Abstract
The egr-type zinc-finger transcription factor encoded by the Drosophila gene stripe (sr) is expressed in a subset of epidermal cells to which muscles attach during late stages of embryogenesis. We report loss-of-function and gain-of-function experiments indicating that sr activity provides ectodermal cells with properties required for the establishment of a normal muscle pattern during embryogenesis and for the differentiation of tendon-like epidermal muscle attachment sites (EMA). Our results show that sr encodes a transcriptional activator which acts as an autoregulated developmental switch gene. sr activity controls the expression of EMA-specific target genes in cells of ectodermal but not of mesodermal origin. sr-expressing ectodermal cells generate long-range signals that interfere with the spatial orientation of the elongating myotubes.
Keywords: epidermal switch gene, muscle pattern, transcriptional activator
The larval body wall musculature of Drosophila is composed of a stereotyped array of segmentally repeated myofibers which anchor at specific ectoderm-derived epidermal muscle attachment (EMA) sites (for review see refs. 1 and 2). Its formation is initiated during midstages of embryogenesis when mesodermal muscle primordia progress from morphologically indistinguishable myoblasts to syncytial myotubes (3). While myotubes enlarge through continued fusion with myoblasts, they stretch and extend growth-cone-like polar processes to anchor at the EMA sites (2, 3). Studies involving transplantation (4, 5) and embryonic tissue culture experiments (6) suggested that epidermal cells may play a vital role in myotube guidance.
Recently, the stripe (sr) gene has been shown to be required for the generation of a normal muscle pattern of the larvae (6, 7). It encodes two differentially spliced transcripts, sr a and sr b, which are expressed in the subset of ectodermal cells that develop into EMA sites (7). Embryos that lack sr activity or express mutant variants of sr develop strong muscle pattern defects (6, 7). The early steps of myogenesis, including myotube formation and the initial stretching of myotubes, appeared normal in such embryos, but the leading edges of the myotubes become misrouted before they encounter the EMA sites (7). This suggested that myotube pathfinding requires the activity of one or both Stripe variants, termed Stripe a and Stripe b, which share a unique DNA-binding domain (7), a triple zinc-finger motif characteristic for vertebrate egr-type transcriptional activators (8), but differ in their N-terminal regions (7).
Here we report loss-of-function and gain-of-function experiments showing that Stripe b acts as a transcriptional activator. Ectopic expression of Stripe b activates the expression of both sr transcripts and causes the activation of EMA-specific target genes in cells of ectodermal origin but not in mesoderm. Ectodermal Stripe b interferes with myotube guidance prior to the binding of the myotubes to the EMA cells. The results suggest that in the developing epidermis, sr acts within a genetic circuitry that directs ectodermal cells to differentiate into EMA sites and causes them to propagate long-range signals that interfere with the orientation of myotubes.
MATERIALS AND METHODS
Plasmid Constructs.
The UAS Striperep construct was cloned by inserting the sequences coding for the sr zinc-finger domain behind the engrailed repressor domain (amino acids 1–298) (9–11). The sr zinc-finger region was amplified by PCR using the oligonucleotides CGCGGATCCGGCGGATCTGGT and GCTCTAGACTAGCCGCGCACCTTTTCC introducing a BamHI site at the 5′ end and an XbaI site at the 3′ end (underlined) and a stop codon (boldface). The PCR fragment was subcloned into pBst (Stratagene), fused with the EcoRI/BamHI fragment of the engrailed cDNA (12), and inserted into pUAST (13) using EcoRI and XbaI. The upstream activating sequence (UAS) Stripe b expression domain contains the EcoRI/XbaI fragment of the sr b cDNA (nucleotides 405-4458) (7) inserted into pUAST (13).
Fly Lines.
Several transgenic lines with an insertion of the pUAS-Striperep and the pUAS-Stripe b on the second and third chromosome were obtained. For heat shock-induced expression, pUAS-Striperep transgene-containing flies were crossed with K25 (obtained from Konrad Basler, Zürich) containing a combination of heat shock and sevenless enhancers. Embryo collections (1 h at 25°C) were kept at 18°C, heat shocked (Striperep for 5 min; Stripe b for 20 min) at 37°C, and returned to 18°C. The single minded (14) and the hairy stripe 6 GAL4 lines were obtained from Christian Klämbt (Münster), the engrailed GAL4 line from Andrea Brand (Cambridge), the patched GAL4 line from Konrad Basler, and the 24B GAL4 line (13) from the Bloomington stock center.
Analysis of Expression Patterns.
In situ hybridizations of whole-mount embryos using digoxigenin-labeled DNA probes and antibody stainings were performed as described (15, 16) using the Vectastain ABC Elite horseradish peroxidase system (Vector) or a directly coupled secondary antibody. Dilutions were as follows: Stripe-specific antiserum (7), 1/200; anti-myosin heavy chain (MHC) antiserum (obtained from D. P. Kiehart, Durham), 1/2000; monoclonal anti-Groovin hybridoma supernatant [obtained from Talila Volk (6)], 1/3. Endogenous sr a and sr b transcripts were detected with specific probes for their 5′ untranslated regions (7) (sr a, nucleotides 281-1289; sr b, nucleotides 1–405) not present in the transgene-derived sr b transcript. The β1-tubulin probe was a 1.5-kb EcoRI cDNA fragment (provided by Detlev Buttgereit, Marburg), the delilah probe was the cDNA (17).
RESULTS AND DISCUSSION
The sr gene of Drosophila encodes two proteins which differ with respect to their N-terminal regions—i.e., the amino acid sequence of the shorter protein is fully contained within the longer (7). The shorter sr protein variant, Stripe b, is expressed in all developing tendon-like EMA cells of the embryo, while the longer variant, Stripe a, is expressed only in a subset of these cells (7). Recent analysis of the phenotype of a strong sr allele suggested that sr wild-type activity is required not only for the normal differentiation of the EMA cells but also for the directed growth of myotubes along the inner surface of the epidermis toward their EMA sites (6, 7).
To interfere with the activity of both proteins at the same time and to show that they normally act as transcriptional activators, we generated a dominant-negative Stripe variant which is a transcriptional repressor that acts from the Stripe binding sites in vivo. For this, we fused the unique Stripe DNA-binding domain (7) to the repressor domain of the transcription factor Engrailed (9, 11, 12). After P-element-mediated integration into the germ line (18), the resulting Striperep transgene was ubiquitously activated by the heat shock-inducible GAL4/UAS expression system (13).
Striperep Expression Causes sr Phenocopies.
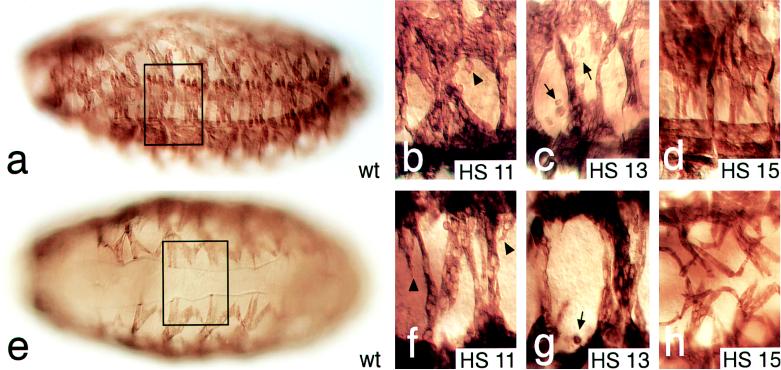
Fig. 1 shows wild-type embryos (Fig. 1 a and e) and enlarged portions of the muscle patterns of embryos (Fig. 1 b–d and f–h) that have received heat shock-induced Striperep expression pulses at stage 11 (Fig. 1 b and f), stage 13 (Fig. 1 c and g), and stage 15 (Fig. 1 d and h), respectively. Striperep expression during embryonic stages 11 to 14 caused strongly disrupted muscle patterns (representative examples are shown Fig. 1 b, c, f, and g) in all of more than 50 embryos analyzed at each of the stages. No other aspects of embryonic development were discernibly affected (not shown). Expression from stage 15 onwards did not interfere significantly with the formation of a normal muscle pattern (Fig. 1 d and h). However, as shown in Fig. 1h, the ventral oblique muscles, which develop normal patterns and morphology, extend closer to the ventral midline than the ventral oblique muscles do normally (compare Fig. 1 e and h). This effect was observed in all of the 40 embryos examined. It suggests a stage-specific change in Striperep-dependent effects on muscle guidance at stage 15 as compared with the earlier stages. At stage 15, the filopodia of the myotubes have just reached the EMA cells but are not yet stably attached (3). Thus, the late effect of Striperep expression at stage 15 might already be linked to EMA differentiation rather than to Stripe-dependent myotube guidance.
Figure 1.
Striperep expression interferes with muscle pattern formation. Lateral (a–d) and ventral (e–h) views of the muscle pattern of stage 16 embryos (staging according to ref. 19) stained with anti-MHC antibodies. Shown are wild-type (a and e) and enlargements (b–d and f–h) (two segments corresponding to the area framed in a and e) of embryos that received pulsed Striperep expression using the GAL4/UAS system with a heat shock-dependent GAL4 driver line (HS). (b and f) Striperep expression at stage 11 (HS 11) results in the fusion of myofibers of abnormal shape with elongated processes (arrowheads); they cross the ventral midline (f) and fail to attach to the epidermis. (c and g) Striperep expression during stage 13 (HS 13) results in the same phenotype. Note residual MHC-expressing myoblasts (arrows in c and g) as observed in sr mutants (7). (d and h) Striperep expression at stage 15 (HS 15) induces only mild defects. Note improper attachment of lateral transverse and ventral muscles (for details see text). Orientation of embryos: dorsal side up and anterior to the left.
The Striperep-dependent generation of a phenocopy of the sr mutant phenotype indicates that Striperep represents indeed a dominant-negative variant of the Stripe proteins by acting as a transcriptional repressor. This implies that Striperep negatively interferes with the control of sr target genes by repressing them through competition with the Stripe wild-type function. More importantly, the results also show that Striperep expression specifically perturbs muscle pattern formation already during an early stage when endogenous sr is first expressed in a subset of ectodermal cells (7) and that the phenocritical period covers the time window when the myotubes normally undergo their oriented growth along the inner surface of the epidermis (3). We also asked whether the expression patterns of putative target genes of sr, such as groovin (6), delilah (17), and β1-tubulin (20), are altered in response to Striperep activity. The results (data not shown) indicated that the levels of expression of these EMA-specific markers were reduced and their patterns were perturbed by Striperep activity as had been described recently for sr mutant embryos (6, 7). The dominant effects of Striperep expression therefore complement recent results obtained with sr mutants (6, 7), and they establish that the early sr activity (stages 11 to 14) in epidermal cells is necessary not only for EMA cell differentiation but also to spatially orient the myotubes through early epidermal signals (2).
Ectopic Stripe b Activates EMA-Specific Gene Expression.
Striperep acts in a dominant-negative fashion, suggesting that wild-type Stripe functions as a transcriptional activator on EMA-specific target genes. To demonstrate this function directly, we monitored the expression of EMA-specific epidermal marker genes such as sr itself, groovin (6), delilah (17), and β1-tubulin (20) in response to ectopic Stripe b (7) expression driven by the GAL4/UAS system (13). For this, Stripe b was expressed either ubiquitously under the control of a heat shock-inducible GAL4 line or in response to GAL4 driver lines that direct Stripe b expression in a series of stripes in the ectoderm (Figs. 2 and 3), in neuroectoderm-derived ventral midline cells of the central nervous system (CNS) and in the developing mesoderm, respectively (Figs. 3 and 4; for details on the corresponding driver lines see Materials and Methods).
Figure 2.
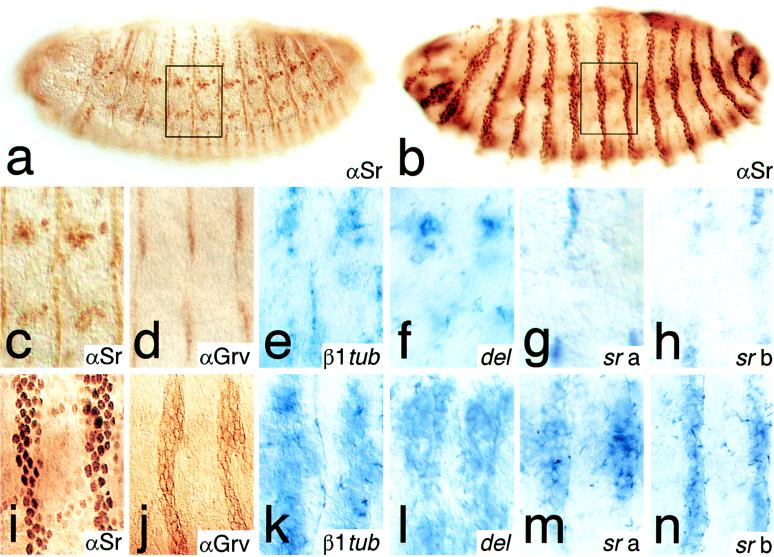
Ectopic Stripe b expression in the epidermis activates EMA-specific target genes. The Stripe b transgene was expressed in the epidermis using an engrailed GAL4 driver line. Shown are lateral views of embryos (late stage 14); enlarged area is indicated by frames. Expression of Stripe in a wild-type embryo (a and c) as visualized by antibodies (αSr) that detect both Stripe variants. (b and i) Ectopic expression of Stripe b in the engrailed pattern. Expression of EMA-specific genes in wild-type embryos (d–h) and in embryos expressing the Stripe b transgene (j–n) in the engrailed pattern is shown. (d and j) Groovin expression as detected by a monoclonal antibody (αGrv). (e and k) β1-tubulin (β1tub) and (f and l) delilah (del) expression as detected by whole-mount in situ hybridization. (g and m) sr a and (h and n) sr b expression as detected by transcript-specific probes (7); see Materials and Methods. Note that the induction of the sr target genes was 100% penetrant.
Figure 3.
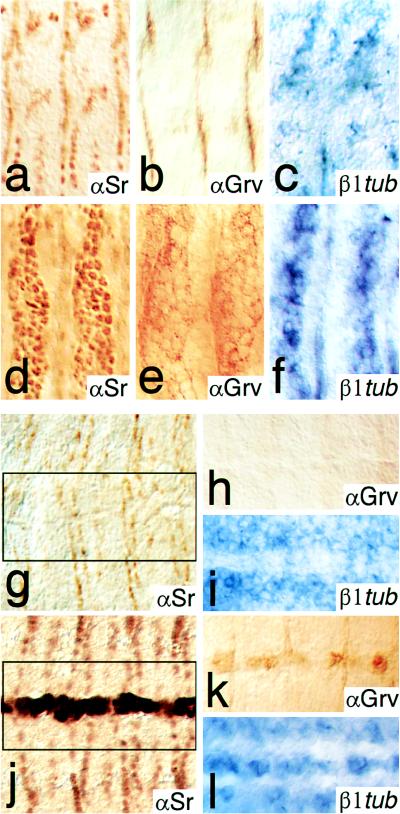
Ectoderm-specific activation of EMA target genes in response to Stripe b expression. Lateral views of stage 15 embryos (a–f) and ventral views of stage 14 embryos (g–l) are shown. (a and g) Expression of Stripe is shown by anti-Stripe (αSr) antibody stainings of wild-type embryos, of embryos ectopically expressing Stripe b in the patched expression pattern (d) and in the ventral midline using a patched and a single minded GAL4 driver line (j), respectively. Groovin expression is detected by a monoclonal antibody (αGrv) in wild-type embryos (b and h) and in embryos ectopically expressing Stripe b in the patched (e) or the single minded (k) pattern. β1-tubulin expression (β1tub) is detected by whole-mount in situ hybridizations in wild-type (c and i) and in embryos expressing Stripe b in the patched (f) or the single minded (l) domain. Note that in wild-type embryos Stripe, Groovin, and β1-tubulin are not expressed in midline cells (g–i).
Figure 4.
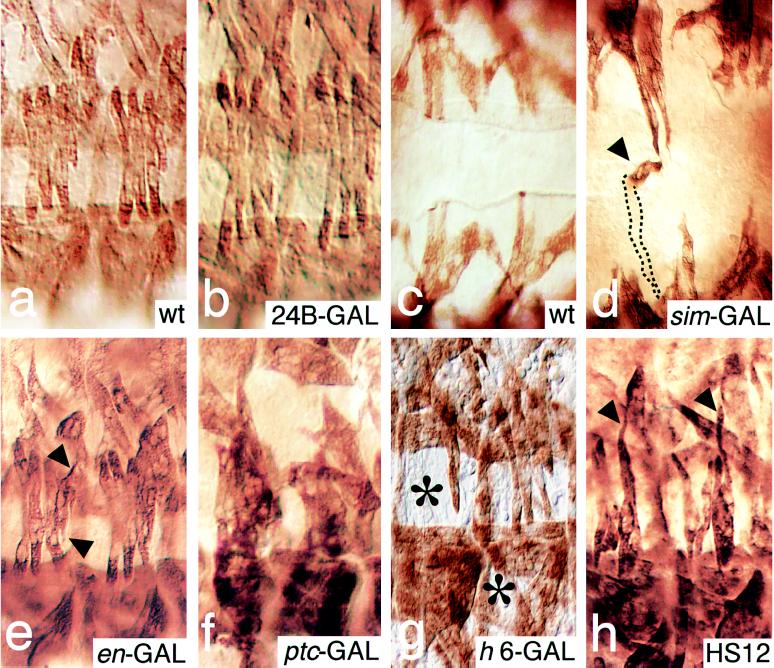
Examples showing that ectopic Stripe b expression in the epidermis interferes with myotube guidance. Enlarged lateral (a, b, and e–h) and ventral (c and d) views of muscle pattern (stage 16) as detected with anti-MHC antibodies. (a and c) Lateral and ventral view of wild-type (wt) embryos. (b) Expression of Stripe b in the developing mesoderm under the control of the 24B GAL4 driver line (24B-GAL) results in no pattern defects. (d) Expression in the ventral midline using the single minded GAL4 driver line (sim-GAL) results in a strong distortion of the ventral muscles, which frequently cross the midline. The example marked by an arrowhead shows an elongated ventral muscle which reaches the midline, takes a 90° turn, extends anteriorly along the midline (arrowhead), leaves the optical plane of the section, and extends internally, crossing the midline to eventually fuse with muscles on the other side of the midline. Out-of-focus information is indicated by dotted lines. (e) Expression of Stripe b in the engrailed pattern (en-GAL). Note the elongated processes (arrowheads) of lateral transverse muscles. (f) Expression of Stripe b in the patched pattern (ptc-GAL). Note that myotubes of abnormal shape are fused. (g) Expression of Stripe b in the hairy stripe 6 domain (h6-GAL) results in the absence (indicated by the asterisks in abdominal segments 5 and 6) of ventral longitudinal and lateral transversal muscles in the abdominal segments 5 to 7. Note that the absence of a muscle does not imply that the corresponding muscle is missing, rather that it had been misguided and thus taken another position within the defective muscle pattern. (h) Ubiquitous heat-shock-induced Stripe b expression during stage 12 (HS12) causes elongated processes. Arrowheads mark examples of lateral transverse muscles. Such defects were observed only in response to ectopic Stripe b expression during stages 11–14. For details on the generation of the ectopic Stripe b patterns see Materials and Methods.
Fig. 2 summarizes the wild-type patterns of the EMA cell marker genes sr, groovin, β1-tubulin, and delilah (Fig. 2 a and c–h) and their Stripe b-induced activation in response to an engrailed GAL4 driver line (Fig. 2 b and i–n) or a patched GAL4 driver line (Fig. 3 d–f) in the developing epidermis. Note that Stripe b also activates sr a and sr b transcription (Fig. 2 m and n), indicating that sr acts in a positive autoregulatory loop. These results establish that sr activity is sufficient to activate EMA-specific target genes in the developing epidermis. The same results were obtained when Stripe b was ectopically expressed in ectoderm-derived ventral midline cells (Fig. 3 j–l), but not in Stripe b-expressing mesodermal cells (not shown). Thus, EMA-specific gene activation in response to Stripe appears to be limited to cells of ectodermal origin, suggesting that those cells are preconditioned for Stripe action. However, recent results have shown that EMA-specific β1-tubulin expression is initiated in response to the muscle fibers attachment to the EMA cells (20). This indicates that sr expression or the preconditioning of ectodermal cells may depend on signals of mesodermal origin. In this view, the autoregulatory feedback loop on sr expression, as well as the ectoderm-dependent activation of EAM-specific genes, may depend on signals from somatic mesoderm, a possibility that is not excluded by our experiments.
Epidermal Stripe b Expression Interferes with Myotube Guidance.
Stripe b-dependent EMA marker gene expression indicates that Stripe b activity alters the fate of ectoderm-derived cells. Are those cells able to provide spatial cues that interfere with the orientation of myotubes toward the epidermis? We addressed this question by examining the muscle pattern in embryos in which Stripe b was expressed in different ectopic locations (Fig. 4).
No muscle pattern defects were observed when Stripe b was expressed in the developing mesoderm under the control of the 24B GAL4 driver line. In contrast, ectopic Stripe b expression in the ventral midline cells did interfere with the orientation of myotubes, whereby the effects on the muscle pattern were restricted to muscles of the ventral half of the embryo. Ventral oblique muscles were frequently found to extend over the ventral midline, being in close contact with the ectopic Stripe b-expressing cells, but never attached to them (compare Fig. 4 c and d). Stronger defects and more generally disturbed muscle patterns were observed when Stripe b was expressed in the pattern of engrailed stripes in the epidermis (Fig. 4e). The most severe pattern defects were observed when Stripe b expression was under the control of the patched GAL4 line (Fig. 4f), causing a broader ectodermal sr expression domain in the center of each segment. The defective muscle patterns summarized in Fig. 4 were observed in all embryos that had received ectopic stripe expression under the conditions described. However, the defects were not specific for distinct sets of muscle fibers and varied from hemisegment to hemisegment and from embryo to embryo with respect to which muscles and to what degree the muscle patterns were affected.
Due to the narrow spacing of ectopic Stripe expression along the developing epidermis, it was impossible to determine whether Stripe-dependent myotube misguidance was restricted to the region of Stripe b expression and whether the weaker defects observed between the rows of Stripe b-expressing cells were a secondary consequence of a local effect (Fig. 4f). To address this question, we ectopically expressed Stripe b in a single ectodermal band corresponding to the stripe 6 domain of the pair rule gene hairy (21, 22). As shown in Fig. 4g, strong muscle pattern defects were found not only in the segment that received Stripe b expression through the hairy stripe 6 GAL4 driver line but also in the directly adjacent segments and to a low degree in the segments next to them. However, no effects have been noted in segments that are more than two segments apart from the Stripe b expression domain. These results establish that the ectopic Stripe b not only affects the epidermal cells in an autonomous manner but also induces a long-range signal, likely to involve a diffusible factor, which acts in a non-cell-autonomous manner by interfering with the orientation of myotubes before their encounter the EMA cells.
We next asked whether such a sr-dependent long-range signal might be required for myotube guidance. We found that heat shock-induced ubiquitous Stripe b expression causes an abnormal muscle pattern only when expressed prior to stage 15, when the muscle fibers were not yet attached to the EMA cells (3). Fig. 4h shows that Stripe b expression at stage 12 results in an abnormal outgrowth of muscles, indicating that it interferes with myotube guidance already at the time when the myotubes form growth-cone-like processes at their leading edges (3). We cannot decide whether Stripe-dependent induction of EMA differentiation, as suggested by the ectopic activation of the EMA marker genes, and the Stripe-dependent interference with myotube guidance are linked processes or whether they occur in parallel.
Recent analysis of sr function by mutant analysis has shown that sr is necessary for EMA differentiation and myotube guidance (6, 7). The results reported here indicate that sr expression is sufficient to provide both functions in a selective manner. It activates EAM marker gene expression in ectoderm-derived but not in mesoderm-derived cells. This suggests that sr functions as a developmental switch gene in preconditioned ectodermal cells which requires at least one additional transcription factor, a cofactor, and/or an ectoderm-specific protein modification of Stripe to provide its germ layer-restricted function. It appears that sr carries a cell-autonomous function in ectodermal cells that causes EMA cell development as indicated by the activation of specific marker genes and a non-cell-autonomous function likely to be mediated by a long-range signal that emanates from the Stripe-expressing epidermal cells.
Acknowledgments
We thank G. Frommer, C. Klämbt, T. Volk, K. Basler, D. Wilkinson, A. Ferrús, A. Spradling, M. Cole, D. Kiehart, M. Bate, A. Michelson, and D. Buttgereit for their various and important contributions, S. Elend for technical assistance, G. Dowe for sequencing and oligonucleotide synthesis, and H. Taubert for P-element transformations. G.V. was supported by a postdoctoral fellowship of the Deutsche Forschungsgemeinschaft. The work was supported by the Max Planck Society and the Sonderforschungsbereich 271 of the Deutsche Forschungsgemeinschaft (H.J.).
ABBREVIATIONS
- EMA
epidermal muscle attachment
- MHC
myosin heavy chain
References
- 1.Abmayr S M, Erickson M S, Bour B A. Trends Genet. 1995;11:153–159. doi: 10.1016/s0168-9525(00)89030-7. [DOI] [PubMed] [Google Scholar]
- 2.Bate M. In: The Mesoderm and Its Derivatives. Bate M, Martinez Arias A, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1993. pp. 1013–1090. [Google Scholar]
- 3.Bate M. Development (Cambridge, UK) 1990;110:791–804. doi: 10.1242/dev.110.3.791. [DOI] [PubMed] [Google Scholar]
- 4.Williams G J A, Caverney S. J Embryol Exp Morphol. 1980;58:13–33. [PubMed] [Google Scholar]
- 5.Williams G J A, Caveney S. J Embryol Exp Morphol. 1980;58:35–61. [PubMed] [Google Scholar]
- 6.Volk T, VijayRaghavan K. Development (Cambridge, UK) 1994;120:59–70. doi: 10.1242/dev.120.1.59. [DOI] [PubMed] [Google Scholar]
- 7.Frommer G, Vorbrüggen G, Pasca G, Jäckle H, Volk T. EMBO J. 1996;15:1642–1649. [PMC free article] [PubMed] [Google Scholar]
- 8.Madden S M, Rauscher F J. In: Positive and Negative Regulation of Transcription and Cell Growth Mediated by the EGR Family of Zinc-Finger Gene Products. Sluyer M, Ab G, Brinkmann A O, Blankenstein R A, editors. New York: New York Acad. Sci.; 1993. pp. 75–84. [DOI] [PubMed] [Google Scholar]
- 9.Han K, Manley J L. EMBO J. 1993;12:2723–2733. doi: 10.1002/j.1460-2075.1993.tb05934.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jaynes J B, O’Farrell P H. EMBO J. 1991;10:1427–1433. doi: 10.1002/j.1460-2075.1991.tb07663.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.John A, Smith S T, Jaynes J B. Development (Cambridge, UK) 1995;121:1801–1813. doi: 10.1242/dev.121.6.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Poole S J, Kauvar L M, Drees B, Kornberg T B. Cell. 1985;40:37–43. doi: 10.1016/0092-8674(85)90306-x. [DOI] [PubMed] [Google Scholar]
- 13.Brand A H, Perrimon N. Development (Cambridge, UK) 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 14.Golembo M, Raz E, Shilo B Z. Development (Cambridge, UK) 1997;122:3363–3370. doi: 10.1242/dev.122.11.3363. [DOI] [PubMed] [Google Scholar]
- 15.Tautz D, Pfeifle C. Chromosoma. 1989;98:81–85. doi: 10.1007/BF00291041. [DOI] [PubMed] [Google Scholar]
- 16.Macdonald P M, Struhl G. Nature (London) 1986;324:537–545. doi: 10.1038/324537a0. [DOI] [PubMed] [Google Scholar]
- 17.Armand P, Knapp A C, Hirsch A J, Wieschaus E F, Cole M D. Mol Cell Biol. 1994;14:4145–4154. doi: 10.1128/mcb.14.6.4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rubin G M, Spradling A C. Science. 1982;218:348–353. doi: 10.1126/science.6289436. [DOI] [PubMed] [Google Scholar]
- 19.Campos-Ortega J A, Hartenstein V. The Embryonic Development of Drosophila melanogaster. Berlin: Springer; 1985. [Google Scholar]
- 20.Buttgereit D, Leiss D, Michiels F, Renkawitz-Pohl R. Mech Dev. 1991;33:107–118. doi: 10.1016/0925-4773(91)90077-j. [DOI] [PubMed] [Google Scholar]
- 21.Pankratz M J, Seifert E, Gerwin N, Billi B, Nauber U, Jäckle H. Cell. 1990;61:309–317. doi: 10.1016/0092-8674(90)90811-r. [DOI] [PubMed] [Google Scholar]
- 22.Riddihough G, Ish-Horowicz D. Genes Dev. 1991;5:840–854. doi: 10.1101/gad.5.5.840. [DOI] [PubMed] [Google Scholar]