SDG110 and SDG127, 4α-methyl-24,25-epoxy-cholesta-7-en-3-one may form by incorporation of the diepoxide into the sterol metabolic pathway. The ability of squalene dioxide to function as a substrate for oxidosqualene cyclase and to be converted to 24,25-epoxy-lanosterol or 24,25-epoxy-cholesterol is known in both animals and fungi (25–28). Finally, the epoxy-sterone is not found in an extract from an erg1erg27 (SDG105) strain in which the squalene epoxidase has been disrupted (Table 2).
Table 4.
Comparison of 1H NMR spectra data (δ, CDCl3) of sterone X and 4α-methyl-fecosterone
| Assignment | Chemical shift
|
|
|---|---|---|
| 4α-Methyl-24,25-epoxy-cholesta-7- en-3-one (sterone X) | 4α-Methyl-fecosterone | |
| H-2α | 2.45 (ddd, J = 13.97, 13.97, 6.62 Hz) | 2.45 (ddd, J = 14.71, 14.71, 6.62 Hz) |
| H-2β | 2.35 (unresolved ddd) | 2.34 (ddd, J = 14.71, 5.15, 2.2 Hz) |
| H-4β | 2.30 (unresolved dq) | 2.29 (unresolved dq) |
| C-4α-CH3 | 1.00 (d, J = 6.62 Hz) | 1.01 (d, J = 7.36 Hz) |
| H-7 | 5.32 (m) | — |
| H-9 | 2.67 (dd, J = 6.62, 6.62 Hz) | — |
| H-19 | 1.17 (s) | 1.18 (s) |
| H-18 | 0.62 (s) | 0.62 (s) |
| H-21 | 0.94 (d, J = 6.62 Hz) | 0.94 (d, J = 6.62 Hz) |
| H-24 | 2.33 (t, J = 7.35 Hz) | — |
| C-24-exo-methylene | — | 4.67, 4.64 (d, J = 1.5 Hz) |
| H-25 | — | 2.21 (septet, J = 7.35 Hz) |
| H-26, H-27 | 1.29 (s), 1.25 (s) | 1.00 (d, J = 6.62 Hz) |
Decreased Lanosterol Synthesis in erg27 Strains.
Although little is known as to how the various sterol proteins interact, this study indeed suggests an interaction between the Erg7 and Erg27 gene products. Loss of 3-ketoreductase enzyme activity results in a marked loss of all sterols. This is surprising, given that lesions in virtually all other genes in the postsqualene pathway give rise to the accumulation of immediate precursors or even downstream sterol products. The accumulation of sterones could only be observed when sterol substrates such as lanosterol were added to the growth medium. Instead of accumulating sterone intermediates, cells grown on ergosterol or cholesterol accumulated squalene and squalene oxides, suggesting that the cyclization of squalene epoxide was compromised. There are several lines of evidence to indicate that loss of the ERG27 gene product results in a marked decrease of squalene epoxide cyclization to lanosterol and that a separate erg7 lesion does not exist in our strains. (i) The disruption of ERG27 in several different genetic backgrounds gave rise to mutants that accumulated sterones and were severely compromised in the conversion of squalene epoxide to lanosterol (Table 2, compare strains SDG110 and SDG115). Genetic analysis of 30 tetrads obtained from the diploid WA1/6 made heterozygous for erg27 indicated single-gene segregation for sterol auxotrophy. (ii) Integration of a plasmid containing ERG27 into SDG110 restores a wild-type sterol profile (SDG127) and also the ability to grow without exogenous sterol. (iii) Overproduction of ERG7 plasmid by transforming into SDG110 with pDG7 (strain SDG116), although increasing the level of accumulating lanosterol, still resulted in a marked accumulation of squalene, squalene epoxide, and squalene diepoxide, indicating that conversion to lanosterol was still compromised even when the enzyme responsible for this conversion was overexpressed.
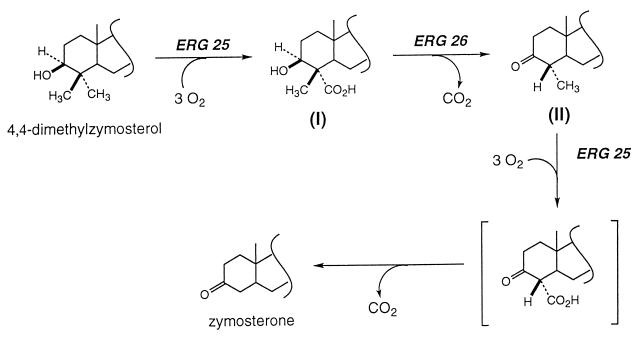
Another interesting observation arising from this study is that desmethyl sterols can accumulate even in the absence of 3-ketoreductase enzyme activity. In yeast, the two C-4 methyl groups are sequentially removed by the C-4 methyloxidase, the C-3 dehydrogenase, and the 3-ketoreductase. It is commonly assumed that interruption of the last step would prevent the second round of demethylation, and only 4 methyl-3-ketosterols would accumulate. Clearly, although such sterols were observed, two desmethyl sterones also accumulated. Reduction of the 3-keto sterols is, therefore, not required for removal of the second C-4 methyl group. In a scheme consistent with the above results (Fig. 4) in the erg27 mutant strain, dimethylzymosterol is converted to a β-methyl-α-carboxylic acid sterol (structure I; see ref. 19). Dehydrogenation of I results in decarboxylation with inversion of configuration at C-4, as in 6-phosphogluconate dehydrogenase (29), to produce 4α-methyl sterols (structure II, Tables 3 and 4). Another cycle of methyl oxidation and decarboxylation leads to zymosterone. The facile spontaneous decarboxylation of 3-keto-4-carboxysterols was reported by Bloch and collaborators (30). The isolation of a 3-ketoreductase mutant in yeast and the cloning of the wild-type gene represents the complete elucidation of a sterol pathway in a eukaryotic organism; however, this does not imply that structural proteins that contribute to sterol biosynthesis will not be found, such as proteins required for the functioning and maintenance of the sterol multienzyme complex. Now that all of the biosynthetic steps appear to be accounted for, the nature of the multienzyme complex and its component interactions are more amenable to biochemical characterization.
Figure 4.
Proposed reactions leading to the biosynthesis of zymosterone from 4,4-dimethylzymosterol in an erg27 strain. See text for details.
Acknowledgments
We thank Dr. James L. Gaylor for many useful discussions. We thank the Indiana University-Purdue University at Indianapolis Department of Physics for the generous use of its NMR facility. This work was supported by National Institutes of Health Grant 1R01 AI38598 to M.B.
Abbreviations
- sterones
3-keto-sterols
- kb
kilobase
- YPD
yeast extract/peptone/glucose
- ergosterol
ergosta-5,7,22-trien-3β-ol
- 4,4-dimethylzymosterol
4,4-dimethyl-cholesta-8,24-dien-3β-ol
- lanosterol
4,4,14-trimethyl-cholesta-8,24-dien-3β-ol
- 4α-methyl-zymosterone
4α-methyl-cholesta-8,24-dien-3-one
- zymosterone
cholesta-8,24-dien-3-one
- 4α-methyl-fecosterone
4α-methyl-ergosta-8,24(28)-dien-3-one
References
- 1.Daum G, Lees N D, Bard M, Dickson R. Yeast. 1998;14:1471–1510. doi: 10.1002/(SICI)1097-0061(199812)14:16<1471::AID-YEA353>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 2.Trazaskos J M, Gaylor J L. In: The Enzymes of Biological Membranes. Martinosi A N, editor. Vol. 2. New York: Plenum; 1985. pp. 177–204. [Google Scholar]
- 3.Benveniste P. Annu Rev Plant Physiol. 1986;37:275–308. [Google Scholar]
- 4.Pascal S, Taton M, Rahier A. Arch Biochem Biophys. 1994;312:260–271. doi: 10.1006/abbi.1994.1308. [DOI] [PubMed] [Google Scholar]
- 5.Taton M, Salmon F, Pascal S, Rahier A. Plant Physiol Biochem. 1994;32:751–760. [Google Scholar]
- 6.Kuchta T, Bartkova K, Kubinec R. Biochem Biophys Res Commun. 1992;189:85–91. doi: 10.1016/0006-291x(92)91529-y. [DOI] [PubMed] [Google Scholar]
- 7.Bard M, Bruner D A, Pierson C A, Lees N D, Biermann B, Frye L, Koegel C, Barbuch R. Proc Natl Acad Sci USA. 1996;93:186–190. doi: 10.1073/pnas.93.1.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shanklin J, Whittle E, Fox B G. Biochemistry. 1994;33:12784–12794. doi: 10.1021/bi00209a009. [DOI] [PubMed] [Google Scholar]
- 9.Gachotte D, Barbuch R, Gaylor J, Nickel E, Bard M. Proc Natl Acad Sci USA. 1998;95:13794–9. doi: 10.1073/pnas.95.23.13794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bilheimer J T, Alcorn M, Gaylor J L. Arch Biochem Biophys. 1981;211:430–438. doi: 10.1016/0003-9861(81)90474-4. [DOI] [PubMed] [Google Scholar]
- 11.Billheimer J T, Chamoun D, Esfahani M. J Lipid Res. 1987;28:704–709. [PubMed] [Google Scholar]
- 12.Vanden Bossche H, Marichal P, LeJeune L, Coene M-C, Gorrens J, Cools W. Antimicrob Agents Chemother. 1993;37:2101–2105. doi: 10.1128/aac.37.10.2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gachotte D, Pierson C A, Lees N D, Barbuch R, Koegel C, Bard M. Proc Natl Acad Sci USA. 1997;94:11173–11178. doi: 10.1073/pnas.94.21.11173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baudin A, Ozier-Kalogeropoulos O, Denouel A, Lacroute F, Cullin C. Nucleic Acids Res. 1993;21:3329–3330. doi: 10.1093/nar/21.14.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sikorski R S, Hieter P. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rose M D, Novick P, Thomas J H, Botstein D, Fink G R. Gene. 1987;60:237–243. doi: 10.1016/0378-1119(87)90232-0. [DOI] [PubMed] [Google Scholar]
- 17.Maniatis T, Fritsch E F, Sambrook J. Molecular Cloning. Plainview, NY: Cold Spring Harbor Lab. Press; 1982. [Google Scholar]
- 18.Bligh E G, Dyer W J. Can J Biochem. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 19.Plemenitas A, Havel C M, Watson J A. J Biol Chem. 1985;265:17012–17017. [PubMed] [Google Scholar]
- 20.Karst F, Lacroute F. Mol Gen Genet. 1977;154:269–277. doi: 10.1007/BF00571282. [DOI] [PubMed] [Google Scholar]
- 21.Lewis T L, Keesler G A, Fenner G P, Parks L W. Yeast. 1988;4:93–106. doi: 10.1002/yea.320040203. [DOI] [PubMed] [Google Scholar]
- 22.Harvey R, Bradshaw S A, O’Hara S C M, Eglinton G, Corner E D S. Phytochemistry. 1988;27:1723–1729. [Google Scholar]
- 23.Barrero A F, Oltra J E, Poyatos J A, Jimenez D, Oliver E. J Nat Prod. 1998;61:1491–1496. doi: 10.1021/np980199h. [DOI] [PubMed] [Google Scholar]
- 24.Barton D H R, Harrison D M, Widdowson D A. Chem. Commun. 1968. 17–19. [Google Scholar]
- 25.Boutard O, Dolis D, Schuber F. Biochem Biophys Res Commun. 1992;188:898–904. doi: 10.1016/0006-291x(92)91140-l. [DOI] [PubMed] [Google Scholar]
- 26.Nelson J A, Steckbeck S, Spencer T A. J Biol Chem. 1981;256:1067–1068. [PubMed] [Google Scholar]
- 27.Field R B, Holmlund C E. Arch Biochem Biophys. 1977;180:465–471. doi: 10.1016/0003-9861(77)90061-3. [DOI] [PubMed] [Google Scholar]
- 28.Taylor F R, Kandutsch A A, Gayen A K, Nelson J A, Nelson S S, Phirwa S, Spencer T A. J Biol Chem. 1986;261:15039–15044. [PubMed] [Google Scholar]
- 29.Lienhard G E, Rose I A. Biochemistry. 1964;3:190–195. doi: 10.1021/bi00890a009. [DOI] [PubMed] [Google Scholar]
- 30.Lindberg M, Gautschi F, Bloch K. J Biol Chem. 1963;238:1661–1664. [PubMed] [Google Scholar]