Abstract
Traditionally, the structure and properties of natural products have been determined by total synthesis and comparison with authentic samples. We have now applied this procedure to the first nonproteinaceous ion channel, isolated from bacterial plasma membranes, and consisting of a complex of poly(3-hydroxybutyrate) and calcium polyphosphate. To this end, we have now synthesized the 128-mer of hydroxybutanoic acid and prepared a complex with inorganic calcium polyphosphate (average 65-mer), which was incorporated into a planar lipid bilayer of synthetic phospholipids. We herewith present data that demonstrate unambiguously that the completely synthetic complex forms channels that are indistinguishable in their voltage-dependent conductance, in their selectivity for divalent cations, and in their blocking behavior (by La3+) from channels isolated from Escherichia coli. The implications of our finding for prebiotic chemistry, biochemistry, and biology are discussed.
Keywords: [bacterial calcium channel, polymer electrolyte complex, planar bilayer, poly3-hydroxybutyrate]
Synthetic and naturally occurring ion channels are amphiphilic structures with an outer coat of nonpolar residues and a lining of polar and charged residues (1–7). In this report, we demonstrate the formation of synthetic nonproteinaceous ion channels from two structurally distinct polymers that share the above attributes in a cooperative fashion. The polymers in question, poly(3-hydroxybutyrate) (PHB) and inorganic polyphosphate (polyP), are ubiquitous constituents of biological cells (8–12). PHB is an amphiphilic homopolymer of (R)-3-hydroxybutanoic acid, which in bacteria is synthesized from acetyl-CoA in three steps: dimerization to form acetoacetyl-CoA, reduction by NADPH to form 3-hydroxybutyryl-CoA, and polymerization to form PHB (13, 14). Under growth-limiting conditions, some bacteria produce high-molecular-mass PHB (60,000 to >1,000,000 Da) in amounts of up to 90% of the cell dry weight. PolyP, a polyanion composed of phosphate residues linked by anhydride bonds, is formed by repetitive phosphoryl transfer from ATP or other high-energy phosphates (15–17).
Both PHB and polyP have molecular characteristics that are consistent with a role in ion conduction. PHB has salt-solvating properties that derive from the recurrence of electron-donating ester carbonyl oxygens at regular and frequent intervals along its flexible backbone (18, 19). The solvating abilities of PHB are illustrated by studies that demonstrate its capacity to transport cations across methylene chloride layers in U-tubes (20), form ion-conducting complexes with lithium perchlorate (21), or large-conductance nonselective ion channels in planar lipid bilayers (22). PolyP has a flexible backbone and high density of monovalent negative charges that create a large capacity for ion exchange and a stronger affinity for multivalent over monovalent cations (23).
Plasma membrane vesicles of Escherichia coli contain complexes of PHB and polyP that function as calcium-selective channels in planar lipid bilayers (24). In vivo, such channels may control the influx of calcium to maintain homeostasis or effect transient increases to trigger certain physiological responses. The importance of calcium regulation in prokaryotes is becoming evident as calcium is increasingly implicated in bacterial functions, including chemotaxis, cell division, differentiation, and pathogenesis (25). The channel complexes can be extracted from the membranes into chloroform and purified by HPLC nonaqueous size-exclusion chromatography (24). Analysis of purified channel extracts indicated that the channels were composed of PHB (130–140 residues) and polyP (55–65 residues). No protein was detected. When the complexes were reconstituted from purified E. coli PHB and synthetic calcium polyphosphate, the same channel activity was observed; however, inasmuch as hydrophobic proteins bind very tightly to PHB, the presence of trace protein or peptide contaminants in PHB of cellular origin could not be ruled out. Consequently, doubt remained as to whether the observed channel activity was entirely attributable to complexes of these two simple polymers. In this study, we prove by total synthesis of the channel complexes that protein is not essential to the observed ion channel activity.
METHODS
Preparation of HB128/polyP Complexes.
Ca(polyP) was prepared from sodium polyphosphate glass (Av residue number 65; Sigma) and calcium chloride as previously described (24). A chloroform solution of HB128 (1 μg/ml) was added to dry, finely pulverized Ca(polyP). The chloroform was removed by evaporation with a stream of dry nitrogen, and the mixture was heated in a microwave oven for 0.5 min (3×). A dry, cold chloroform solution of synthetic 1-palmitoyl, 2-oleoyl, phosphatidylcholine (10 μg/ml) was added to the dry blend of HB128 and Ca(polyP), and the mixture was sonicated in a ultrasonication bath (47 kHz, Model 2210, Branson) for ca. 20 min. The temperature was 0°C at the beginning of sonication and ≈22°C at the end. The supernatant containing the complexes was filtered with a Teflon syringe filter (0.2 μm) and stored at −20°C.
Isolation of PHB/CapolyP Complexes from E. coli.
Complexes were isolated from competent cells of E. coli DH5α as previously described (24). E. coli DH5α cells were made genetically competent by a variation of the method of Hanahan (26) as previously described (27). Briefly, cells were cultured in SOB medium [2% Bacto-tryptone (Difco)/0.5% yeast extract (Difco)/10 mM NaCl/2.5 mM KCl] to an absorbance at 550 nm of ca. 0.4. The cells were pelleted at low centrifugal speed (800 g) at 4°C and gently resuspended in 1/3 volume of 100 mM KCl/45 mM MnCl2/10 mM CaCl2 in 4-morpholineethanesulfonic acid (Mes) (Sigma), which was neutralized to pH 6.3 with KOH, for 30–60 min at 4°C. The cells were then collected under the same conditions as above and washed sequentially with methanol (2×), methanol:acetone (1:1) (2×), and acetone (2×) and then extracted with dry, cold chloroform. The extract was filtered and stored as above.
Determination of PHB.
PHB was measured by chemical assay by a variation of the method of Karr et al. (28) as previously described (24). Briefly, the dry sample was hydrolyzed in concentrated sulfuric acid at 92°C for 30 min, and the resulting crotonic acid was extracted, separated by HPLC chromatography using a Spectra-Physics SP8700 Chromatography System with a Bio-Rad Organic Acids Column HPX87H, and quantitated by comparison of peak area, measured on a Shimadzu C-R3A Chromatopac Integrator, with that of crotonic acid standards.
Size Determination of PolyP.
PolyP was extracted from the filtered chloroform solution containing HB128/polyP complexes with 10 mM Tris⋅EDTA (TE buffer), pH 7.5. The average chain length of polyP was determined by polyacrylamide gel electrophoresis as previously described (29) using polyP standards prepared as described by Clark and Wood (29).
Incorporation of HB128/CapolyP and E. coli PHB/CapolyP Complexes in Planar Bilayers.
A portion of the chloroform solution containing the complexes was added to a solution of synthetic 1-palmitoyl, 2-oleoyl, phosphatidylcholine (Avanti Polar Lipids) and cholesterol (5:1; wt/wt) in decane (40 mg/ml). Final concentrations of HB128 and phospholipid were in a weight ratio of 1:10,000. The chloroform was evaporated, and the remaining decane solution was used to paint a bilayer in a small aperture (0.2 mm diameter) in a partition between two aqueous bathing solutions in a Delrin cuvette (Warner Instruments, Hamden, CT) (31).
Planar Bilayer Measurements.
The aqueous compartments were connected to an integrating patch-clamp amplifier (Axopatch 200A, Axon Instruments) via a matched pair of Ag/AgCl electrodes; the cis compartment was connected to the head-stage (CV 201A) of the amplifier, and the trans compartment was held at virtual ground. Channel currents were recorded at full bandwidth on a video cassette recorder (Sony) after digitization through an A-D converter (VR-10B, Instrutech, Mineola, NY). Data was filtered through an eight-pole Bessel filter (Model 200, Frequency Devices, Haverhill, MA), sampled at a rate equal to or greater than the corner frequency using a Labmaster Tl-1 DMA interface (Axon Instruments) connected to a Pentium microcomputer. All experiments were performed at 22°C.
RESULTS AND DISCUSSION
To resolve the question of whether PHB/CapolyP complexes alone are capable of forming calcium-selective channels in planar lipid bilayers, we carried out a total synthesis of the channel complex from (R)-3-hydroxybutanoic acid, sodium polyphosphate, and calcium chloride. An exponential fragment-coupling strategy was employed to prepare the 128-mer of 3-hydroxybutyrate (HB128) (32). This synthetic polymer has a molecular mass of 11,000 kDa, which is close to that of E. coli PHB (ca. 12 kDa), as estimated by size-exclusion chromatography (11). Calcium polyphosphate was prepared from sodium polyphosphate glass (Av residue number 65) and calcium chloride. A mixture of the two dry polymers was briefly heated and then sonicated in a dry, cold chloroform solution of synthetic 1-palmitoyl, 2-oleoyl, phosphatidylcholine (see Methods). Because polyP is itself highly insoluble in chloroform, only PHB-complexed polyP is present in the filtered supernatant. The chain length of polyP in the chloroform filtrate was determined by acrylamide gel electrophoresis to be in the same range (55–65) as found in the E. coli complexes (30).
Incorporation of HB128/polyP complexes into planar bilayers was accomplished by direct addition of a chloroform solution of the complexes to synthetic 1-palmitoyl, 2-oleoyl, phosphatidylcholine and cholesterol (5:1; wt/wt) in decane (40 mg/ml), evaporating the chloroform, and using the resulting decane solution to paint a bilayer in a 200-μm aperture in a partition between two symmetrical bathing solutions. The aqueous compartments were connected to an integrating patch-clamp amplifier, and channel currents were recorded (see Methods). PHB/CapolyP complexes were isolated from E. coli DH5α as previously described (24) and incorporated into planar bilayers of the same lipids in the same manner.
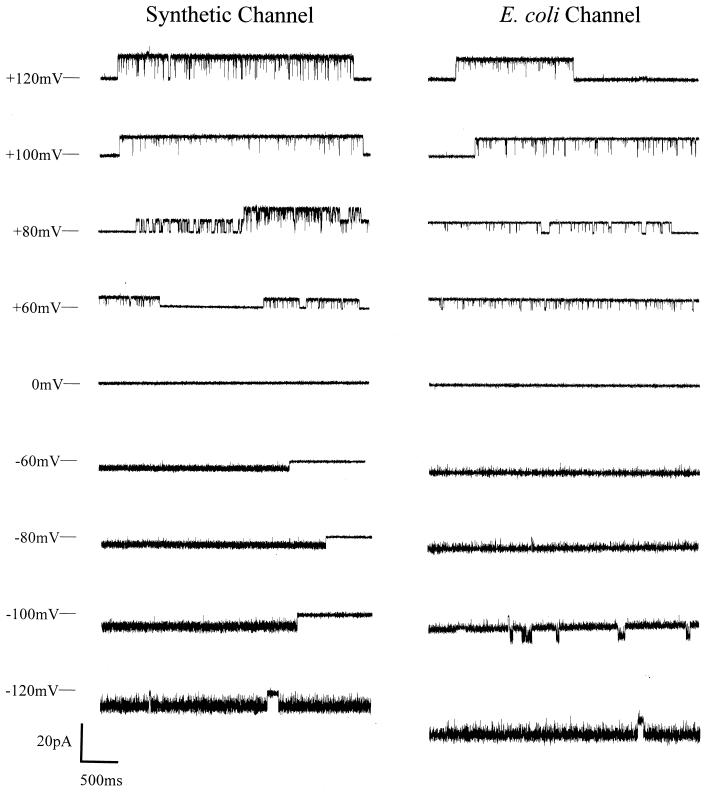
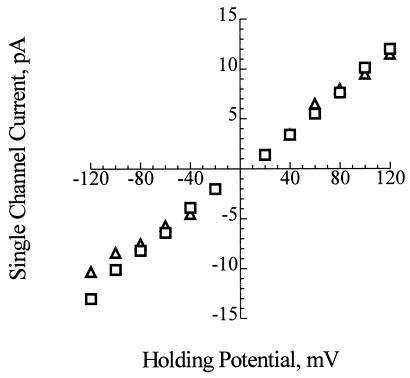
As shown in Fig. 1, the channels formed in planar bilayers between symmetrical bathing solutions of 200 mM CaCl2, 5 mM MgCl2, 10 mM Tris⋅Hepes, pH 7.4, by the synthetic complexes are virtually identical to those formed by PHB/polyP complexes extracted from E. coli. The conductances of the synthetic and E. coli channels are indistinguishable (Fig. 2), and their gating profiles are also similar (Fig. 1). The conductance of the fully open synthetic channels is estimated to be 101 ± 6 pS (n = 10) and that of the E. coli channel is 104 ± 12 pS (n = 10).
Figure 1.
Representative single-channel current fluctuations at various clamping potentials. (Left) Synthetic HB128/polyP. (Right) Channels extracted from competent cells of E. coli DH5α. Complexes were incorporated into planar lipid bilayers composed of synthetic 1-palmitoyl, 2-oleoyl, phosphatidylcholine and cholesterol (5:1; wt/wt) (see Methods) with symmetrical bathing solutions of 200 mM CaCl2, 5 mM MgCl2, 10 mM Tris⋅Hepes, pH 7.4, at 22°C. The bars at the side of each profile indicate the fully closed state of the channel. Clamping potentials with respect to ground are indicated along the left side of the figure. Data shown were filtered at 1 kHz.
Figure 2.
Current-voltage relations for synthetic HB128/polyP (□) and E. coli PHB/polyP channel complexes (▵). The conductance of the channel for Ca2+ in symmetric solutions under the same experimental conditions (described in Fig. 1) is 101 ± 6 pS for the synthetic channels and 104 ± 12 pS for the E. coli channels. The data points represent mean values of 10 observations.
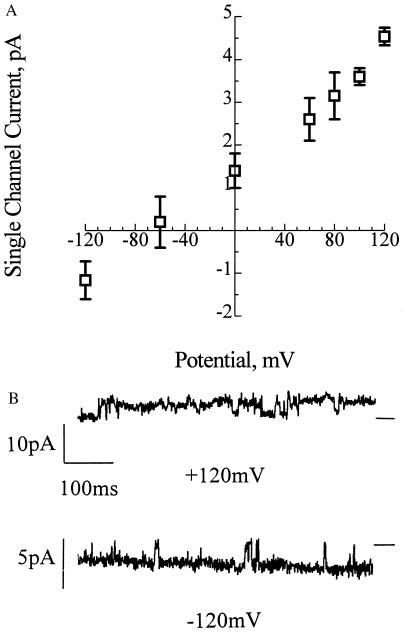
The synthetic channels exhibit the same selectivity for divalent over monovalent cations as the E. coli channels (24). Fig. 3 shows the single-channel current-voltage relations with 65 mM CaCl2, 10 mM NaCl, 5 mM MgCl2, 10 mM Tris⋅Hepes, pH 7.4, cis and 200 mM NaCl, 0.05 mM CaCl2, 5 mM MgCl2, 10 mM Tris⋅Hepes, pH 7.4 trans. The reversal potential was close to the equilibrium potential for calcium of −86 mV calculated from the Nernst equation, whereas the calculated equilibrium potential for chloride was +9 mV and that for sodium was +76 mV. Such selectivity was not observed in channels composed of PHB alone, either with small synthetic PHBs (16–96 residues) in earlier studies (22) or with the 128-mer in this study. The selectivity of PHB/polyP complexes for divalent cations may be largely attributed to the inherent molecular characteristics of polyP, whose well-known preference for binding divalent over monovalent cations has commercial applications in water softening (23). Like most protein calcium channels, the synthetic and E. coli PHB/polyP channels are permeant to the alkaline earth metal cations Ca2+, Sr2+, and Ba2+.
Figure 3.
Selectivity of synthetic HB128/polyP complexes for divalent cations over monovalent cations. (A) The figure shows selectivity of HB128/polyP channels for Ca2+ over Na+. Single-channel current-voltage relations of HB128/polyP complexes incorporated in planar lipid bilayers composed of synthetic 1-palmitoyl, 2-oleoyl, phosphatidylcholine and cholesterol (5:1; wt/wt) between asymmetric solutions: cis (ground) (65 mM CaCl2, 10 mM NaCl, 5 mM MgCl2, 10 mM Tris⋅Hepes, pH 7.4), and trans (ground) (200 mM NaCl, 0.1 mM CaCl2, 5 mM MgCl2, 10 mM Tris⋅Hepes, pH 7.4). The equilibrium potentials (calculated from concentration) were ECa = −82 mV, ECl = +9 mV, and ENa is +76 mV. Error bars indicate standard deviation of the mean (n = 3). (B) Representative current traces of HB128/polyP channels at +120 mV and −120 mV under the experimental conditions detailed in A. Data were filtered at 300 Hz. Note the difference in the current scales. The bars at the side of each profile indicate the fully closed state of the channel.
Both synthetic and E. coli PHB/polyP channels exhibit multiple subconductance states and complex gating kinetics. As shown in Fig. 1, the channel conductances fluctuate between two major modes of gating: (i) in mode 1, illustrated at positive clamping potentials, the channel openings are several seconds long with few complete closures, occasionally interspersed by long closed states; and (ii) in mode 2, shown at negative clamping potentials, the channels open in long bursts with fast-flickering closures to an intermediate closed state as well as to the main closed state. Switching between modes 1 and 2 can be observed both at negative and positive clamping potentials.
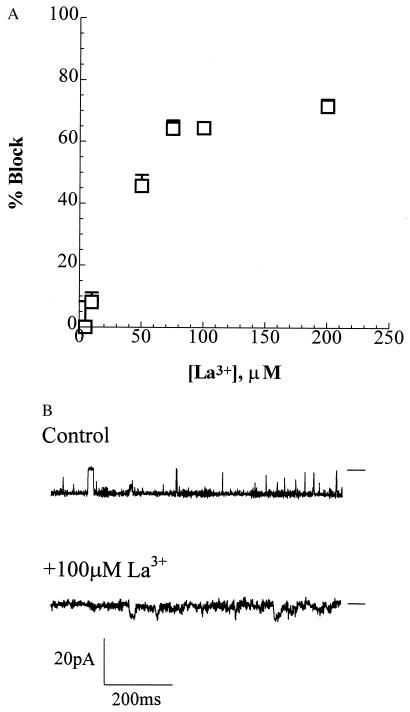
The synthetic and E. coli channels both demonstrate sensitivity to blocking by La3+. The macroscopic currents with several channels in the bilayer show concentration dependent blocking. Currents in the synthetic channels were reduced 50% at concentrations of 50 μM (Fig. 4). In comparison, 50% inhibition of single-channel currents in the E. coli channel was achieved at <100 μM (24). The blockade of PHB/polyP channel complexes by La3+ is an expected consequence of the strong attraction and tight binding of trivalent cations to polyP.
Figure 4.
Block of synthetic HB128/polyP channel by transition metal cation La3+. (A) The bilayer was formed in symmetrical bathing solutions of 200 mM CaCl2, 5 mM MgCl2, 10 mM Tris⋅Hepes, pH 7.4. The clamping potential was −80 mV. After incorporation of the channel, activities were recorded for 5 min. Then, La3+ was added to the trans compartment to achieve the indicated concentrations. The bath was stirred and activities were recorded after 1 min of addition of La3+. Data points represent mean values of the amplitude histograms at −80 mV clamping potential; error bars show the standard deviation from the mean. Data were filtered at 500 Hz. (B) Blockade by 100 μM La3+ under the experimental conditions detailed in A. Data were filtered at 1 kHz. The bars at the side of each profile indicate the fully closed state of the channel.
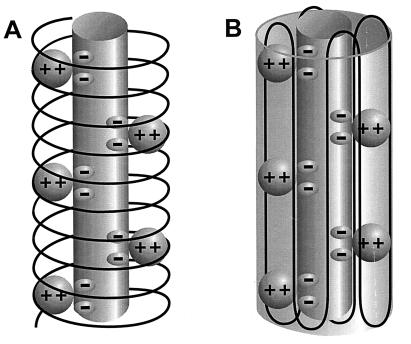
The molecular conformation of the complexes is not yet known. However, the physical properties of the two polymers point toward a structure in which polyP forms an ion-binding column across the hydrophobic region of the bilayer, shielded from the low dielectric environment by amphiphilic PHB. Two models have been proposed for the spatial arrangement of PHB in the complexes. Reusch et al. (24, 33) developed a model based on the physical properties of the two polymers, the coordination preferences of Ca2+, and computer-assisted molecular modeling. They postulate that PHB wraps around Ca(polyP), forming a helix with an external diameter of ca. 24 Å and length of ca. 45 Å. The geometrical arrangement of the ester carbonyl oxygens of PHB and the phosphoryl oxygens of polyP along the channel walls corresponds to the preferred coordination geometry of Ca2+, Sr2+, and Ba2+ (Fig. 5A). Seebach et al. (34, 35) proposed that the PHB molecules are oriented as 21-helices of 6 Å pitch, as determined for the bulk polymer by fiber x-ray scattering measurements (36–39), and fold in the same lamellar morphology as PHB crystallites (40). Each molecule of ca. 140 monomer residues would fold in 16 residues per turn across the bilayer. Eight consecutive antiparallel helices then form an ellipsoid arrangement of ca. 20 Å diameter, reminiscent of a β-sheet, surrounding the Ca(polyP) (Fig. 5B).
Figure 5.
Diagrammatic representations of PHB/PPi channels. The central cylinder represents the polyP helix, which contains pairs of closely spaced monovalent negative charges that provide a ladder of binding sites for Ca2+. The Ca(polyP) is surrounded and solvated by the amphiphilic molecule, PHB. (A) PHB forms a helix encircling Ca(polyP). (B) PHB forms a β-like sheet surrounding Ca(polyP). More detailed molecular representations of the two structures may be found in refs. 24 and 33 for A and refs. 34 and 35 for B.
The models postulated for PHB/polyP bear a remarkable resemblance to the single-file, multiple-site structures described for protein calcium channels (41–43), and, despite their simplicity, the complexes mimic the behavior of the protein channels in many ways. It is conceivable that PHB and polyP were accessible to the earliest cells. PolyP is found in volcanic condensates and is considered to be prebiotic (15–17, 44), and the synthesis of PHB requires only acetate and reducing potential (13). Hence, it is intriguing to postulate that PHB/polyP channels date from primordial times and are the progenitors of eukaryotic calcium channels.
Acknowledgments
We thank W. H. Reusch and G. Huisman for critical reading of the manuscript. This work was supported by a grant from the Division of Molecular and Cellular Biosciences at the National Science Foundation to R.N.R.
ABBREVIATIONS
- PHB
poly(3-hydroxybutyrate)
- polyP
polyphosphate
References
- 1.Epand R M. The Amphipathic Helix. Ann Arbor, MI: CRC; 1993. pp. 222–246. [Google Scholar]
- 2.Sansom M S P. Prog Biophys Mol Biol. 1991;55:139–151. doi: 10.1016/0079-6107(91)90004-c. [DOI] [PubMed] [Google Scholar]
- 3.Urry D W. Top Curr Chem. 1985;128:175–185. [Google Scholar]
- 4.Fyles T M, James T D, Kaye K C. J Am Chem Soc. 1993;115:12315–12321. [Google Scholar]
- 5.Kobuke Y, Ueda K, Sokabe M. J Am Chem Soc. 1992;114:7618–7622. [Google Scholar]
- 6.Christensen B, Fink J, Merrifield R B, Mauzerall D. Proc Natl Acad Sci USA. 1988;85:5072–5076. doi: 10.1073/pnas.85.14.5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lehn J M. Struct Bonding (Berlin) 1973;16:1–69. [Google Scholar]
- 8.Reusch R N. Proc Soc Exp Biol Med. 1989;191:377–381. doi: 10.3181/00379727-191-42936. [DOI] [PubMed] [Google Scholar]
- 9.Reusch R N. FEMS Rev. 1992;103:119–130. doi: 10.1111/j.1574-6968.1992.tb05829.x. [DOI] [PubMed] [Google Scholar]
- 10.Müller H M, Seebach D. Angew Chem. 1994;32:477–502. [Google Scholar]
- 11.Seebach D, Brunner A, Bürger H M, Schneider J, Reusch R N. Eur J Biochem. 1994;224:317–328. doi: 10.1111/j.1432-1033.1994.00317.x. [DOI] [PubMed] [Google Scholar]
- 12.Huang R, Reusch R N. J Biol Chem. 1996;271:22196–22202. doi: 10.1074/jbc.271.36.22196. [DOI] [PubMed] [Google Scholar]
- 13.Dawes E A, Senior P J. Adv Microb Physiol. 1973;10:135–206. doi: 10.1016/s0065-2911(08)60088-0. [DOI] [PubMed] [Google Scholar]
- 14.Anderson A J, Dawes E A. Microbiol Rev. 1990;54:450–472. doi: 10.1128/mr.54.4.450-472.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kulaev I S. In: The Biochemistry of Inorganic Polyphosphates. Brookes R F, editor. New York: Wiley; 1979. [Google Scholar]
- 16.Kulaev I S, Vagabov V M. Adv Microb Phys. 1983;24:83–171. doi: 10.1016/s0065-2911(08)60385-9. [DOI] [PubMed] [Google Scholar]
- 17.Kornberg A. J Bacteriol. 1995;177:491–495. doi: 10.1128/jb.177.3.491-496.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Armand M B. In: Polymer Electrolyte Reviews-1. MacCallum J R, Vincent C A, editors. New York: Elsevier; 1987. pp. 1–22. [Google Scholar]
- 19.Gray F M. Solid Polymer Electrolytes. New York: VCH; 1992. pp. pp.1–3. [Google Scholar]
- 20.Bürger H M, Seebach D. Helv Chim Acta. 1993;76:2570–2580. [Google Scholar]
- 21.Reusch, R. N. & Reusch, W. H. (1993) U.S. Patent No. 5,266,422.
- 22.Seebach D, Brunner A, Bürger H M, Reusch R N, Bramble L L. Helv Chim Acta. 1996;79:507–517. [Google Scholar]
- 23.Corbridge D E C. Stud Inorg Chem. 1985;6:170–178. [Google Scholar]
- 24.Reusch R N, Huang R, Bramble L L. Biophys J. 1995;69:754–766. doi: 10.1016/S0006-3495(95)79958-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Norris V, Grant S, Freestone P, Canvin J, Sheikh F N, Toth I, Trinel M, Modha K, Norman R I. J Bacteriol. 1996;178:3677–3682. doi: 10.1128/jb.178.13.3677-3682.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hanahan D. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 27.Reusch R N, Hiske T W, Sadoff H L. J Bacteriol. 1986;168:553–562. doi: 10.1128/jb.168.2.553-562.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karr D B, Waters J K, Emerich D W. Appl Environ Microbiol. 1983;46:1339–1344. doi: 10.1128/aem.46.6.1339-1344.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Clark J E, Wood H G. Anal Biochem. 1987;16:280–290. doi: 10.1016/0003-2697(87)90452-0. [DOI] [PubMed] [Google Scholar]
- 30.Castuma C E, Huang R, Kornberg A, Reusch R N. J Biol Chem. 1995;270:12980–12983. doi: 10.1074/jbc.270.22.12980. [DOI] [PubMed] [Google Scholar]
- 31.Mueller P, Rudin D O. Curr Top Bioeng. 1969;3:157–249. [Google Scholar]
- 32.Lengweiler U D, Fritz M G, Seebach D. Helv Chim Acta. 1996;79:670–701. [Google Scholar]
- 33.Reusch R N, Sadoff H L. Proc Natl Acad Sci USA. 1988;85:4176–4180. doi: 10.1073/pnas.85.12.4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seebach D, Bürger H M, Müller H M, Lengweiler U D, Beck A K. Helv Chim Acta. 1994;77:1099–1123. [Google Scholar]
- 35.Seebach D, Brunner A, Bachmann B M, Hoffmann T, Kühnle F N, Lengweiler U D. Ernst Shering Res Found. 1995;28:7–98. [Google Scholar]
- 36.Cornibert J, Marchessault R H. J Mol Biol. 1972;71:735–756. doi: 10.1016/s0022-2836(72)80035-4. [DOI] [PubMed] [Google Scholar]
- 37.Yokuchi M, Chaitani Y, Tadokoror H, Teranishi K, Tani H. Polymer. 1973;14:267–272. [Google Scholar]
- 38.Cornibert J, Marchessault H. Macromolecules. 1975;8:296–305. [Google Scholar]
- 39.Brückner S, Meille A V, Malpezzi L, Cesaro A, Navarini L, Tombolini R. Macromolecules. 1988;21:967–972. [Google Scholar]
- 40.Welland E L, Stejny J, Halter A, Keller A. Polym Commun. 1989;30:302–304. [Google Scholar]
- 41.Almers W, McCleskey E W, Palade P T. J Physiol. 1984;353:585–608. doi: 10.1113/jphysiol.1984.sp015352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hess P, Tsien R W. Nature (London) 1984;309:453–456. doi: 10.1038/309453a0. [DOI] [PubMed] [Google Scholar]
- 43.Tsien R W, Hess P, McCleskey E W, Rosenberg R L. Annu Rev Biophys Biophys Chem. 1987;16:265–290. doi: 10.1146/annurev.bb.16.060187.001405. [DOI] [PubMed] [Google Scholar]
- 44.Yamagata Y, Watanabe H, Saitoh M, Namba T. Nature (London) 1991;352:516–519. doi: 10.1038/352516a0. [DOI] [PubMed] [Google Scholar]