Abstract
Aberrant DNA methylation is a common phenomenon in human cancer, but its patterns, causes, and consequences are poorly defined. Promoter methylation of the DNA mismatch repair gene MutL homologue (MLH1) has been implicated in the subset of colorectal cancers that shows microsatellite instability (MSI). The present analysis of four MspI/HpaII sites at the MLH1 promoter region in a series of 89 sporadic colorectal cancers revealed two main methylation patterns that closely correlated with the MSI status of the tumors. These sites were hypermethylated in tumor tissue relative to normal mucosa in most MSI(+) cases (31/51, 61%). By contrast, in the majority of MSI(−) cases (20/38, 53%) the same sites showed methylation in normal mucosa and hypomethylation in tumor tissue. Hypermethylation displayed a direct correlation with increasing age and proximal location in the bowel and was accompanied by immunohistochemically documented loss of MLH1 protein both in tumors and in normal tissue. Similar patterns of methylation were observed in the promoter region of the calcitonin gene that does not have a known functional role in tumorigenesis. We propose a model of carcinogenesis where different epigenetic phenotypes distinguish the colonic mucosa in individuals who develop MSI(+) and MSI(−) tumors. These phenotypes may underlie the different developmental pathways that are known to occur in these tumors.
Colon cancer is thought to arise through the progressive accumulation of defects in various tumor-suppressor genes and oncogenes that first cause hyperproliferation of the normal epithelium, followed by adenoma formation and subsequent malignant transformation (1). The process of transformation is accelerated in patients with hereditary nonpolyposis colorectal cancer (HNPCC) in whom inherited mutations in the MutS homologue 2, MutL homologue 1 (MLH1), or other genes cause a profound DNA mismatch repair defect giving rise to microsatellite instability (MSI) in tumors. Apart from HNPCC, the MSI(+) phenotype occurs in some 15% of unselected colorectal cancers (2). However, structural changes of DNA mismatch repair genes are detected much less frequently in these sporadic colorectal cancers as compared with HNPCC (3), which has prompted the search for additional mechanisms for DNA mismatch repair gene inactivation.
Widespread changes in DNA methylation have long been observed in colon cancer, ranging from decreased genomic methylation in tumor tissue (4) to targeted de novo methylation of promoter regions of tumor-suppressor genes (5, 6). Recently, it was shown that the MLH1 gene (but not the MutS homologue 2 gene) was prone to inactivation by promoter hypermethylation (6–8). These observations have raised several important questions. Are methylation changes affecting the MLH1 promoter specific to this gene, or alternatively, indicative of more generalized patterns that might be defined, for example, by the involved loci or the type of tumors? Is altered methylation restricted to neoplastic tissue or does it occur in normal tissues as well? What is the etiology and functional significance of DNA methylation changes in colon cancer development?
To address some of these questions, we investigated a large series of colorectal tumors and report that hypermethylation of one or several CCGG sites at the MLH1 promoter region is a feature of MSI(+) tumors, whereas hypomethylation of the same sites relative to normal mucosa characterizes MSI(−) tumors. We provide evidence that these methylation changes are likely to represent more generalized patterns involving hypermethylation in MSI(+) tumors and hypomethylation in MSI(−) tumors.
Materials and Methods
Tissue Samples.
Paired fresh-frozen normal colonic mucosa and colorectal tumor samples of 89 individuals were studied for methylation changes, representing a prospective collection of 509 unselected tumors previously analyzed for MSI (9). Of these, all available tumors that had shown MSI but no germline mutations of the MutS homologue 2 or MLH1 genes by direct sequencing (n = 51) were included. Additionally, 38 MSI(−) tumors were included on the basis of random selection, except that equal numbers of proximal and distal tumors were chosen. All patients gave informed consent before sample collection, according to institutional guidelines.
DNA Methylation Analysis.
A PCR-based assay was applied that relies on the inability of the HpaII restriction enzyme to cut CCGG sequences with the internal cytosine methylated. The methylation status of four HpaII sites contained at the MLH1 promoter region (located at nucleotide positions −567, −527, −347, and −341, GenBank accession no. U83845) was studied by first digesting the samples with HpaII and thereafter amplifying each region individually with flanking primers. The two adjacent HpaII sites at positions −347 and −341 were amplified in a single reaction (MLH1/3). A similar assay was previously used to study all sites simultaneously (7) or the two sites contained in MLH1/3 (8). The primer sequences were as follows: MLH1/1, sense 5′-AGTATTCGTGCTCAGCCTC-3′, antisense 5′-CCAGCGTTATTTGGTGGTG-3′; MLH1/2, sense 5′CACCACCAAATAACGCTGG-3′, antisense 5′-TTGGCGCTTCTCAGGCTC-3′; and MLH1/3, sense 5′-AGCCTGAGAAGCGCCAAG-3′, antisense 5′-TCCGCTCTTCCTATTGGTTC-3′. The optimal cycle number for PCR was determined to be that which produced a detectable band from the undigested template but did not yield any product from DNA digested with MspI (a methylation-insensitive isoschizomer of HpaII). For MLH1/1 35 cycles were used whereas MLH1/2 and MLH1/3 had 27 cycles. Annealing temperature was 58°C for all three fragments. An undigested DNA control (to show that amplification was successful) and a MspI-digested DNA control (to verify that the CCGG site was present and accessible to digestion) were included for every site examined, and the assays were performed at least twice to ensure reproducibility of results. The calcitonin gene promoter (region V) was studied for comparison as described by Heiskanen et al. (10).
Immunohistochemistry.
Six-micrometer sections from paraffin blocks mounted on superfrost slides were incubated for 1 hr at 60°C, deparaffinized by using standardized methods (11), and placed in Tris-buffered saline (TBS) (Dako, code no. S3001). Antigen retrieval was performed in 0.01 M citrate-buffered solution (pH 6.0) heated at 120°C for 2 min. The slides were subsequently cooled to room temperature and transferred to TBS. Endogenous peroxidase activity was blocked by incubating slides for 5 min in peroxidase blocking reagent containing H2O2 and NaN3 (Dako, code no S2001). Nonspecific antibody binding was quenched by incubation of the sections for 30 min with 10% goat serum (Dako, code No. X0907) in Tris-buffered antibody diluent (Dako, code no. S2022). Primary anti-MLH1 antibody (clone G168–728, PharMingen) was incubated with the sections for 2 hr at room temperature at the concentration of 1.25 μg/ml antibody diluent (Tris buffer, pH 7.2, containing 15 mM NaH3 protein, Dako code no. S2022). After a 10-min wash in TBS, secondary antibody was applied for 30 min in a two-step visualization system (EnVisionTM + System, Dako code no. K4006). Sections were counterstained with hematoxylin and photographed.
Statistical Analysis.
Fisher’s exact test was used to assess differences between the groups.
Results
DNA Methylation Patterns at the MLH1 Promoter and Correlation with Microsatellite Instability.
A majority (65/89, 73%) of all sporadic colorectal tumors showed alterations in DNA methylation relative to paired normal mucosa (Table 1). Two main patterns were distinguishable that closely correlated with the microsatellite instability status of tumors. Although most MSI(+) cases (31/51, 61%) showed the absence of methylation in normal DNA and presence of methylation at the same sites in tumor DNA (i.e., the tumor DNA showed “hypermethylation”), the opposite was true for MSI(−) cases, in which 53% (20/38) displayed methylation in normal mucosa and absence of methylation in tumor DNA (i.e., “hypomethylation” in tumor DNA). Most tumors associated with hypermethylation showed methylation in all three fragments, whereas methylation in normal mucosa more frequently affected only one or two fragments (especially fragment 3). Normal mucosa samples with hypermethylation located adjacent to MSI(−) tumors were directly tested for MSI by using the highly sensitive marker BAT26 that is constitutionally monomorphic in most Caucasians (12, 13). No MSI was found, which confirmed the absence of single prominent DNA mismatch repair deficient clones in the paired mucosa samples and was in contrast to tumors showing MLH1 promoter hypermethylation (analyzed for MSI in ref. 9).
Table 1.
Summary of methylation changes at the MLH1 promoter region as evaluated in tumor DNA relative to paired normal DNA
| Normal mucosa
|
Tumor
|
Number of cases
|
|||||
|---|---|---|---|---|---|---|---|
| MLH1/
|
MLH1/
|
MSI(+) | MSI(−) | ||||
| 1 | 2 | 3 | 1 | 2 | 3 | (n = 51) | (n = 38) |
| Hypermethylation in tumor DNA | |||||||
| − | − | − | + | + | + | 17 | 1 |
| − | − | − | − | + | + | 1 | |
| − | − | − | + | − | + | 5 | |
| − | − | − | + | − | − | 1 | 3 |
| − | − | − | − | + | − | 1 | |
| − | − | + | + | − | + | 2 | |
| − | − | + | + | + | + | 1 | |
| − | + | + | + | + | + | 1 | |
| + | − | + | + | + | + | 2 | |
| Total | 31 (61%) | 4 (11%) | |||||
| P < 0.001 | |||||||
| Hypomethylation in tumor DNA | |||||||
| + | + | + | − | − | − | 1 | 5 |
| + | + | + | + | − | − | 1 | |
| + | + | + | − | − | + | 1 | |
| + | − | + | − | − | − | 2 | |
| − | + | + | − | − | − | 1 | |
| + | − | + | − | − | + | 1 | |
| + | − | − | − | − | − | 1 | 1 |
| − | + | − | − | − | − | 2 | |
| − | − | + | − | − | − | 6 | |
| Total | 2 (4%) | 20 (53%) | |||||
| P < 0.001 | |||||||
| Mixed pattern | |||||||
| + | − | − | − | + | − | 1 | |
| + | − | − | − | − | + | 1 | |
| − | − | + | + | − | − | 2 | |
| + | − | + | − | + | + | 2 | |
| + | − | + | + | + | − | 2 | |
| Total | 5 (10%) | 3 (8%) | |||||
| NS | |||||||
| No change in tumor vs. normal DNA | |||||||
| − | − | − | − | − | − | 12 | 9 |
| − | − | + | − | − | + | 2 | |
| + | + | + | + | + | + | 1 | |
| Total | 13 (25%) | 11 (38%) | |||||
| NS | |||||||
The presence (+) vs. absence (−) of methylation at four HpaII sites contained in fragments 1–3 is shown, on the basis of the presence vs. absence of an amplification product from HpaII-digested DNA. Fragment 3 contains two HpaII sites and is located closest to the translation initiation site. Values for the statistical significance of the differences between groups are also given (NS, nonsignificant).
In occasional cases, one of the three fragments displayed hypermethylation, whereas another fragment showed simultaneous hypomethylation; these cases were considered to have a “mixed” DNA methylation pattern. Although the technique used does not allow us to distinguish whether hyper- and hypomethylation occurred in the same vs. separate alleles or the same vs. separate cells, the phenomenon could reflect intratumoral heterogeneity that was recently proposed to be characteristic of tumorigenesis associated with mismatch repair deficiency (14).
Repeated DNA is known to trigger methylation as a silencing mechanism (15), and it follows that alterations in microsatellite length and/or configuration resulting from DNA mismatch repair deficiency might induce hypermethylation per se. If microsatellite instability indeed were the primary inducing factor, then hypermethylation would be expected to be the predominant pattern in all MSI(+) tumors regardless of the etiology of MSI in these tumors. To gain further insight into the relationship between MSI and altered DNA methylation, a cohort (n = 26) of HNPCC patients with germline mutations in the MLH1 gene (16, 17) and a profuse MSI(+) phenotype in tumor DNA were evaluated for methylation changes. In these tumors, the frequencies for hypermethylation, hypomethylation, mixed pattern, and no change were 23%, 19%, 23%, and 35%, respectively, which does not support a primary role of MSI in methylation changes. Instead, these results suggest that among colorectal carcinomas, promoter hypermethylation is a special characteristic of the truly sporadic MSI(+) subset.
Specificity of DNA Methylation Changes.
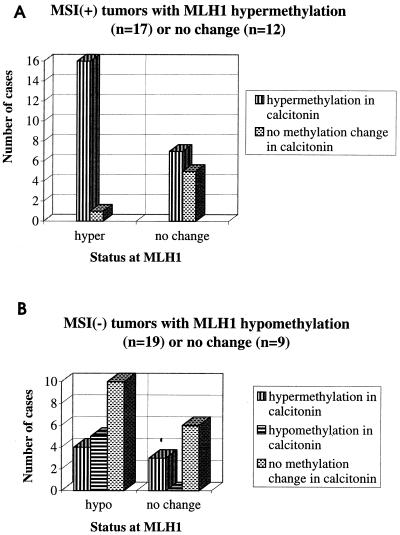
To see whether the methylation patterns observed at the MLH1 promoter were restricted to this gene or, alternatively, were more widespread, we chose to study the calcitonin gene for comparison. The calcitonin gene is a well characterized methylation target in various malignancies including colon cancer, but, unlike MLH1, is not known to be associated with any selective advantage (10, 18–20). As shown in Fig. 1, among MSI(+) tumors with MLH1 promoter hypermethylation, the calcitonin gene promoter was hypermethylated in all cases except one. Consistent with this gene being a known methylation target, it was also hypermethylated in a proportion of tumors not showing MLH1 promoter hypermethylation. Importantly, a significant fraction (5/19, 26%) of MSI(−) tumors with MLH1 promoter hypomethylation showed hypomethylation at the calcitonin gene promoter as well. Although confirmation by techniques that allow genome-wide screening of CpG islands for methylation (21, 22) is necessary, our observations are compatible with a distinct epigenetic signature being associated with MSI(+) and MSI(−) tumors and surrounding mucosa. This signature may have a common (as yet unknown) etiology, and it may result in different developmental pathways in these tumors (see below).
Figure 1.
DNA methylation changes in the calcitonin gene promoter in MSI(+) (A) and MSI(−) tumors (B) selected for the presence vs. absence of methylation changes at the MLH1 promoter region in the same tumors. In A, the MLH1 “hypermethylation” group consists of those MSI(+) cases that showed methylation in tumor tissue and absence of methylation in normal mucosa in all three fragments studied (Table 1).
Clinicopathological Correlations.
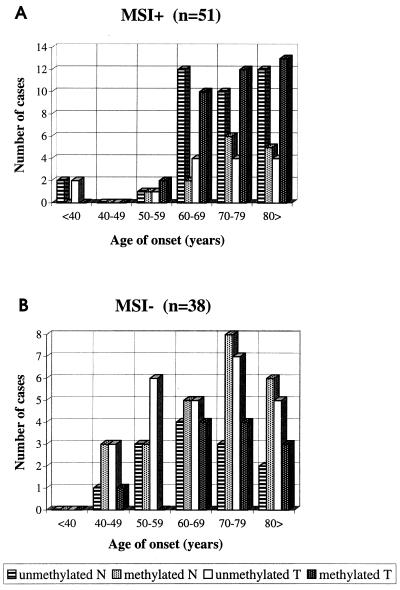
As parameters of potential relevance to the etiology of the observed epigenetic phenotypes, we evaluated the age at diagnosis as well as tumor location. In MSI(+) cases (mean age at diagnosis, 72 yr), the rate of methylation at the MLH1 promoter increased as a function of age in tumor tissue and, to some extent, in normal mucosa as well (Fig. 2A). Yet, when all age categories were combined, most normal mucosa samples remained unmethylated, whereas most tumors were methylated in agreement with “hypermethylation” in tumors relative to normal mucosa. Among MSI(−) cases (average age at diagnosis, 69 yr), the methylation rate in normal mucosa and, to some extent, in cancer likewise increased with age, but here a majority of all normal mucosa samples showed methylation, whereas most tumors were unmethylated, explaining “hypomethylation” in these tumors (Fig. 2B). Our findings justify the addition of MLH1 to the list of loci whose methylation status may be modified by age (23).
Figure 2.
Occurrence of methylation at (one or several) HpaII sites at the MLH1 promoter region in individual tumor (T) and normal mucosa (N) DNA samples from sporadic MSI(+) (A) and MSI(−) cases (B), grouped according to the age at diagnosis of the respective patients.
Besides age, developmental and biologic differences in the proximal vs. distal colon may influence the susceptibility to neoplastic transformation (24). Thus, the inherent differences that are known to occur in the molecular pathogenesis (25, 26) and prognosis (27) of MSI(+) vs. MSI(−) tumors may in part reflect their preferential locations in the bowel (proximal vs. distal, respectively). Among MSI(+) cases, MLH1 promoter methylation was more common among proximal than distal tumors (79% vs. 44%, respectively; P = 0.05), whereas the methylation rates in normal mucosa were less than 30% irrespective of location (Table 2). In the MSI(−) group, the proportion of normal mucosa samples with promoter methylation increased from distal (55%) to proximal (78%), although statistically not significant, whereas the occurrence of methylation in tumor tissue was similar (around 30%). In conclusion, we demonstrate that the hypermethylation phenotype, whether occurring in tumor tissue or normal mucosa, is associated with proximal location. The extent to which this association reflects a pathological process vs. a physiological difference between proximal and distal parts of the large bowel is presently unknown. However, the fact that normal mucosa from MSI(+) cases, in contrast to MSI(−) cases, showed comparable rates of methylation in proximal vs. distal locations argues against a simply physiological basis and implies that the methylation tendency, MSI status, and proximal location might all have a common etiologic denominator.
Table 2.
Incidence of the MLH1 promoter methylation in normal colon and tumor tissue, according to the site of origin
| Number of cases with methylation
|
||||
|---|---|---|---|---|
| Proximal
|
Distal
|
|||
| Normal | Tumor | Normal | Tumor | |
| MSI(+) | 12/42 | 33/42 | 2/9 | 4/9 |
| (29%) | (79%) | (22%) | (44%) | |
| MSI(−) | 14/18 | 6/18 | 11/20 | 6/20 |
| (78%) | (33%) | (55%) | (30%) | |
Sites before vs. after the splenic flexure were classified as proximal vs. distal, respectively.
Functional Significance.
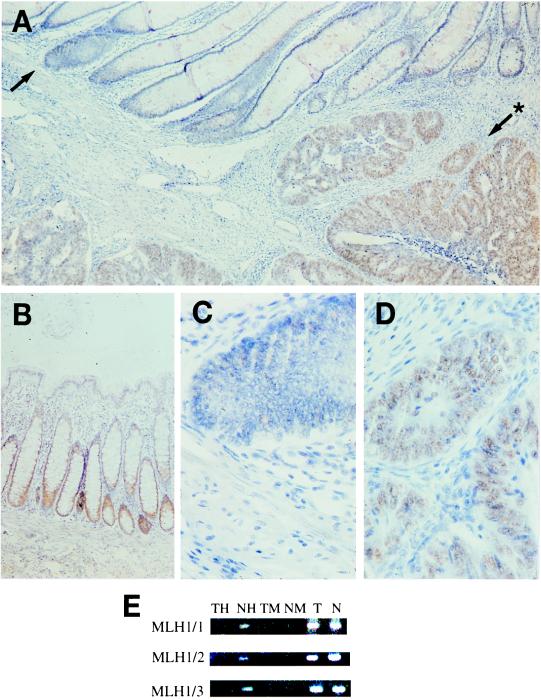
The MLH1 promoter contains 23 CpG sites (28), including those four that constitute HpaII restriction sites, covered by the present investigation. Importantly, only some of these sites regulate MLH1 protein expression through methylation, and it was therefore of interest to see whether promoter methylation affected gene expression in the present tumors and especially normal mucosa. All sporadic MSI(+) tumors of which a paraffin-embedded specimen was available were analyzed for MLH1 protein expression by immunohistochemistry. In accordance with previous reports (6, 8), reduced or lost expression was found, with visible staining in less than 25% of cancer cell nuclei, in a majority of MSI(+) tumors with methylation at one or several of the three MLH1 sites (30/36, 83%). A particular focus in our immunohistochemical studies was those six cases from the “hypomethylation” group in which all three MLH1 sites were methylated in normal mucosa and unmethylated in the adjacent tumor tissue (Table 1). Four of these, all MSI(−) cases, were available for immunohistochemical analysis, and MLH1 promoter hypermethylation was associated with lost or severely reduced protein expression in large regions of all of these normal mucosa tissues, whereas the tumors lacking methylation showed an intense staining (Fig. 3). Loss of MLH1 protein expression thus seemed to accompany promoter methylation both in normal mucosa and in tumors, emphasizing the pathogenetic relevance of the methylation changes.
Figure 3.
Immunohistochemical staining of the MLH1 protein in colon tissues. (A) An overview showing different staining patterns in normal mucosa with MLH1 promoter methylation vs. MSI(−) tumor tissue without such methylation (x68). (B) Normal colonic mucosa from a control individual demonstrating the presence of nuclear staining in the base of the crypts (x68). (C) Higher magnification of the region marked with an arrow in A, showing no MLH1 expression in crypts from the hypermethylated nonneoplastic mucosa (x136). (D) Higher magnification of the region marked with arrow and asterisk in A, showing intense staining in carcinoma cells lacking MLH1 promoter methylation (x136). The specimens were also stained with an antibody against the proliferating cell nuclear antigen (PCNA), which is a marker of cell proliferation; expression was observed both in the bottom of crypts from the mucosa with MLH1 promoter methylation and in tumor cells without methylation (data not shown). (E) Methylation analysis of the MLH1 fragments 1–3 in MSI(−) tumor (T) and paired normal mucosa (N) tissue. The presence of an amplification product in HpaII-digested DNA in normal mucosa (NH) indicates methylation of all HpaII sites contained in fragments 1–3, whereas these sites are not methylated in tumor DNA (TH) on the basis of the absence of an amplification product. Control assays by using MspI-digested DNA (TM, NM) and undigested DNA (T, N) are also shown (see Materials and Methods).
Discussion
The present investigation was conducted to evaluate the role of altered DNA methylation in colon cancer. For this purpose, we chose the DNA mismatch repair gene MLH1 as a determinant of microsatellite instability status and the calcitonin gene as a known methylation target in various malignancies but without a functional role in tumorigenesis. We provide evidence for the existence of two distinct epigenetic phenotypes in sporadic colorectal cancer and adjacent mucosa. The pattern characterized by methylation in tumor tissue and lack of methylation in normal mucosa is consistent with the recently defined “hypermethylator phenotype” typical of MSI(+) tumors (29, 30). The other pattern that is featured by methylation in normal mucosa and lack of methylation in tumor tissue and is a property of sporadic MSI(−) colorectal cancers has, to our knowledge, not been characterized before. Given that 85% of all colorectal cancers belong to the latter group, the pattern that we now describe may constitute a significant epigenetic change.
Although the mechanism by which these methylation changes arise remains unknown, several alternative systems that control the distribution of methylated residues may be involved. Deregulation of enzymes responsible for DNA methylation (methyltransferases) could lead to widespread hypermethylation; however, this possibility was not substantiated in a recent investigation (31). Alternatively, disruption of mechanisms that normally protect CpG islands against the access of DNA methyltransferases could explain hypermethylation (32). In light of recent discoveries, the DNA methylation changes that we observed might reflect as well an imbalance between two opposite systems, one that is responsible for the addition of methyl groups (carried out by DNA methyltransferases) and another one that is responsible for their removal (demethylase activity) (33). In fact, global decreases and increases in DNA methylation are known to occur during development and tissue-specific differentiation (34), and analogous events might be associated with tumorigenesis. Finally, hypermethylation has been reported to occur as a cellular response to environmental carcinogens (35), and in the colon, exogenous compounds from dietary and other sources might therefore be an important modifier of the methylation patterns. Such a connection might also in part explain our observation of the elevated rates of MLH1 promoter methylation with increasing age.
The observed close association between the different DNA methylation patterns and the microsatellite instability status adds another dimension to the discussion about the role of aberrant methylation in colon cancer. In the context of MLH1 alone, MSI might be viewed as a simple consequence of compromised mismatch repair capacity because of MLH1 promoter methylation. The high incidence of MLH1 promoter hypermethylation in MSI(+) tumors and the associated loss of protein expression (refs. 6–8, and this study) as well as the correction of the MSI abnormality after the chemical reversal of methylation (6, 36) indeed support the conclusion that MLH1 promoter hypermethylation can be the cause of deficient DNA mismatch repair and MSI. However, our finding of similar methylation patterns at the MLH1 promoter and the promoter of the functionally neutral calcitonin gene, as well as the occurrence of a methylation “imprint” in normal mucosa, suggests a more primary methylation abnormality whose functional significance extends beyond MLH1. As such, it may precede other genetic or epigenetic events and clonal selection. A related mechanism may be associated with loss of imprinting of the insulin-like growth factor II gene that, being present in both normal and tumor tissues, was recently identified as a marker of colon cancer susceptibility (37).
Our discovery of methylation in CpG islands of autosomal genes in normal colonic mucosa, although unexpected, is not unprecedented. By using a method similar to that applied in the present study, Gonzalez-Zulueta et al. (5) found methylation of the 5′ CpG island of the p16/CDKN2 gene in a high proportion of normal mucosa samples adjacent to colon cancers. The MSI status of the tumors was not reported. As in our case, methylation was associated with gene silencing. However, Toyota et al. (30) reported the absence of both MLH1 and p16 promoter methylation in normal mucosa from colon cancer patients they studied, irrespective of the MSI status, which is in apparent disagreement with our findings. The discrepancy might be because of a different methodology (restriction enzyme assay vs. bisulfite modification), different CpG sites analyzed (cf. ref. 28), or regional differences in the methylation status of the normal colonic mucosa. All four normal mucosa specimens that showed hypermethylation and loss of MLH1 protein expression as shown in Fig. 3 were adjacent to the respective tumors, and it remains to be seen whether this phenomenon was limited to the immediate vicinity of the tumors or was more widespread in the colon.
Methylation of CpG islands is associated with delayed replication, condensed chromatin, and transcriptional repression (20, 34), implying that it may significantly modify cell behavior. Although the timing of the present methylation changes in tumorigenesis remains to be determined, these patterns may confer different developmental pathways on MSI(+) and MSI(−) tumors, depending on the gene targets. In MSI(+) tumors, MLH1 is likely to be among key targets whose inactivation by hypermethylation gives rise to a cascade of mutations in various growth-regulatory genes (38, 39), resulting in the proliferation of mismatch repair deficient clone(s). In contrast, inactivation of other genes, such as p16, that may occur by the same epigenetic mechanism in the nonneoplastic mucosa surrounding MSI(−) tumors may be crucial to the development of these microsatellite-stable tumors. Finally, although the methylated mucosa adjacent to MSI(−) tumors remained BAT26 stable by the present techniques, the MLH1 protein expression was lost, leaving room for the possibility that defective mismatch repair initially played a role in the development of both MSI(+) and MSI(−) tumors, and the different MSI phenotypes were consequent to selection at later stages.
It is remarkable that, by studying only a few genes and CpG sites, clear correlations between methylation of normal mucosa and clinical and biochemical properties of the tumors were evident. It is likely that the abnormalities we detected are the tip of an iceberg, and that profound differences in epigenetic changes distinguish these patients and their tumors. Future studies analyzing additional genes and methylation sites in more detail should be illuminating in this regard.
Acknowledgments
We thank Dr. Bert Vogelstein for critical reading of the manuscript. We also thank Dr. Anu-Liisa Moisio for helpful discussions and Saila Saarinen and Ritva Haider for expert technical assistance. This investigation was supported by grants from the Nordic Cancer Union, Sigrid Juselius Foundation, European Commission (contract BMH4-CT96-0772), National Institutes of Health grants U01 CA67941 and P30 CA16058, and the Ohio Cancer Research Associates.
Abbreviations
- HNPCC
hereditary nonpolyposis colorectal cancer
- MLH1
MutL homologue 1
- MSI
microsatellite instability
References
- 1.Kinzler K W, Vogelstein B. Cell. 1996;87:159–170. doi: 10.1016/s0092-8674(00)81333-1. [DOI] [PubMed] [Google Scholar]
- 2.Boland C R, Thibodeau S N, Hamilton S R, Sidransky D, Eshleman J R, Burt R W, Meltzer S J, Rodriguez-Bigas M A, Fodde R, Ranzani G N, et al. Cancer Res. 1998;58:5248–5257. [PubMed] [Google Scholar]
- 3.Wu Y, Nyström-Lahti M, Osinga J, Looman M W G, Peltomäki P, Aaltonen L, de la Chapelle A, Hofstra R M W, Buys C H C M. Genes Chromosomes Cancer. 1997;18:269–278. [PubMed] [Google Scholar]
- 4.Feinberg A P, Vogelstein B. Nature (London) 1983;301:89–92. doi: 10.1038/301089a0. [DOI] [PubMed] [Google Scholar]
- 5.Gonzalez-Zulueta M, Bender C M, Yang A S, Nguyen T, Beart R W, Van Tornout J M, Jones P A. Cancer Res. 1995;55:4531–4535. [PubMed] [Google Scholar]
- 6.Herman J G, Umar A, Polyak K, Graff J R, Ahuja N, Issa J-P, Markowitz S, Willson J K V, Hamilton S R, Kinzler K W, et al. Proc Natl Acad Sci USA. 1998;95:6870–6875. doi: 10.1073/pnas.95.12.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kane M F, Loda M, Gaida G M, Lipman J, Mishra R, Goldman H, Jessup J M, Kolodner R. Cancer Res. 1997;57:808–811. [PubMed] [Google Scholar]
- 8.Cunningham J M, Christensen E R, Tester D J, Kim C-Y, Roche P C, Burgart L J, Thibodeau S N. Cancer Res. 1998;58:3455–3460. [PubMed] [Google Scholar]
- 9.Aaltonen L A, Salovaara R, Kristo P, Canzian F, Hemminki A, Peltomäki P, Chadwick R B, Kääriäinen H, Eskelinen M, Järvinen H, et al. N Engl J Med. 1998;338:1481–1487. doi: 10.1056/NEJM199805213382101. [DOI] [PubMed] [Google Scholar]
- 10.Heiskanen M, Syvänen A-C, Siitari H, Laine S, Palotie A. PCR Methods Appl. 1994;4:26–30. doi: 10.1101/gr.4.1.26. [DOI] [PubMed] [Google Scholar]
- 11.Bancroft J D, Sevens A, Turner, editors. Theory and Practice of Histological Techniques. 4th Ed. New York: Churchill Livingstone; 1996. [Google Scholar]
- 12.Hoang J-M, Cottu P H, Thuille B, Salmon R J, Thomas G, Hamelin R. Cancer Res. 1997;57:300–303. [PubMed] [Google Scholar]
- 13.de la Chapelle A. Eur J Hum Genet. 1999;7:407–408. doi: 10.1038/sj.ejhg.5200335. [DOI] [PubMed] [Google Scholar]
- 14.Habano W, Sugai T, Nakamura S. Oncogene. 1998;16:1259–1265. doi: 10.1038/sj.onc.1201651. , 1998. [DOI] [PubMed] [Google Scholar]
- 15.Selker E U. Cell. 1999;97:157–160. doi: 10.1016/s0092-8674(00)80725-4. [DOI] [PubMed] [Google Scholar]
- 16.Nyström-Lahti M, Wu Y, Moisio A-L, Hofstra R M W, Osinga J, Mecklin J-P, Järvinen H J, Leisti J, Buys C H C M, de la Chapelle A, et al. Hum Mol Genet. 1996;5:763–769. doi: 10.1093/hmg/5.6.763. [DOI] [PubMed] [Google Scholar]
- 17.Holmberg M, Kristo P, Chadwick R B, Mecklin J-P, Järvinen H, de la Chapelle A, Nyström-Lahti M, Peltomäki P. Hum. Mutat., Mutation in Brief #144 (Online) website address: http://journals.wiley.com/humanmutation. 1997. [DOI] [PubMed] [Google Scholar]
- 18.Baylin S B, Fearon E R, Vogelstein B, deBustros A, Sharkis S J, Burke P J, Staal S P, Nelkin B D. Blood. 1987;70:412–417. [PubMed] [Google Scholar]
- 19.Silverman A L, Park J-G, Hamilton S R, Gazdar A F, Luk G D, Baylin S B. Cancer Res. 1989;49:3468–3473. [PubMed] [Google Scholar]
- 20.Baylin S B, Herman J G, Graff J R, Vertino P M, Issa J-P. Adv Cancer Res. 1998;72:141–196. [PubMed] [Google Scholar]
- 21.Liang G, Salem C E, Yu M C, Nguyen H D, Gonzalez F A, Nguyen T T, Nichols P W, Jones P A. Genomics. 1998;53:260–268. doi: 10.1006/geno.1998.5502. [DOI] [PubMed] [Google Scholar]
- 22.Smiraglia D J F, Frühwald M C, Costello J F, McCormick S P, Peltomäki P, O’Dorisio M S, Cavenee W K, Plass C. Genomics. 1999;58:254–262. doi: 10.1006/geno.1999.5840. [DOI] [PubMed] [Google Scholar]
- 23.Ahuja N, Li Q, Mohan A L, Baylin S B, Issa J-P. Cancer Res. 1998;58:5489–5494. [PubMed] [Google Scholar]
- 24.Bufill J A. Ann Intern Med. 1990;113:779–788. doi: 10.7326/0003-4819-113-10-779. [DOI] [PubMed] [Google Scholar]
- 25.Olschwang S, Hamelin R, Laurent-Puig P, Thuille B, De Rycke Y, Li Y J, Muzeau F, Girodet J, Salmon R-J, Thomas G. Proc Natl Acad Sci USA. 1997;94:12122–12127. doi: 10.1073/pnas.94.22.12122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lengauer C, Kinzler K, Vogelstein B. Nature (London) 1998;396:643–649. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- 27.Lothe R A, Peltomäki P, Meling G I, Aaltonen L A, Nyström-Lahti M, Pylkkänen L, Heimdal K, Andersen T I, Møller P, Rognum T O, et al. Cancer Res. 1993;53:5849–5852. [PubMed] [Google Scholar]
- 28.Deng G, Chen A, Hong J, Chae H S, Kim Y S. Cancer Res. 1999;59:2029–2033. [PubMed] [Google Scholar]
- 29.Ahuja N, Mohan A L, Li Q, Stolker J M, Herman J G, Hamilton S R, Baylin S B, Issa J-P. Cancer Res. 1997;57:3370–3374. [PubMed] [Google Scholar]
- 30.Toyota M, Ahuja N, Ohe-Toyota M, Herman J G, Baylin S B, Issa J-P. Proc Natl Acad Sci USA. 1999;96:8681–8686. doi: 10.1073/pnas.96.15.8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eads C A, Danenberg K D, Kawakami K, Saltz L B, Danenberg P V, Laird P W. Cancer Res. 1999;59:2302–2306. [PubMed] [Google Scholar]
- 32.Macleod D, Charlton J, Mullins J, Bird A P. Genes Dev. 1994;8:2282–2292. doi: 10.1101/gad.8.19.2282. [DOI] [PubMed] [Google Scholar]
- 33.Ramchandani S, Bhattacharya S K, Cervoni N, Szyf M. Proc Natl Acad Sci USA. 1999;96:6107–6112. doi: 10.1073/pnas.96.11.6107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Singal R, Ginder G D. Blood. 1999;93:4059–4070. [PubMed] [Google Scholar]
- 35.Issa J P, Baylin S B, Belinsky S A. Cancer Res. 1996;56:3655–3658. [PubMed] [Google Scholar]
- 36.Veigl M, Kasturi L, Olechnowicz J, Ma A, Lutterbaugh J D, Periyasamy S, Li G-M, Drummond J, Modrich P L, Sedwick W D, et al. Proc Natl Acad Sci USA. 1998;95:8698–8702. doi: 10.1073/pnas.95.15.8698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cui H, Horon I L, Ohlsson R, Hamilton S, Feinberg A. Nat Med. 1998;4:1276–1280. doi: 10.1038/3260. [DOI] [PubMed] [Google Scholar]
- 38.Malkhosyan S, Rampino N, Yamamoto H, Perucho M. Nature (London) 1996;382:499–500. doi: 10.1038/382499a0. [DOI] [PubMed] [Google Scholar]
- 39.Percesepe A, Kristo P, Aaltonen L A, Ponz de Leon M, de la Chapelle A, Peltomäki P. Oncogene. 1998;17:157–163. doi: 10.1038/sj.onc.1201944. [DOI] [PubMed] [Google Scholar]