Abstract
Sec13 is a constituent of the endoplasmic reticulum and the nuclear pore complex (NPC). At the endoplasmic reticulum, Sec13 is involved in the biogenesis of COPII-coated vesicles, whereas at the NPC its function is unknown. We show here, by yeast two-hybrid screenings and biochemical assays, that a region at the amino terminus of the human nuclear pore complex protein Nup96 interacts with the WD (Trp-Asp) repeat region of human Sec13. By using immunofluorescence and confocal and immunoelectron microscopy, we found that in interphase, Sec13 and Nup96 are localized at both sides of the NPC in addition to other intracellular sites. In mitosis, Sec13 was found dispersed throughout the cell, whereas a pool of Nup96 colocalized with the spindle apparatus. Photobleaching experiments showed that Sec13 shuttles between intranuclear sites and the cytoplasm, and a fraction of Sec13 is stably associated with NPCs. Cotransfection of Sec13 and the Sec13 binding site of Nup96 decreased the mobile pool of Sec13, demonstrating the interaction of Sec13 and Nup96 in vivo. Targeting studies showed that Sec13 is actively transported into the nucleus and contains a nuclear localization signal. These results indicate that Sec13 stably interacts with Nup96 at the NPC during interphase and that the shuttling of Sec13 between the nucleus and the cytoplasm may couple and regulate functions between these two compartments.
The traffic of molecules between the nucleus and the cytoplasm of eukaryotic cells occurs through nuclear pore complexes (NPCs) by multiple transport pathways, which control nuclear entry and exit of molecules such as transcription factors, RNAs, kinases, and viral particles (3, 34, 37, 48). The mammalian NPC is constituted of approximately 30 proteins termed nucleoporins or Nups (5, 34, 37, 48). Two subsets of nucleoporins containing peptide repeats have been identified. The first subset includes Nups containing FG (Phe-Gly) repeats, and the second, more recently identified, includes WD (Trp-Asp) repeat-containing Nups (5, 34, 37, 48). Among the FG Nups, the p62 complex formed by four Nups (p62, p58, p54, and p45) has been well characterized and is localized at the central channel of the NPC at both the cytoplasmic and nucleoplasmic sides (13, 19, 26). Other FG Nups, such as Nup358 and Nup153, have an asymmetrical distribution, being localized either at the cytoplasmic or at the nucleoplasmic side of the NPC (44, 52, 55). FG Nups are known to be docking sites for receptor-cargo complexes at the NPC, whereas the WD repeat Nups (Nup37, Nup43, Seh1, ALADIN, RAE, and Sec13) are thought to be involved in the assembly of structural domains of the NPC (5, 34, 37, 48). However, only Sec13 has been reported to be a constituent of a partially characterized NPC subcomplex containing Nup107, Nup160, Nup133, Nup85, and Nup96, termed the Nup107-160 complex (2, 14, 21, 47, 49).
Although progress has been made in the characterization of the Nup107-160 complex, the exact function(s) of this subcomplex and the interactions of Nups within the subcomplex remain to be elucidated. Recently, the Nup107-160 complex was shown to be critical for nuclear pore complex assembly (21, 49). The Saccharomyces cerevisiae Nup84p complex is homologous to the vertebrate Nup107-160 complex and forms a Y-shaped multiprotein complex containing seven nucleoporins which have recently been assembled in vitro (28, 41). Previous studies suggest a potential role for the Nup84p complex in NPC structure and mRNA export (1, 8, 10, 18, 38, 40, 42, 46). C-Nup145p, the yeast homologue of Nup96 and also a constituent of the Nup84p complex, has been shown to have functions related to mRNA export and chromatin metabolism (8, 10, 17, 46).
Both Nup96 in vertebrates and C-Nup145p in S. cerevisiae are derived from autocleavage of precursor proteins, which is an important mechanism for correct intracellular targeting (8, 10, 14, 33, 46). Furthermore, C-Nup145p was found to form a complex with yeast Sec13 (28), which is a component of the Nup84p complex and COPII-coated vesicles (25). These vesicles mediate anterograde transport from the endoplasmic reticulum to the Golgi apparatus, requiring assembly of a coat constituted of cytosolic components, including Sar1 and the heterodimers Sec23-Sec24 and Sec13-Sec31 (25). Assembly of the Sec13-Sec31 heterodimer into the coat complex results in membrane deformation and budding of COPII vesicles (25). At the yeast NPC, Sec13 has been shown to have functions related to nuclear pore complex structure and organization (42); however, the molecular mechanisms involved in these processes are not known. Interestingly, a connection between the NPC and the endoplasmic reticulum has recently been demonstrated. Sec mutants, including Sec13, which are defective in endoplasmic reticulum structure and function, were shown to mislocalize Nups (31, 36). These findings established a cross talk between the endoplasmic reticulum and the NPC.
Here we report the interaction of human Sec13 with an amino-terminal region of human Nup96 in vitro and in vivo and further analyzed the intracellular localization of both proteins during interphase and mitosis. Moreover, we studied the intracellular dynamics and targeting of Sec13 between different cellular compartments. We have shown a stable association of Sec13 with the NPC in contrast to more mobile pools found in the cytoplasm and in the nucleus. In addition, we found that Sec13 is exchanged between the intranuclear and cytoplasmic pools, is imported into the nucleus by active transport, and contains a nuclear localization signal.
MATERIALS AND METHODS
Yeast two-hybrid screening.
The yeast two-hybrid screenings (12) were performed in the Molecular Interaction Facility, University of Wisconsin-Madison, according to their protocols. The yeast strains and vectors have been described previously (23). For bait preparation, Nup96 (amino acids 1 to 378) was cloned in frame with the GAL4 DNA-binding domain of bait vector pBUTE (a kanamycin-resistant version of GAL4 bait vector pGBDUC1). The resulting vector was sequenced to confirm an in-frame fusion, then transformed into the α mating type of S. cerevisiae strain PJ694, and tested for autoactivation of the β-galactosidase reporter gene.
Library screenings were conducted with the Molecular Interaction Facility human library collection representing cDNAs from B-cell, breast, prostate, and placenta tissues. Approximately 50 million clones were screened by mating. Of these, six yeast clones tested positive for interaction by selection on histidine drop-out and β-galactosidase assays. Plasmids were rescued and analyzed by restriction digest. Six isolated prey plasmids were retransformed into the α mating type of PJ694 and validated in a mating and selection assay with the Nup96 bait, the empty bait vector, and unrelated baits. Five clones were positive (they grew in interaction selection medium and were β-galactosidase positive) in the validation test and subsequently were identified by sequencing.
Plasmid construction.
Wild-type Nup98 and Nup96 were cloned into Myc-pAlter-MAX as described previously (14). The Nup96 truncation mutants (amino acids 1 to 427, 53 to 427, 1 to 200, 53 to 200, 201 to 427, and 428 to 849) were generated by PCR using the wild-type Nup96 cDNA as the template and cloned into the SalI and NotI sites of Myc-pAlter-MAX (Promega).
The glutathione S-transferase (GST)-Sec13 fusion encoding full-length Sec13 (amino acids 1 to 322) or Sec13 mutants (amino acids 244 to 322, 1 to 302, and 301 to 322) were generated by PCR with wild-type Sec13 cDNA as the template (kindly provided by D. I. Smith, University of Michigan, Ann Arbor) and cloned into the BamHI and NotI sites of pGEX4T3 (Amersham). A Sec13-green fluorescent protein (GFP) fusion encoding the full-length Sec13 (amino acids 1 to 322) was generated by PCR with full-length Sec13 as the template and cloned into the EcoRI and BamHI sites of pEGFP N1 (Clontech). The Myc-tagged pyruvate kinase-Sec13 fusions (amino acids 1 to 322, 1 to 243, 244 to 322, 1 to 302, and 301 to 322) were generated by PCR using full-length Sec13 as the template and cloned into the EcoRI and XhoI sites of pcDNA1-mycPK (kindly provided M. Michael, University of California, San Diego). All constructs were sequenced. The PML-enhanced green fluorescent protein (EGFP) plasmid was kindly provided by P. P. Pandolfi (Memorial Sloan Kettering Cancer Center, New York, N.Y.).
Protein expression, antibody production, and in vitro binding assays.
All wild-type and mutant Nup98 and Nup96 proteins were in vitro transcribed and translated by a coupled reticulocyte lysate transcription-translation system (Promega Corp., Madison, Wis.) in the presence of [35S]methionine, according to the manufacturer's instructions. GST fused with full-length Sec13 (amino acids 1 to 322) or fused with Sec13 mutants (amino acids 244 to 322, 1 to 302, and 301 to 322) were expressed as described previously (15, 54). For antibody production, purified GST-Sec13 (amino acids 244 to 322) was injected into rabbits, and serum was collected after an appropriate response had been elicited. Antibodies were affinity purified as previously described (11).
Binding reactions were carried out as described previously (15, 53) with recombinantly expressed GST-Sec13 fusion proteins and in vitro transcribed and translated Nup96 proteins as indicated in the figure legends. Bound and unbound fractions were separated on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and analyzed by autoradiography.
Cell culture.
HeLa cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum and antibiotics. For transfections and immunofluorescence, HeLa cells were grown on coverslips placed on 35-mm dishes and transfected with Effectene (Qiagen) following the manufacturer's instructions. After 24 h, cells were assayed for immunofluorescence studies. For live cell imaging, HeLa cells were grown and transfected with Effectene in LabTek II-chambered coverslips (Nalgene). The medium was changed 1 h before the assays, and cells were incubated at 37°C in HEPES-buffered, CO2-independent medium 199 (Gibco-BRL).
Immunoblot analysis.
Cells were lysed in 0.4 N NaOH and harvested, and the cell extracts were immediately neutralized with 0.4 N HCl. Protein concentrations were determined by the Bradford assay (Bio-Rad). Immunoblots were performed as described previously (14). Blots were probed with affinity-purified anti-Sec13 antibodies diluted 1:250 in PBS containing 2% bovine serum albumin, anti-Nup96 antibodies diluted 1:100, or affinity-purified anti-Nup96/p87 antibodies diluted 1:300. Anticalnexin antibody was obtained from Affinity Bioreagents, Inc. Antibodies were detected with the luminol-based chemiluminescence SuperSignal West Femto Maximum kit (Pierce).
Immunofluorescence and confocal microscopy.
HeLa cells grown on coverslips were washed in phosphate-buffered saline, fixed in 2% formaldehyde-phosphate-buffered saline for 30 min at room temperature, and permeabilized with 0.2% Triton X-100-phosphate-buffered saline for 5 min at room temperature or with 0.5% saponin in phosphate-buffered saline for 10 min at room temperature. In cells treated with Triton X-100 prior to fixation, cells were first incubated with 0.5% Triton X-100 for 3 min. Affinity-purified anti-Sec13 antibodies (diluted 1:100), affinity-purified anti-Nup96 antibodies (diluted 1:50), anti-Nup358 antibodies (diluted 1:2,000), monoclonal anti-c-Myc antibody 9E10 (diluted 1:40) (Boehringer Mannheim), and monoclonal antibody 414 (mAb414) (diluted 1:1,000) (7) were used. The following steps have been described previously (11). Samples were examined on a Zeiss LSM510 confocal microscope.
Isolation and fractionation of nuclear envelopes.
Nuclear envelopes were prepared from rat liver nuclei as described previously (14) except that heparin treatment was not performed.
Immunogold electron microscopy.
Isolated nuclear envelopes were isolated and processed as previously described (14). Nuclear envelopes were incubated with affinity-purified anti-Sec13 antibodies diluted 1:100, affinity-purified anti-Nup96/p87 antibodies diluted 1:300 (14), affinity-purified anti-Nup96 antibodies diluted 1:50 (11), and mouse monoclonal antibody 19C7 (specific for RanGAP1) (29). Bound antibodies were detected with goat anti-rabbit immunoglobulin G conjugated with 10-nm gold or goat anti-mouse immunoglobulin G conjugated with 5-nm gold (Amersham Life Science Inc.).
FRAP and FLIP.
Experiments were performed on a Zeiss LSM510 confocal microscope equipped with an air stream incubator as previously described (6, 9). HeLa cells expressing low levels of Sec13-EGFP or EGFP alone were selected for the experiments and monitored with a 488-nm Kr/Ar laser line at 75% laser power and 2% transmission (imaging intensity). For fluorescence recovery after photobleaching (FRAP) analyses, eight imaging scans of the area of interest were performed, and then a specific region was selected for bleaching. Twenty bleaching iterations were performed with 75% laser power and 100% transmittance. Then, scans were taken every 15 s or 45 s during the course of fluorescence recovery until the fluorescence intensity reached a plateau. FRAP recovery curves were generated from background subtracted images, and fluorescence was normalized by measuring the fluorescence intensity of an unbleached adjacent cell. The normalized fluorescence was determined for each image and compared with the initial normalized fluorescence to determine the amount of signal lost during the bleach pulse and during imaging. The equation used for these calculations has been described previously (32).
Fluorescence loss in photobleaching (FLIP) experiments were performed by bleaching an endoplasmic reticulum region of cells expressing Sec13-GFP or PML-EGFP every 4 min followed by imaging scans every 30 s to monitor recovery. In the case of EGFP alone, bleaching was performed every 30 s. This procedure was repeated until the nuclear fluorescence was significantly reduced. The selected areas chosen in Fig. 6 were bleached 10 times, which resulted in reduction of the nuclear fluorescence by approximately 80%. For plotting the data, measurements were normalized as described previously for the FRAP experiments. All FRAP and FLIP experiments were repeated five times. For quantification, three experiments were analyzed with the LSM510 software package and Excel. Videos of the experiments are added as supplements.
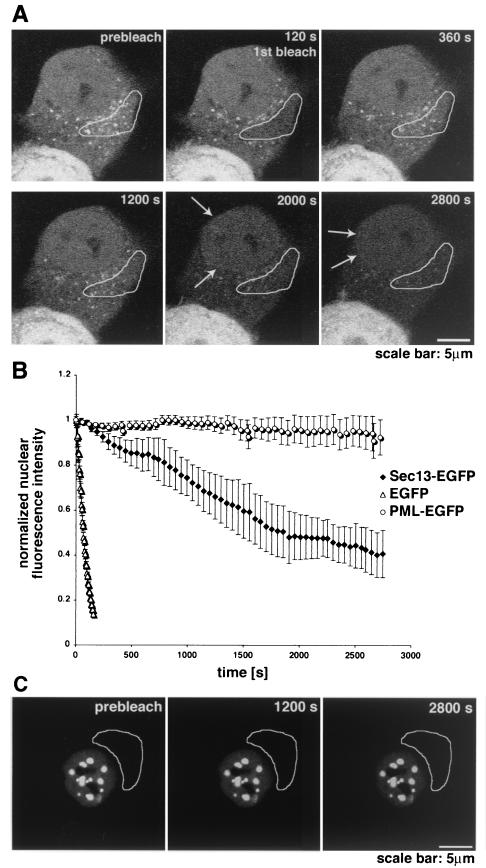
FIG. 6.
Shuttling of Sec13 between the nucleus and the cytoplasm. (A) FLIP experiments in HeLa cells transfected with Sec13-GFP. The bleached region highlighted includes the endoplasmic reticulum and cytosol. Photobleaching was performed every 4 min, and images were scanned every 30 s. Note the instant recovery of fluorescence at the endoplasmic reticulum after bleaching in contrast to the sequential loss of fluorescence observed inside the nucleus and not at the NPC. Arrows show NPC labeling, which is better visualized in the last panels. (B) Plot of fluorescence loss in the nucleus over time in cells transfected with Sec13-GFP, EGFP alone, or PML-EGFP. Error bars are standard deviations (n = 3). (C) FLIP experiments in HeLa cells transfected with PML-EGFP performed as in A. Fluorescence loss in the nucleus was not observed after photobleaching the cytoplasm 10 times.
RESULTS
Specific interaction of Sec13 with an amino-terminal region of Nup96.
To identify Nup96-interacting partners, we used yeast two-hybrid screenings and biochemical assays. Based on the predicted secondary structure of Nup96 determined by the PHD program (4), we chose an amino-terminal region of Nup96 (amino acids 1 to 378) as bait to perform yeast two-hybrid screenings. Yeast cells expressing the GAL4 DNA-binding domain fused with the amino-terminal region of Nup96 were mated with yeast cells expressing GAL4 activation domain-library protein fusions. Three different libraries derived from B cells, breast, and placenta were used. Upon protein-protein interaction, reporter genes (HIS, ADE, and LACZ) were transcribed. Interactors were selected by growth on yeast drop-out medium and confirmed by assaying for β-galactosidase activity. Plasmids encoding putative interactors were isolated from S. cerevisiae, analyzed by restriction digest, and reassessed for interaction against an array of target and nontarget bait constructs. This validation step helps to eliminate false positives that arise in a yeast two-hybrid screening.
Approximately 50 million clones were screened by mating. Of these, six yeast clones tested positive for interaction by selection on histidine drop-out medium and β-galactosidase assay. Plasmids were isolated and analyzed by restriction digest. From these six colonies, five clones listed in Fig. 1A to C grew in interaction selection medium and were β-galactosidase positive in the validation test and therefore were strong interactors. These clones were subsequently identified by sequencing as Sec13 (Sec13R).
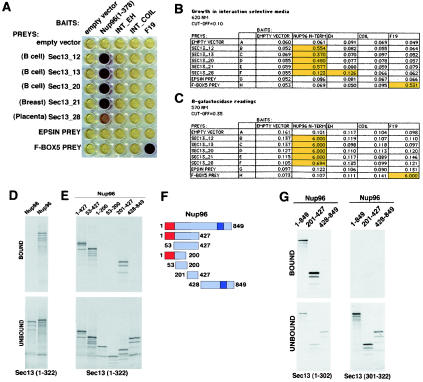
FIG. 1.
Interaction of Sec13 with an amino-terminal region of Nup96 in yeast two-hybrid screenings performed with multiple libraries. (A) β-Galactosidase assays demonstrate interaction between Sec13 cDNAs (Sec13_12, Sec13_13, Sec13_20, Sec13_21, and Sec13_28) isolated independently in yeast two-hybrid screenings and Nup96 (amino acids 1 to 378) as bait. Baits include the empty vector (pBUTE) and the following proteins cloned in frame with the GAL4 DNA binding domain: Nup96 N terminus (amino acids 1 to 378), the EF hand domain of intersectin (INT EH), the coiled-coiled domain of intersectin (INT COIL), and the human SKP1 homologue (F19). Preys include the empty prey (activation domain) vector (pGADC1), Sec13 cDNAs isolated independently from different screenings, mouse epsin, and human Fbox5. β-Galactosidase activity was assayed in cultures expressing the bait and prey following mating and selection in interaction selection medium. Purple indicates a positive signal; yellow is negative. (B) The table shows baits and preys as in A. Growth was measured by optical density at 620 nm readings 48 h after dilution of mating mixtures into interaction selection medium. (C) β-Galactosidase readings from A. High optical density values marked in yellow represent positive interactions. (D and E) Wild-type Nup98 and Nup96 and truncated mutants of Nup96 were transcribed and translated in vitro in the presence of [35S]methionine and incubated with immobilized recombinant GST-Sec13 (1 μg) as described in Materials and Methods. Bound and unbound fractions were analyzed by SDS-PAGE and autoradiography. (F) Schematic representation of wild-type and truncated mutants of Nup96. Shown in red is the 6-kDa region that is present in all isoforms of the Nup98-Nup96 gene. Shown in purple is the 9-kDa region unique to the Nup96 isoform. (G) Wild-type Nup96 and truncated mutants of Nup96 were transcribed and translated in vitro in the presence of [35S]methionine and incubated with 1 μg of immobilized recombinant GST-Sec13 mutants (amino acids 1 to 302 and 301 to 322) as described in Materials and Methods. Bound and unbound fractions were analyzed by SDS-PAGE and autoradiography.
To confirm our findings obtained with the yeast two-hybrid screenings, we performed in vitro binding studies with recombinantly expressed Sec13 and in vitro-expressed full-length Nup96, Nup98, and mutants of Nup96. As shown in Fig. 1D, full-length Nup96 interacted with Sec13, as opposed to Nup98, which did not show any significant interaction with Sec13. To map the Sec13 binding site on Nup96, we generated truncated mutants of Nup96 and again performed in vitro binding assays (Fig. 1E and F). The amino-terminal region of Nup96 (amino acids 1 to 427), which contains the sequence used as bait in the yeast two-hybrid screening, interacted with Sec13. In contrast, the carboxyl-terminal region of Nup96 (amino acids 428 to 849) did not bind Sec13.
By generating additional mutants of the amino-terminal region of Nup96 and based on the yeast two-hybrid results, the Sec13 binding site on Nup96 was mapped between residues 201 and 378. To map the domain of Sec13 that interacts with Nup96, we expressed the recombinant WD repeat region of Sec13 (amino acids 1 to 302), which is predicted to form a β-propeller structure (39), and the carboxy-terminal region of Sec13 (amino acids 301 to 322). Nup96 and the Sec13 binding site of Nup96 interacted with the WD repeat region of Sec13, whereas the carboxy terminus of Nup96 did not show any significant binding (Fig. 1G). In addition, no significant interaction was observed between full-length Nup96 or Nup96 mutants with the carboxy-terminal region of Sec13 (amino acids 301 to 322) (Fig. 1G). We also generated truncated mutants of the Sec13 propeller region, which disturbed the structure resulting in nonspecific interactions. Therefore, these mutants could not be used for binding assays (data not shown). Altogether, these results show specific interaction of the Sec13 propeller domain with an amino-terminal region of Nup96.
In interphase, Sec13 and Nup96 are localized at both sides of the NPC in addition to other intracellular sites.
To correlate our genetic and biochemical findings on the Sec13-Nup96 interaction with in situ localization data, we developed an antibody against a carboxyl-terminal region of Sec13 (amino acids 244 to 322). As shown by immunoblot analysis in Fig. 2A, affinity-purified antibodies raised against the carboxyl-terminal region of Sec13 specifically recognized Sec13 in HeLa cell lysates. We then carried out double immunofluorescence and confocal microscopy on HeLa cells with these anti-Sec13 antibodies. Sec13 was localized at the endoplasmic reticulum, cytosol, nuclear rim, and intranuclear sites (Fig. 2B to D). Sec13 colocalized with Nups that are recognized by the monoclonal antibody mAb414 (Fig. 2B), and the Sec13-GFP fusion protein showed similar localization as the endogenous proteins (Fig. 2C). In addition, Sec13 partially colocalized with calnexin at the endoplasmic reticulum (Fig. 2D).
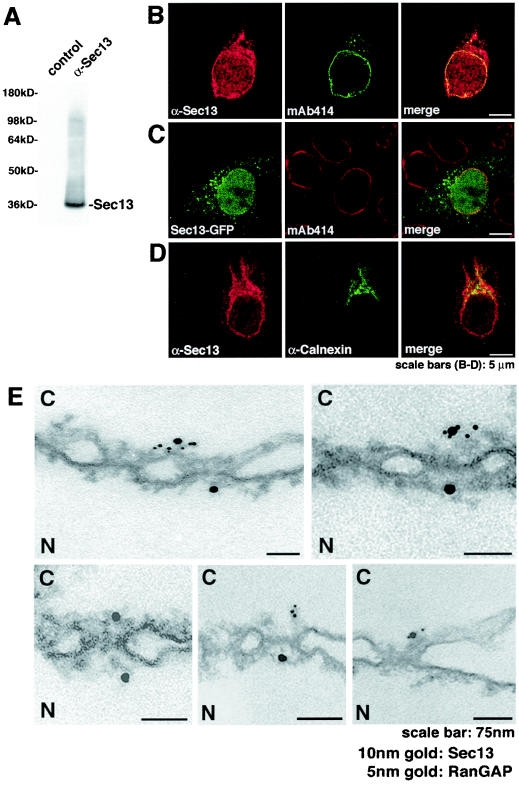
FIG. 2.
Immunolocalization of Sec13 to both sides of the NPC in addition to cytoplasmic and intranuclear sites. (A) Immunoblot analysis of HeLa cell lysates performed with anti-Sec13 antibodies and probed with either preimmune serum (control, lane 1) or antibodies against Sec13 (lane 2). (B) Immunofluorescence and confocal microscopy of HeLa cells, which were fixed, permeabilized with saponin, and labeled with anti-Sec13 antibodies and mAb414 (labels some FG repeat-containing Nups). (C) HeLa cells transfected with GFP-Sec13 and analyzed by confocal microscopy show staining similar to the observed labeling of endogenous Sec13 shown in B and D. (D) HeLa cells were processed as in B except that anticalnexin antibodies were used. Endoplasmic reticulum labeling is observed. The merged image shows partial colocalization of Sec13 with calnexin at the endoplasmic reticulum. (E) Isolated rat liver nuclear envelopes were probed with rabbit anti-Sec13 antibodies and mouse anti-RanGAP1 monoclonal antibody 19C7. Rabbit and mouse antibodies were detected with 10-nm and 5-nm gold-coupled secondary antibodies, respectively. Envelopes were processed for thin sectioning and observed by electron microscopy. C, cytoplasmic side; N, nucleoplasmic side.
In order to analyze the NPC localization of Sec13 at the ultrastructural level, we performed double immunoelectron microscopy in isolated nuclear envelopes with our newly developed anti-Sec13 antibodies and the previously characterized anti-RanGAP1 antibodies, which recognize RanGAP1 at the cytoplasmic side of the NPC (29). As shown in Fig. 2E, Sec13 localized at both the cytoplasmic and nucleoplasmic sides of the NPC, as opposed to RanGAP1, which localized at the cytoplasmic side of the NPC.
The localization of Sec13 at both sides of the NPC and its interaction with Nup96 led us to further analyze the intracellular localization of Nup96. We performed immunofluorescence and confocal microscopy with antibodies that can differentially recognize alternatively spliced forms of the Nup96 gene. Previously, we reported the identification of three precursor proteins which are derived from alternative splicing of a primary transcript termed Nup98-Nup96 and are autocatalytically proteolysed, yielding mature Nups (14, 33). The Nup98-6kD precursor yields Nup98 and a 6-kDa polypeptide, the Nup98-Nup96 precursor generates Nup98 and Nup96, and the Nup98-p87 precursor yields Nup98 and the yet uncharacterized p87 protein (14).
We previously developed antibodies that can differentially recognize the Nup96 and p87 isoforms. The difference between Nup96 and p87 is an additional 9-kDa region present at the carboxy terminus of Nup96 which is absent on p87. Therefore, we developed anti-Nup96 antibodies against this 9-kDa region (amino acids 1485 to 1559) (11) and anti-Nup96/p87 antibodies generated against a region upstream of the 9-kDa sequence (amino acids 1291 to 1482) that is common to both Nup96 and p87 (14). Expression of the p87 isoform is dependent on specific cellular environments such as interferon treatment (11). We recently reported induction of the p88 mRNA (here termed p87, based on the cDNA sequence) by reverse transcription-PCR which showed that it is considerably less abundant than Nup96 (11).
By using both anti-Nup96 and anti-Nup96/p87 antibodies in immunoblot analysis, we showed here that in U937 cells that were not treated with interferon gamma, only the Nup96 isoform was detected (Fig. 3A). To demonstrate the specificity of both antibodies to these different isoforms, we performed in vitro expression of Nup96 and p87 followed by immunoprecipitation of both proteins with anti-Nup96 and anti-Nup96/p87 antibodies (Fig. 3B). The anti-Nup96/p87 antibodies immunoprecipitated both Nup96 and p87 isoforms (Fig. 3B). In contrast, the anti-Nup96 antibodies immunoprecipitated exclusively the Nup96 isoform (Fig. 3B). The fast migrating bands obtained during in vitro translation most probably represent protein products resulting from downstream initiation of translation. Immunoblot analysis with both anti-Nup96 and anti-Nup96/p87 antibodies was also performed with U937 cells treated with 100 U of interferon gamma per ml for 12 h (Fig. 3C). In U937 cells that were incubated with interferon gamma, both Nup96 and p87 were detected, although p87 was expressed in much lower amounts than Nup96 (Fig. 3C).
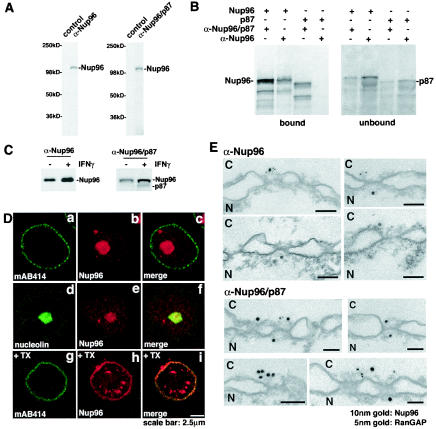
FIG. 3.
Alternatively spliced forms of the Nup98-Nup96 gene. (A) Immunoblot analysis of cell extracts obtained from U937 cells performed with preimmune serum (control), anti-Nup96 antibodies, or anti-Nup96/p87 antibodies. (B) Differential recognition of Nup96 and p87 by the anti-Nup96 and anti-Nup96/p87 antibodies. The anti-Nup96 antibodies were developed against the 9-kDa polypeptide region which is unique to the Nup96 isoform (amino acids 1485 to 1559), and the anti-Nup96/p87 antibodies were developed against a region which is common to Nup96 and p87 (amino acids 1291 to 1482). Nup96 and p87 were in vitro transcribed and translated in a reticulocyte lysate system and immunoprecipitated with the indicated antibodies to determine specificity. (C) Immunoblot analysis of cell extracts obtained from U937 cells either treated or untreated with interferon gamma performed with anti-Nup96 antibodies or anti-Nup96/p87 antibodies. Nup96 is expressed in higher amounts compared to the p87 isoform. (D) Nup96 is localized at the NPC, intranuclear sites, and nucleolus. (a to f) U937 cells were first fixed with formaldehyde, permeabilized with Triton X-100, and immunolabeled with anti-Nup96 and mAb414 antibodies (a to c) or anti-Nup96 and antinucleolin antibodies (d to f). (g to i) U937 cells were treated with Triton X-100 prior to fixation and labeled with anti-Nup96 and mAb414 antibodies. (E) Immunolocalization of Nup96 to both the cytoplasmic and nucleoplasmic sides of the NPC. Isolated rat liver nuclear envelopes were probed with rabbit anti-Nup96/p87 or anti-Nup96 antibodies and mouse anti-RanGAP1 monoclonal antibody 19C7. Rabbit and mouse antibodies were detected with 10-nm and 5-nm gold-coupled secondary antibodies, respectively. Envelopes were processed for thin sectioning and observed by electron microscopy. C, cytoplasmic side; N, nucleoplasmic side.
Immunolocalization studies previously performed with the Nup96/p87 antibodies showed labeling at the nuclear rim and intranuclear sites (14). In the absence of interferon, these cells expressed only the Nup96 isoform, indicating that the labeling observed with the Nup96/p87 antibodies reflected the intracellular localization of Nup96. We show here labeling of cells with anti-Nup96 antibodies, which localized Nup96 at the nucleolus in addition to the nuclear rim and intranuclear sites (Fig. 3D). Thus, both anti-Nup96/p87 and anti-Nup96 antibodies detected Nup96 at the nuclear rim and intranuclear sites, but only the latter labeled the nucleolus. These results indicate differences in epitope exposure. In addition, the nuclear rim could only be labeled with anti-Nup96 antibodies when cells were treated with Triton X-100 prior to fixation, which allowed epitope exposure (Fig. 3D, bottom panel).
To further analyze the NPC localization of Nup96 at the ultrastructural level, we carried out double immunoelectron microscopy with the two anti-Nup96 antibodies described above. As shown in Fig. 3E, Nup96 localized at both the nucleoplasmic and cytoplasmic sides of the NPC. In contrast, RanGAP1 was localized exclusively to the cytoplasmic side of the NPC. We previously reported the localization of Nup96 at the nucleoplasmic side of the NPC with the anti-Nup96/p87 antibodies and nuclear envelopes prepared in the presence of heparin (14). Recently, we observed that heparin used in the preparation can dissociate some Nups from the NPC, indicating that the association of Nup96 with the cytoplasmic side of the NPC is sensitive to heparin treatment, as opposed to its association with the nucleoplasmic side, which was clearly resistant to heparin treatment (14). Thus, Nup96 and Sec13 are localized at the nucleoplasmic and cytoplasmic sides of the NPC.
In mitosis, Nup98 and Sec13 are diffused throughout the cell, whereas a significant fraction of Nup96 colocalizes with the spindle apparatus.
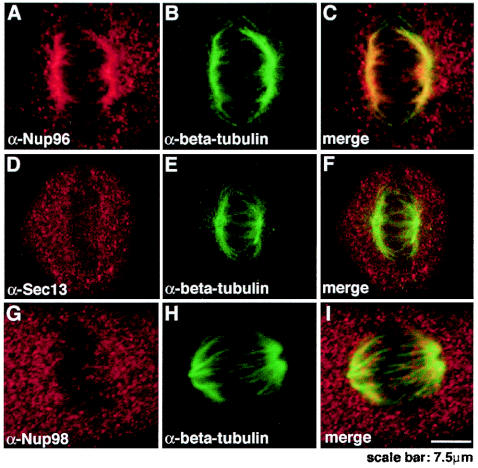
A potential role for Nups in mitosis has been reported (2). During mitosis, a fraction of Nup133 and Nup107 is localized to the kinetochores (2), raising a potential role for Nups on kinetochores or as “kinetochore-associated passenger proteins,” which would be involved in the positioning and assembly of NPCs during late mitotic processes. Nup358 has also been localized at the spindles and kinetochores (24). Therefore, to determine the localization of Nup96 and its interacting partners, Nup98 (16) and Sec13, during mitosis, we performed double immunofluorescence and confocal microscopy with anti-Nup96, anti-Nup98, and anti-Sec13 antibodies in HeLa cells. Both anti-Nup96 (Fig. 4A) and anti-Nup96/p87 antibodies (data not shown) showed colocalization of a major fraction of Nup96 with β-tubulin, indicating association of Nup96 with the spindle apparatus. Another fraction of Nup96 localized in a diffuse pattern throughout the cell. In contrast, when HeLa cells were labeled with anti-Nup98 (Fig. 4G) or anti-Sec13 antibodies (Fig. 4D) and anti-β-tubulin antibodies, Sec13 and Nup98 did not colocalize with the spindle apparatus and were distributed in a diffuse pattern throughout the cell. Thus, Nup96 and Sec13 are distributed differently throughout the cell during mitosis.
FIG. 4.
Nup96 colocalized with the spindle apparatus during mitosis, whereas Sec13 and Nup98 were distributed throughout the cell. Immunofluorescence and confocal microscopy were performed with HeLa cells at mitosis. Cells were fixed with formaldehyde, permeabilized with Triton X-100, and labeled with anti-Nup96 and anti-β-tubulin antibodies (A to C), with anti-Sec13 and anti-β-tubulin antibodies (D to F), or with anti-Nup98 and anti-β-tubulin antibodies (G to I). Merged images showed partial colocalization of Nup96 and β-tubulin at the spindle apparatus, whereas Sec13 and Nup98 did not colocalize with the spindles and was dispersed throughout the cell.
Sec13 shuttles between the nucleus and the cytoplasm and stably associates with NPCs.
The localization of Sec13 at different intracellular compartments led us to investigate the dynamics of Sec13 in the cytoplasm, endoplasmic reticulum, nuclear rim, and nucleus. Full-length Sec13 fused with GFP at the carboxy terminus was transfected into HeLa cells and subjected to fluorescence recovery after photobleaching (FRAP) and fluorescence loss in photobleaching (FLIP) experiments. Sec13-GFP strongly labeled the endoplasmic reticulum, and a less intense staining was detected at the nuclear rim, cytoplasm, and the nucleus. The same results were obtained when Sec13 was fused with GFP at the amino terminus (data not shown). This intracellular localization of Sec13-GFP is in agreement with the localization of the endogenous protein observed in Fig. 2 and also with previously published results (20, 45).
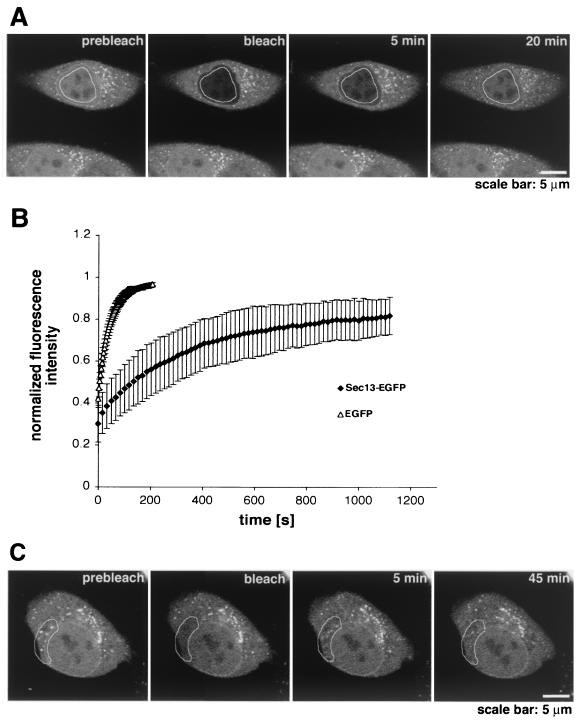
To analyze the dynamics of the intranuclear pool of Sec13, half of the nuclear area of a HeLa cell was photobleached, and fluorescence recovery was detected after 15 s, indicating very rapid movement of Sec13 between intranuclear sites (data not shown). When most of the area of a HeLa cell nucleus was photobleached, a significant recovery of intranuclear Sec13-GFP was detected after 10 min, with a half-time of 3.5 min (Fig. 5A and B). Altogether, these results suggest exchange of Sec13-GFP molecules between the nucleus and other compartments (see Fig. 6 below). Control experiments with GFP showed immediate recovery after photobleaching, with a half-time of approximately 15 s (Fig. 5B).
FIG. 5.
Dynamics of Sec13 in the nucleus and in the cytoplasm. (A) FRAP analysis of HeLa cells transfected with Sec13-GFP. The nucleus was bleached, and the fluorescence recovery in the nucleus was monitored. The image shows a confocal section at the equatorial plane of the nucleus. Images were acquired every 15 s. (B) Plots show fluorescence recovery of nuclear Sec13-GFP in the bleached area. Measurements of fluorescence intensity were subtracted from background fluorescence and normalized from loss of fluorescence during bleaching and imaging. Error bars are standard deviations (n = 3). (C) FRAP analysis of HeLa cells transfected with Sec13-GFP where the NPC, endoplasmic reticulum, and cytosol were bleached and fluorescence recovery was followed. Note the instant fluorescence recovery at the endoplasmic reticulum after 5 min as opposed to the NPC, which had still not recovered after 45 min.
When FRAP analysis was performed by photobleaching the nuclear envelope, the recovery of Sec13 at the nuclear rim was still not observed after 45 min, indicating that the pool of Sec13 localized at the nuclear rim is stably associated with NPCs (Fig. 5C). In this experiment, the selected bleached area included a region of the nuclear envelope, part of the endoplasmic reticulum, cytosol, and intranuclear sites, which was chosen to compare simultaneously the dynamics of Sec13 in these different compartments. As shown in Fig. 5C and as reported previously (50), the pools of Sec13 localized in the cytosol and endoplasmic reticulum were recovered very rapidly after photobleaching, indicating rapid movement of Sec13 between these two compartments. The equatorial section was chosen because sections closer to the top of the nucleus are in proximity to the endoplasmic reticulum, where Sec13 is rapidly exchanged, and could then result in misinterpretations. Interestingly, in FLIP experiments where the endoplasmic reticulum and cytosol were photobleached several times (Fig. 6A), a sequential loss of fluorescence intensity was observed in the nucleoplasm (Fig. 6A and B), indicating that there is exchange of Sec13 between the endoplasmic reticulum and cytosol with the nucleoplasm. Exchange of Sec13 between the endoplasmic reticulum and cytosol was also observed and occurred almost instantaneously (Fig. 6A). As a control for the FLIP experiments, we used PML-GFP, which is localized in nuclear bodies and diffusely in the nucleoplasm and has been shown to move very slowly inside the nucleus (30, 51). As shown in Fig. 6B and C, when the cytoplasm of a HeLa cell transfected with PML-GFP was photobleached several times, the fluorescence intensity of intranuclear PML-GFP was not reduced, indicating that there is no exchange of PML between the nucleus and cytoplasm as previously reported (30, 51). This result also shows the specificity of the findings obtained with Sec13, which, in contrast, is exchanged between the nucleus and cytoplasm. The FRAP and FLIP experiments with Sec13-GFP were also performed in the presence of cycloheximide and leptomycin B (data not shown). No changes in the results were observed, indicating that the intracellular dynamics of Sec13 is not dependent on ongoing protein synthesis and that its nuclear export is probably not mediated by the Crm1 pathway.
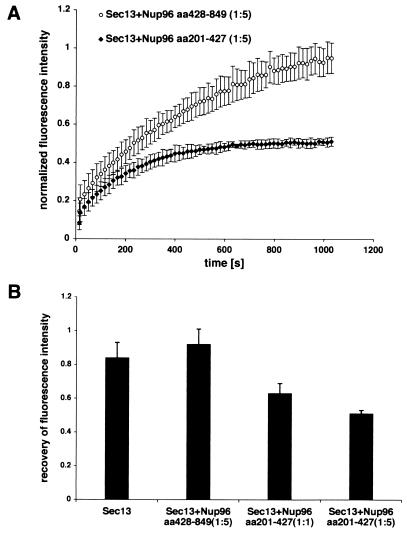
To demonstrate the interaction of Nup96 and Sec13 in vivo and its potential role in regulating Sec13 dynamics, we performed FRAP assays on HeLa cells cotransfected with Sec13-GFP and the Sec13-interacting region of Nup96 (amino acids 201 to 427) or with a carboxyl-terminal region of Nup96 (amino acids 428 to 849) which did not interact with Sec13 (Fig. 7). Strikingly, the Sec13-interacting domain of Nup96 decreased the number of Sec13-GFP molecules exchanged between the nucleus and the cytoplasm, resulting in an increase of the immobile pool of Sec13-GFP (Fig. 7). In contrast, the Sec13-GFP dynamics was not altered in the presence of a carboxyl-terminal region of Nup96 which does not interact with Sec13 (Fig. 7). These results demonstrated a specific interaction of Sec13 with an amino-terminal region of Nup96 in vivo and a potential role in regulating the intracellular dynamics of Sec13. These findings also corroborate the interaction of Sec13 with Nup96 shown by yeast two-hybrid screenings and biochemical assays (Fig. 1). In summary, Sec13 is mobile, shuttles between the nucleus and the cytoplasm, and associates stably with NPCs.
FIG. 7.
Interference with Sec13 dynamics by the Sec13-binding site of Nup96. (A) Plots show FRAP assays performed with HeLa cells cotransfected with Sec13-GFP and Nup96 (amino acids 201 to 427; the Sec13 binding site of Nup96), or Sec13-GFP and Nup96 (amino acids 428 to 849, which does not interact with Sec13). Measurements were performed as in Fig. 5 (n = 6). (B) Histograms show measurements by FRAP assays as in A of the total number of mobile Sec13-GFP molecules in the absence or presence of the Sec13-binding site of Nup96 at ratios of 1:1 or 1:5 or the carboxyl-terminal region of Nup96 (amino acids 428 to 849) at a ratio of 1:5.
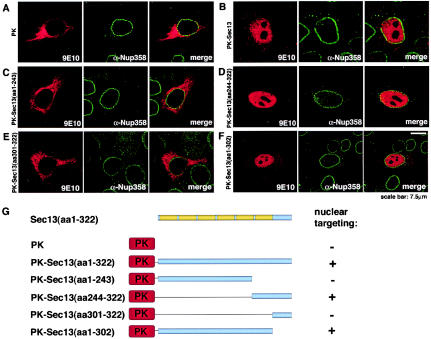
To analyze the mechanism(s) by which Sec13 can be targeted to different intracellular compartments, fusion proteins containing Myc-pyruvate kinase fused with full-length Sec13 or different regions of Sec13 were constructed, expressed in HeLa cells, and observed in a confocal microscope. As shown in Fig. 8A, the Myc-pyruvate kinase fusion protein, which lacks a nuclear localization signal, is known to be localized in the cytoplasm, but when Myc-pyruvate kinase was fused with full-length Sec13, a significant amount of Myc-pyruvate kinase-Sec13 was detected in the nucleus and a smaller pool in the cytoplasm (Fig. 8B), indicating that Sec13 has a nuclear localization signal and is actively transported into the nucleus.
FIG. 8.
The carboxy terminus of Sec13 contains a nuclear localization signal(s). HeLa cells were transfected with Myc-pyruvate kinase alone (A), Myc-pyruvate kinase fused with full-length Sec13 (amino acids 1 to 322) (B), or Myc-pyruvate kinase fused with different regions of Sec13 (amino acids 1 to 243; 244 to 322; 1 to 302; and 301 to 322) (C to F). Transiently expressed proteins were detected by immunofluorescence and confocal microscopy with anti-Myc (9E10) and anti-Nup358 antibodies, which were labeled with secondary antibodies conjugated with indocarbocyanine and fluorescein isothiocyanate, respectively. (G) Schematic representation of the constructs.
To further analyze the intranuclear targeting of Sec13, we fused several different regions of Sec13 to pyruvate kinase and determined their intracellular localizations (Fig. 8C to F and schematics in Fig. 8G). A truncated mutant containing the WD repeat domain, which is predicted to form a β-propeller structure (amino acids 1 to 302), targeted pyruvate kinase to the nucleus (Fig. 8F). Further analysis of mutants showed that the nuclear localization signal was localized between amino acids 244 and 302 (Fig. 8C to F). This region contains the sixth WD repeats of Sec13. In contrast, the region at the very carboxy terminus of Sec13 (amino acids 301 to 322) did not alter the cytoplasmic localization of pyruvate kinase (Fig. 8E). Together, these findings demonstrate a potential role of specific regions of Sec13 in actively targeting the full-length protein to different intracellular compartments.
DISCUSSION
We identified Sec13 as a Nup96-interacting partner during interphase, when both proteins colocalized at the nucleoplasmic and cytoplasmic sides of the nuclear pore complex. To investigate Nup96-interacting partners, we performed yeast two-hybrid screenings with three different libraries, including B cells, breast, and placenta. Strikingly, we pulled out Sec13 in all screenings (Fig. 1A to C). In addition, we confirmed this interaction by in vitro binding assays and mapped the Sec13 binding site to a specific amino-terminal region of Nup96 (amino acids 201 to 378) (Fig. 1D to F). This region is also present in another isoform of the Nup96 gene denominated p87, whose function is not known and which is only expressed in very small amounts under specific conditions, such as interferon treatment (11). Furthermore, we showed that Nup96 interacts with the WD repeat region of Sec13, which is predicted to form a β-propeller structure that is usually involved in protein-protein interactions (39, 43).
The interaction of Nup96 with Sec13 was corroborated by our previous findings, where we identified a complex of mammalian nucleoporins which contains Nup96, Nup107, mammalian Sec13, at least one Sec13-related protein, and at least two additional proteins of ≈150 kDa (14). Also in agreement with these results, studies with Xenopus egg extracts found Sec13 in a complex containing several Nups, including Nup160, Nup133, Nup107, Nup96, and Nup85, although the direct Sec13-interacting partner was not determined (21, 47). This complex has also been identified by Belgareh et al. (2) and termed the Nup107-160 complex.
In S. cerevisiae, Sec13 was shown to be a constituent of the Nup84 complex, which is homologous to the Nup107-160 complex in vertebrates (28, 41, 42). Furthermore, interaction of C-Nup145p, the yeast homologue of Nup96, with Sec13 has been demonstrated to be an integral part of a Y-shaped multiprotein complex, the Nup84p complex, constituted of seven nucleoporins which has recently been assembled in vitro (28, 41). The Y-shaped fork of the complex is formed by two contacts of Nup120p with the Nup85p arm and with C-Nup145p, and the elongated fibril or stalk of the Y is constituted by C-Nup145p-Sec13p-Nup84-Nup133 (28). In conclusion, Sec13 interacts with Nup96, and they are both constituents of a highly conserved NPC subcomplex.
These findings on the interaction of Sec13 with Nup96 support our results on the sublocalization of Sec13 and Nup96 to both sides of the NPC (Fig. 2E and 3E). To perform these studies, we developed a new antibody against a carboxy-terminal region of Sec13. Two different antibodies that specifically recognize Nup96 were used in these experiments (Fig. 3A and B). We previously localized Nup96 to the nucleoplasmic side of the NPC in nuclear envelopes prepared in the presence of heparin (14). In the absence of heparin, we were able to detect Nup96 at both sides of the NPC (Fig. 3E), indicating that the association of Nup96 with the cytoplasmic side of the NPC is sensitive to heparin treatment, whereas its association with the nucleoplasmic side was resistant. These results suggest that the interaction(s) of the Nup96-Sec13 complex and/or of Nup96 with the NPC may differ depending on the side of the NPC where it is localized.
Our findings on the sublocalization of Nup96 and Sec13 at both sides of the NPC are corroborated by the studies of Belgareh et al., who showed that Nup107 and Nup133, which are in the same subcomplex as Sec13, are localized at both the cytoplasmic and nucleoplasmic sides of the NPC (2). Furthermore, the yeast homologue of Nup96, C-Nup145p, was also localized at both sides of the NPC (35). Together, these studies indicate that the Nup107-160 and Nup84p complexes are localized at the cytoplasmic and nucleoplasmic sides of the NPC.
In addition to its localization at the NPC, Sec13 is also localized at the endoplasmic reticulum, cytosol, and intranuclear sites (Fig. 2). Although a small intranuclear pool of Sec13 has been observed previously, it has not been studied (20, 45). With our newly developed antibody against the carboxy-terminal region of Sec13, we clearly demonstrated that a major pool of Sec13 is also present in the nucleus (Fig. 2), which has not been fully demonstrated previously, probably due to differences in epitope exposure. Furthermore, Sec13-GFP also localized at intranuclear sites in addition to the NPC and cytoplasm (Fig. 2), and full-length Sec13 was able to target a cytoplasmic protein, pyruvate kinase, into the nucleus (Fig. 8). In the cytosol, Sec13p interacts with Sec31p, and together with Sar1p and the Sec23p-Sec24p complex, they are recruited to the endoplasmic reticulum membrane for biogenesis of the COPII coat complex (25). This complex is known to mediate vesicle budding from the endoplasmic reticulum (25). Thus, Sec13 is part of at least two distinct multiprotein complexes localized at the NPC and endoplasmic reticulum.
The presence of a WD repeat region on Sec13, which interacts with Nup96 (Fig. 1G) and is usually involved in protein-protein interactions within macromolecular complexes (43), is consistent with Sec13's being a constituent of both the NPC and COPII multiprotein complexes. It is possible that different WD repeats are involved in the interaction of Nup96 at the NPC and with Sec31 in the cytoplasm. In addition, we found that the region containing the sixth WD repeat of Sec13 also has a nuclear localization signal(s) which mediates active import of Sec13 into the nucleus (Fig. 8).
The localization of Sec13 at multiple intracellular sites led us to investigate the intracellular dynamics of Sec13. Photobleaching experiments performed with Sec13-GFP showed that there is a pool of Sec13 which is stably associated with NPCs, whereas the cytoplasmic and intranuclear pools are very dynamic, and Sec13 is exchanged between the nucleus and the endoplasmic reticulum and cytosol (Fig. 5 and 6). The stable association of Sec13 with the NPC is compatible with a putative structural role in the Y-shaped NPC subcomplex and is also in agreement with the stable association of Nup133 and Nup107 with the NPC (2), which are constituents of the same subcomplex. In contrast, the pool localized at the endoplasmic reticulum was rapidly exchanged with the cytosolic pool (Fig. 5C) (50).
Strikingly, when the Sec13 binding site of Nup96 was cotransfected with Sec13-GFP, the mobile pool of Sec13 was reduced, demonstrating in vivo the interaction of Sec13 with the Sec13 binding site of Nup96 and its potential role in regulating Sec13 dynamics (Fig. 7). Interestingly, it has been reported that the arrest of secretion response in some sec mutants inhibits nuclear import and relocates nuclear proteins and Nups to the cytoplasm as a potential mechanism for “biosynthetic economy,” which allows the cell to handle the secretion defect (31). These findings established a cross talk between the endoplasmic reticulum, NPC, and the nucleus. Although Sec13 was not shown to be part of this specific pathway, it could potentially be a constituent of similar mechanisms of other stress responses, which would be involved in coupling nuclear and cytoplasmic functions. Consistent with this mechanism, it was recently shown that mutant alleles of all COPII constituents, including Sec13, affected Nup localization (36). In these studies, it was proposed that blockage of the secretory pathway could titrate away the pool of Sec13 that is associated with the NPC or that the abnormalities of the nuclear envelope found in these mutants could prevent NPC assembly (36). We also showed that the targeting of Sec13 to the nucleus or cytoplasm may be regulated by specific regions of the Sec13 protein. Whereas the sixth WD repeat region contains a nuclear localization signal(s), other regions of Sec13 may retain it in the cytoplasm or contain a nuclear export signal(s) (Fig. 8). Thus, it would be important to dissect the molecular mechanisms that couple the endoplasmic reticulum with the NPC and nucleus.
Moreover, we showed that, during mitosis, Sec13 was dispersed throughout the cell, whereas Nup96 was partially colocalized with the spindle apparatus (Fig. 4). In addition, Nup98, which can interact with Nup96 after cleavage of the Nup98-Nup96 precursor, was also dispersed throughout the cell during mitosis. The localization of Nup96 with the mitotic spindles suggests a potential role for Nup96 at this phase of the cell cycle. A role(s) for Nups during mitosis has been proposed based on the association of Nup107 and Nup133 with kinetochores (2) and also on the localization of Nup358 at the kinetochores and spindles (24). Although the function(s) of these Nups during mitosis has not been elucidated, the Nup107-160 complex was recently shown to have a crucial role in NPC assembly (21, 49). Another possibility is that Nups are constituents and play a role in the kinetochore and/or spindle machinery during mitosis. In this regard, other components of the nuclear transport machinery, including Ran, RanGEF, and karyopherins α and β, are known to be key regulators of spindle assembly (22, 27).
In summary, Sec13 interacts with Nup96 and forms a stable complex that is localized at both sides of the NPC. Other pools of Sec13 are targeted to different cellular compartments in a sequence-specific manner. The movement of Sec13 between the nucleus and the cytoplasm may regulate the coupling of functions exerted by these two major cellular compartments.
Acknowledgments
We thank Gunter Blobel for helpful discussions and support. We thank Carlos Arana, Papia Chakraborty, and Helene Valentine for assistance in some of the experiments. We thank Helen Shio for technical support of EM samples. We thank Pier P. Pandolfi for the generous gift of the PML-EGFP plasmid. We thank Aurelian Radu, Michel C. Nussenzweig, and Victor Nussenzweig for critical reading of the manuscript and Jurgen Helmers for image processing.
This work was supported by Florida biomedical research grant BM025 (to B.F.), National Institutes of Health grant RO1 GM67159-01 (to B.F.), American Cancer Society institutional grant G98-277 (to B.F.), and by Stanley J. Glaser biomedical research grant 700393 (to B.F.).
REFERENCES
- 1.Aitchison, J. D., G. Blobel, and M. P. Rout. 1995. Nup120p: a yeast nucleoporin required for NPC distribution and mRNA transport. J. Cell Biol. 131:1659-1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belgareh, N., G. Rabut, S. W. Bai, M. van Overbeek, J. Beaudouin, N. Daigle, O. V. Zatsepina, F. Pasteau, V. Labas, M. Fromont-Racine, J. Ellenberg, and V. Doye. 2001. An evolutionary conserved NPC subcomplex, which redistributes in part to kinetochores in mammalian cells. J. Cell Biol. 154:1147-1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carmo-Fonseca, M. 2002. The contribution of nuclear compartmentalization to gene regulation. Cell 108:513-521. [DOI] [PubMed] [Google Scholar]
- 4.Combet, C., C. Blanchet, C. Geourjon, and G. Deleage. 200. NPS@Network Protein Sequence Analysis. Trends Biochem. Sci. 25:147-150. [DOI] [PubMed] [Google Scholar]
- 5.Cronshaw, J. M., A. N. Krutchinsky, W. Zhang, B. T. Chait, and M. J. Matunis. 2002. Proteomic analysis of the mammalian nuclear pore complex. J. Cell Biol. 158:915-927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Daigle, N., J. Beaudouin, L. Hartnell, G. Imreh, E. Hallberg, J. Lippincott-Schwartz, and J. Ellenberg. 2001. Nuclear pore complexes form immobile networks and a very low turnover in live mammalian cells. J. Cell Biol. 154:71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davis, L. I., and G. Blobel. 1986. Identification and characterisation of a nuclear pore complex protein. Cell 45:699-709. [DOI] [PubMed] [Google Scholar]
- 8.Dockendorff, T. C., C. V. Heath, A. L. Goldstein, C. A. Snay, and C. N. Cole. 1997. C-terminal truncations of the yeast nucleoporin Nup145p produce a rapid temperature-conditional mRNA export defect and alterations to nuclear structure. Mol. Cell. Biol. 17:906-920. (Erratum, Mol. Cell. Biol. 17:2347-2350.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ellenberg, J., and J. Lippincott-Schwartz. 1997. Fluorescence photobleaching techniques, p. 79.1-79.23. In D. Spector, R. Goldman, and L. Leinwand (ed.), Cells: a laboratory manual, vol. 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 10.Emtage, J., M. Bucci, J. Watkins, and S. Wente. 1997. Defining the essential functional regions of the nucleoporin Nup145p. J. Cell Sci. 110:911-925. [DOI] [PubMed] [Google Scholar]
- 11.Enninga, J., D. E. Levy, G. Blobel, and B. M. Fontoura. 2002. Role of nucleoporin induction in releasing an mRNA nuclear export block. Science 295:1523-1525. [DOI] [PubMed] [Google Scholar]
- 12.Fields, S., and O. Song. 1989. A novel genetic system to detect protein-protein interactions. Nature 340:245-246. [DOI] [PubMed] [Google Scholar]
- 13.Finlay, D. R., E. Meier, P. Bradley, J. Horecka, and D. J. Forbes. 1991. A complex of nuclear pore proteins required for pore formation. J. Cell Biol. 114:169-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fontoura, B. M., G. Blobel, and M. J. Matunis. 1999. A conserved biogenesis pathway for nucleoporins: proteolytic processing of a 186-kilodalton precursor generates nup98 and the novel nucleoporin, nup96. J. Cell Biol. 144:1097-1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fontoura, B. M., G. Blobel, and N. R. Yaseen. 2000. The nucleoporin nup98 is a site for GDP/GTP exchange on ran and termination of karyopherin beta 2-mediated nuclear import. J. Biol. Chem. 275:31289-31296. [DOI] [PubMed] [Google Scholar]
- 16.Fontoura, B. M. A., S. Dales, G. Blobel, and H. Zhong. 2001. The nucleoporin Nup98 associates with the intranucear filamentous protein network of TPR. Proc. Natl. Acad. Sci. USA 98:3208-3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Galy, V., J. C. Olivo-Marin, H. Scherthan, V. Doye, N. Rascalou, and U. Nehrbass. 2000. Nuclear pore complexes in the organization of silent telomeric chromatin. Nature 403:108-112. [DOI] [PubMed] [Google Scholar]
- 18.Goldstein, A. L., C. A. Snay, C. V. Heath, and C. N. Cole. 1996. Pleiotropic nuclear defects associated with a conditional allele of the novel nucleoporin Rat9p/Nup85p. Mol. Biol. Cell 7:917-934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guan, T., S. Muller, G. Klier, N. Pante, J. M. Blevitt, M. Haner, B. Paschal, U. Aebi, and L. Gerace. 1995. Structural analysis of the p62 complex, an assembly of O-linked glycoproteins that localizes near the central gated channel of the nuclear pore complex. Mol. Biol. Cell 6:1591-1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hammond, A. T., and B. S. Glick. 2000. Dynamics of transitional endoplasmic reticulum sites in vertebrate cells. Mol. Biol. Cell 11:3013-3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harel, A., A. V. Orjalo, T. Vincent, A. Lachish-Zalait, S. Vasu, S. Shah, E. Zimmerman, M. Elbaum, and D. J. Forbes. 2003. Removal of a single pore subcomplex results in vertebrate nuclei devoid of nuclear pores. Mol. Cell 11:853-864. [DOI] [PubMed] [Google Scholar]
- 22.Hetzer, M., O. J. Gruss, and I. W. Mattaj. 2002. The Ran GTPase as a marker of chromosome position in spindle formation and nuclear envelope assembly. Nat. Cell Biol. 4:E177-E184. [DOI] [PubMed] [Google Scholar]
- 23.James, P., J. Halladay, and E. A. Craig. 1996. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144:1425-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Joseph, J., S. H. Tan, T. S. Karpova, J. G. McNally, and M. Dasso. 2002. SUMO-1 targets RanGAP1 to kinetochores and mitotic spindles. J. Cell Biol. 156:595-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaiser, C., and S. Ferro-Novick. 1998. Transport from the endoplasmic reticulum to the Golgi. Curr. Opin. Cell Biol. 10:477-482. [DOI] [PubMed] [Google Scholar]
- 26.Kita, K., S. Omata, and T. Horigome. 1993. Purification and characterization of a nuclear pore glycoprotein complex containing p62. J. Biol. Chem. 113:377-382. [DOI] [PubMed] [Google Scholar]
- 27.Kunzler, M., and E. Hurt. 2001. Targeting of Ran: variation on a common theme? J. Cell Sci. 114:3233-3241. [DOI] [PubMed] [Google Scholar]
- 28.Lutzmann, M., R. Kunze, A. Buerer, U. Aebi, and E. Hurt. 2002. Modular self-assembly of a Y-shaped multiprotein complex from seven nucleoporins. EMBO J. 21:387-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matunis, M. J., E. Coutavas, and G. Blobel. 1996. A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J. Cell Biol. 135:1457-1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Muratani, M., D. Gerlich, S. M. Janicki, M. Gebhard, R. Eils, and D. L. Spector. 2002. Metabolic-energy-dependent movement of PML bodies within the mammalian cell nucleus. Nat. Cell Biol. 4:106-110. [DOI] [PubMed] [Google Scholar]
- 31.Nanduri, J., and A. M. Tartakoff. 2001. The arrest of secretion response in yeast: signaling from the secretory path to the nucleus by Wsc proteins and Pkc1p. Mol. Cell 8:281-289. [DOI] [PubMed] [Google Scholar]
- 32.Phair, R. D., and T. Misteli. 2000. High mobility of proteins in the mammalian cell nucleus. Nature 404:604-609. [DOI] [PubMed] [Google Scholar]
- 33.Rosenblum, J. S., and G. Blobel. 1999. Autoproteolysis in nucleoporin biogenesis. Proc. Natl. Acad. Sci. USA 96:11370-11375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rout, M. P., and J. D. Aitchison. 2001. The nuclear pore complex as a transport machine. J. Biol. Chem. 276:16593-16596. [DOI] [PubMed] [Google Scholar]
- 35.Rout, M. P., J. D. Aitchison, A. Suprapto, K. Hjertaas, Y. Zhao, and B. T. Chait. 2000. The yeast nuclear pore complex: composition, architecture, and transport mechanism. J. Cell Biol. 148:635-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ryan, K. J., and S. R. Wente. 2002. Isolation and characterization of new Saccharomyces cerevisiae mutants perturbed in nuclear pore complex assembly. BMC Genet. 3:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ryan, K. J., and S. R. Wente. 2000. The nuclear pore complex: a protein machine bridging the nucleus and cytoplasm. Curr. Opin. Cell Biol. 12:361-371. [DOI] [PubMed] [Google Scholar]
- 38.Santos-Rosa, H., H. Moreno, G. Simos, A. Segref, B. Fahrenkrog, N. Pante, and E. Hurt. 1998. Nuclear mRNA export requires complex formation between Mex67p and Mtr2p at the nuclear pores. Mol. Cell. Biol. 18:6826-6838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saxena, K., C. Gaitatzes, M. T. Walsh, M. Eck, E. J. Neer, and T. F. Smith. 1996. Analysis of the physical properties and molecular modeling of Sec13: a WD repeat protein involved in vesicular traffic. Biochemistry 35:15215-15221. [DOI] [PubMed] [Google Scholar]
- 40.Segref, A., K. Sharma, V. Doye, A. Hellwig, J. Huber, R. Luhrmann, and E. C. Hurt. 1997. Mex67p which is an essential factor for nuclear mRNA export binds to both poly(A)+ RNA and nuclear pores. EMBO J. 16:3256-3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Siniossoglou, S., M. Lutzmann, H. Santos-Rosa, K. Leonard, S. Mueller, U. Aebi, and E. Hurt. 2000. Structure and assembly of the Nup84p complex. J. Cell Biol. 149:41-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Siniossoglou, S., C. Wimmer, M. Rieger, V. Doye, H. Tekotte, C. Weise, S. Emig, A. Segref, and E. C. Hurt. 1996. A novel complex of nucleoporins, which includes Sec13p and a Sec13p homolog, is essential for normal nuclear pores. Cell 84:265-275. [DOI] [PubMed] [Google Scholar]
- 43.Smith, T. F., C. Gaitatzes, K. Saxena, and E. J. Neer. 1999. The WD repeat: a common architecture for diverse functions. Trends Biochem. Sci. 24:181-185. [DOI] [PubMed] [Google Scholar]
- 44.Sukegawa, J., and G. Blobel. 1993. A nuclear pore complex protein that contains zinc finger motifs, binds DNA, and faces the nucleoplasm. Cell 72:29-38. [DOI] [PubMed] [Google Scholar]
- 45.Tang, B. L., F. Peter, J. Krijnse-Locker, S. H. Low, G. Griffiths, and W. Hong. 1997. The mammalian homolog of yeast Sec13p is enriched in the intermediate compartment and is essential for protein transport from the endoplasmic reticulum to the Golgi apparatus. Mol. Cell. Biol. 17:256-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Teixeira, M. T., S. Siniossoglou, S. Podtelejnikov, J. C. Benichou, M. Mann, B. Dujon, E. Hurt, and E. Fabre. 1997. Two functionally distinct domains generated by in vivo cleavage of Nup145p: a novel biogenesis pathway for nucleoporins. EMBO J. 16:5086-5097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vasu, S., S. Shah, A. Orjalo, M. Park, W. H. Fischer, and D. J. Forbes. 2001. Novel vertebrate nucleoporins Nup133 and Nup160 play a role in mRNA export. J. Cell Biol. 155:339-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vasu, S. K., and D. J. Forbes. 2001. Nuclear pores and nuclear assembly. Curr. Opin. Cell Biol. 13:363-375. [DOI] [PubMed] [Google Scholar]
- 49.Walther, T. C., A. Alves, H. Pickergill, I. Loiodice, M. Hetzer, V. Galy, B. B. Hulsmann, T. Kocher, M. Wilm, T. Allen, I. W. Mattaj, and V. Doye. 2003. The conserved Nup107-160 complex is critical for nuclear pore complex assembly. Cell 113:195-206. [DOI] [PubMed] [Google Scholar]
- 50.Ward, T. H., R. S. Polishchuck, S. Caplan, K. Hirschberg, and J. Lippincott-Schwartz. 2001. Maintenance of Golgi structure and function depends on the integrity of endoplasmic reticulum export. J. Cell Biol. 155:557-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wiesmeijer, K., C. Molenaar, I. M. Bekeer, H. J. Tanke, and R. W. Dirks. 2002. Mobile foci of Sp100 do not contain PML: PML bodies are immobile but PML and Sp100 proteins are not. J. Struct. Biol. 140:180-188. [DOI] [PubMed] [Google Scholar]
- 52.Wu, X., L. H. Kasper, R. T. Mantcheva, G. T. Mantchev, M. J. Springett, and J. M. van Deursen. 2001. Disruption of the FG nucleoporin NUP98 causes selective changes in nuclear pore complex stoichiometry and function. Proc. Natl. Acad. Sci. USA 98:3191-3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yaseen, N. R., and G. Blobel. 1997. Cloning and characterization of human karyopherin beta3. Proc. Natl. Acad. Sci. USA 94:4451-4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yaseen, N. R., and G. Blobel. 274. GTP hydrolysis links initiation and termination of nuclear import on the nucleoporin nup358. J. Biol. Chem. 274:26493-26502. [DOI] [PubMed] [Google Scholar]
- 55.Yokoyama, N., N. Hayashi, T. Seki, N. Pante, T. Ohba, K. Nishii, K. Kuma, T. Hayashida, T. Miyata, U. Aebi, et al. 1995. A giant nucleopore protein that binds Ran/TC4. Nature 376:184-188. [DOI] [PubMed] [Google Scholar]