Abstract
The crystal structures of cytochrome c oxidase from both bovine and Paracoccus denitrificans reveal two putative proton input channels that connect the heme-copper center, where dioxygen is reduced, to the internal aqueous phase. In this work we have examined the role of these two channels, looking at the effects of site-directed mutations of residues observed in each of the channels of the cytochrome c oxidase from Rhodobacter sphaeroides. A photoelectric technique was used to monitor the time-resolved electrogenic proton transfer steps associated with the photo-induced reduction of the ferryl-oxo form of heme a3 (Fe4+ = O2−) to the oxidized form (Fe3+OH−). This redox step requires the delivery of a “chemical” H+ to protonate the reduced oxygen atom and is also coupled to proton pumping. It is found that mutations in the K channel (K362M and T359A) have virtually no effect on the ferryl-oxo-to-oxidized (F-to-Ox) transition, although steady-state turnover is severely limited. In contrast, electrogenic proton transfer at this step is strongly suppressed by mutations in the D channel. The results strongly suggest that the functional roles of the two channels are not the separate delivery of chemical or pumped protons, as proposed recently [Iwata, S., Ostermeier, C., Ludwig, B. & Michel, H. (1995) Nature (London) 376, 660–669]. The D channel is likely to be involved in the uptake of both “chemical” and “pumped” protons in the F-to-Ox transition, whereas the K channel is probably idle at this partial reaction and is likely to be used for loading the enzyme with protons at some earlier steps of the catalytic cycle. This conclusion agrees with different redox states of heme a3 in the K362M and E286Q mutants under aerobic steady-state turnover conditions.
Cytochrome oxidase (COX) is the terminal enzyme in the respiratory chains of mitochondria and of many bacteria. The enzyme is part of a superfamily of respiratory heme-copper oxidases (1, 2) that have in common a bimetallic center, consisting of a high-spin heme (a3, o3, or b3) and associated copper (CuB), which is where dioxygen is reduced to water. These oxidases are redox-driven proton pumps; for each O2 reduced to H2O, 4 H+ are taken up to combine with oxygen to form 2 H2O, and another 4 H+ are pumped electrogenically across the membrane. This generates pH and voltage differences across the membrane (i.e., the protonmotive force), which are used to drive the synthesis of ATP and other energy-requiring processes (3).
The active site where O2 is reduced is buried within the enzyme far from the internal and external aqueous spaces (4–6). Hence, substantial intraprotein movement of protons is necessary not only for those protons that are translocated all the way across the membrane (“pumped” protons), but also for the protons that are used for the protonation of reduced oxygen species (“chemical” protons). How the protons travel inside COX has long remained an intriguing question. It was postulated many years ago that COX must have “proton channels” (7–9) that connect the redox-linked protonatable groups of the enzyme with the positively (P) and negatively (N) charged aqueous phases and provide for the intraprotein transfer of protons. As proposed originally in refs. 10 and 11, there are two separate input channels in COX for the uptake of “pumped” and “chemical” protons. Evidence for electrogenic movement of protons within the channel involved in the protonation of the binuclear center from the mitochondrial matrix was reported (12, 13) and ionization of groups within this channel is apparently coupled to electron transfer between hemes a and a3 (14, 15).
The x-ray structures of COX from bovine (5, 6) and from Paracoccus denitrificans (4) reveal domains within subunit I that have been interpreted as possible pathways for proton uptake from the negative side of the membrane. Several residues within these putative channels had been predicted on the basis of the site-directed mutagenesis studies (16–18). One pathway contains residues in helix VI and helix VIII, including highly conserved polar residues K362, T359, and Y288 (here and below, R. sphaeroides numbering is used if not indicated otherwise) and has been speculated (4) to be specifically involved in delivering protons used chemically in the reduction of dioxygen to water (“chemical channel”). Because its function is not yet established, the term “K channel” (for the lysine component) is perhaps preferable.
A second channel involves residues within helix III and IV and has a highly conserved aspartate and asparagine (D132 and N139) near the entrance, and a hydrogen bond path can be discerned up to S201 in helix IV. In the P. denitrificans COX, the pathway leads via water molecules to a highly conserved glutamic acid residue (E286), at which point the pathway is not clear. Following the original observation on proton pumping being abolished specifically by mutations in the conserved aspartate (D135 in cytochrome bo3 from Escherichia coli) (19), this structure has been speculated to be a “pumping channel” (4), but it would be preferable at this point to refer to this as the “D channel” (for the aspartic acid component) until its role is better defined experimentally.
Mutations in some residues within either the K channel or the D channel result in severe inhibition of steady-state turnover in the bo3 and aa3 oxidases (18–21). The current work is directed at determining which steps of the catalytic cycle are affected by these mutations. We have examined the effects of amino acid replacements in each of the two channels of the COX from R. sphaeroides (K362M and T359A in the K channel; D132N and E286Q in the D channel) on (i) the absorption spectra of hemes a and a3 in the aerobic steady state, and (ii) the time-resolved generation of the membrane potential (Δψ) associated with the isolated redox step of COX conversion from the ferryl-oxo to the ferric (oxidized) state (i.e., delivery of the fourth electron needed to complete the reduction of oxygen) ![]() .
.
The F-to-Ox step requires the delivery of at least one chemical proton to form hydroxide (Fe4+ = O2− + e− + H+ → Fe3+ − OH−) (22) or two to form water (3). It is also associated with the transmembrane pumping of at least one, or probably two, protons (3, 23). Vectorial movements of these protons within the membrane-incorporated enzyme result in the generation of a transmembrane electric potential difference (ΔΨ) that can be monitored electrometrically with submicrosecond time resolution using the method developed by Drachev (24–26).
Under appropriate conditions, the majority of COX can be converted to the ferryl-oxo state in the presence of H2O2 (27–29). By using Ru(II)-tris-bipyridine (RuBpy) as a photo-activated reductant, the transition from F-to-Ox under these conditions can be initiated by a laser flash (30).
The combined technique of the flash-induced reduction by RuBpy and time-resolved electrometric monitoring of ΔΨ has been previously utilized in studies of COX from beef heart (31, 32). Three electrogenic processes with time constants of about 40 μs, 1 ms, and 4 ms were resolved in the F-to-Ox step and identified provisionally as (i) vectorial transfer of the electron from CuA to heme a, (ii) uptake of “chemical” protons, and (iii) transmembrane proton pumping, respectively. The current work applies this approach to both the wild-type and mutant variants of the R. sphaeroides oxidase.
MATERIALS AND METHODS
Materials.
Ni-NTA-resin for histidine-tagged cytochrome oxidase purification was from Qiagen. Ru(II)tris-bipyridyl chloride was obtained from Aldrich. Phosphatidyl choline type II-s for liposome preparation (azolectin) was purchased from Sigma, and egg yolk lecithin for collodion membrane preparation was from either Reakhim (Kharkov, former USSR) or was Grade I lecithin from Lipid Products (Nutley, U.K.). Thirty percent H2O2 Suprapur grade was obtained from Merck.
Site-Directed Mutants.
Site-directed mutants were constructed on a template plasmid (pJS3) containing the ctaD gene, which codes for subunit I of cytochrome oxidase (33). D132N, E286Q, T359A, and K362M were constructed by an extension of a mutagenic single-stranded DNA primer on a single-stranded DNA template, using the method of Vandeyar et al. (34), and have been previously described (18, 21, 35).
Purification.
Purification of wild-type and mutant forms of COX was facilitated by the genetic fusion of a six-histidine affinity tag to the C terminus of subunit I, as previously described (36). This allows the single-step purification of this mutant using a Ni-NTA affinity column. The activity of purified COX was measured following the oxidation of 30 μM cytochrome c spectrophotometrically at 550 nm in 50 mM potassium phosphate buffer, pH 6.5, with 0.02% lauryl maltoside (36).
Electrometric Measurements of ΔΨ Generation.
Time-resolved measurements of ΔΨ generation induced by single-electron photoreduction of COX by RuBpy were made exactly as described previously (31). Briefly, COX was reconstituted in azolectin vesicles by a cholate dialysis method (37). The proteoliposomes were adhered to one side of a collodion film insulating the two compartments of an electrometric cell. The film was presoaked in a decane solution of egg yolk lecithin and stearyl amine (100 and 1 mg/ml, respectively) insulating the two compartments of an electrometric cell. Generation of ΔΨ in proteoliposomes gives rise to an electric potential difference between the two compartments of the electrometric cell that can be monitored with a pair of light-protected Ag/AgCl electrodes in a device constructed by L. A. Drachev in the A. N. Belozersky Institute (Moscow) (25, 26). The electrodes were connected to an operational amplifier (Burr-Brown 3554) and to a PC-interfaced digital transient recorder (Datalab 1080).
The buffer in the electrometric cell contained 5 mM Tris acetate, pH 8. RuBpy (40 μM) was added to the collodion film-bound proteoliposomes as the photoactive electron donor and 10 mM aniline were added as a sacrificial electron donor to rapidly re-reduce the RuBpy transiently photooxidized by COX. This makes the COX photoreduction by RuBpy irreversible, as described by Nilsson (30).
RuBpy was photoexcited by 15-ns flashes from a Quantel (Santa Clara, CA) 471 YAG laser operated at a double-frequency mode (532 nm, 30–50 mJ/flash). The individual photoelectric traces show sufficiently good signal-to-noise ratio and usually are shown as such. On some occasions, two to three transients were averaged.
Kinetic curve fitting was made with a software package, gim, developed by Alexander L. Drachev (subroutine “Discrete”; ref. 38).
RESULTS
Steady-State Activity and Spectra.
All the proton-channel mutants studied in this work exhibit severe inhibition of COX activity (Table 1). The inhibition is virtually complete in the K362M and E286Q mutants.
Table 1.
Turnover rates and steady-state heme a reduction of wild-type and mutant COXs
| Enzyme | Turnover, s−1 (%) | % Heme a reduction |
|---|---|---|
| Wild type | 1,500 (100) | 35 |
| T359A | 250 (17) | 50 |
| D132N | 50 (3.3) | 72 |
| K362M | <5 (<0.3) | >95 |
| E286Q | <5 (<0.3) | >95 |
Steady-state reduction of heme a has been measured in a dual-wavelength mode at 605 nm minus 630 nm following the addition of 2 mM ascorbate and 100 μM TMPD and, after reaching steady state, 1 mM KCN and dithionite to the oxidized COX in 50 mM phosphate buffer, pH 8, with 0.1% lauryl maltoside.
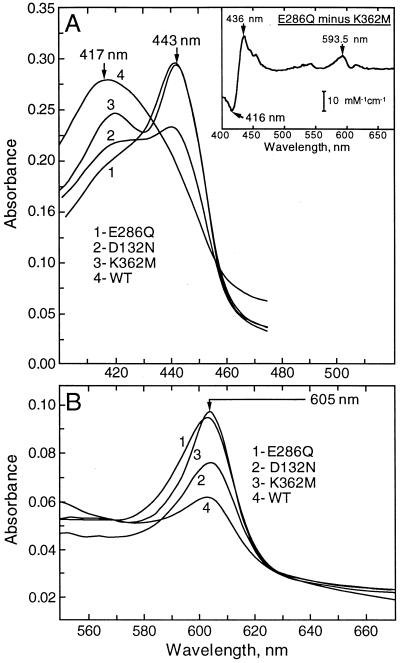
Absorption spectra taken under turnover conditions show that inhibition resulting from each of the four mutations follows the reduction of heme a so that in each of the mutants heme a is reduced more than in the wild-type control (Table 1; Fig. 1). However, the steady-state spectra of the inactive mutants within the K channel (K362M) and the D channel (E286Q) are very different. Whereas the steady-state absolute spectrum of K362M is similar to that of “resting” COX with separate Soret peaks at about 444 nm and 420 nm, corresponding to reduced (ferrous) heme a and oxidized (ferric) high-spin heme a3, respectively (39, 40), the spectrum of E286Q resembles that of fully reduced COX but with ferrous heme a3 bound to a ligand.
Figure 1.
Absolute absorption spectra of the wild-type and mutant forms of COX in the aerobic steady state with ascorbate + TMPD as electron donors. The stirred 1-cm cell contained 0.2–0.5 μM of wild-type or mutant COX in 50 mM potassium phosphate (pH 8.0) and 0.1% lauryl maltoside. The spectra shown were recorded about 3 min after the addition of 1 mM ascorbate and 100 μM TMPD. KCN (1 mM) and then solid dithionite were added subsequently to obtain full reduction of heme a. The spectra have been normalized by the amplitude of the peak of heme a at 605 nm vs. the 630-nm reference point in the dithionite-reduced form of the enzyme. Slit width = 2 nm. (A) Soret band spectra. (B) Visible spectra. Inset in A gives a difference between the normalized absolute “steady-state” spectra of the E286Q and K362M mutants.
This is best illustrated by the difference spectrum E286Q minus K362M (Fig. 1 Inset). The lineshape and magnitude (red shift of the Soret band, peak-to-trough size 40–45 mM−1 cm−1; 6–7 mM−1 cm−1 for the peak at 594 nm) indicate that the major part of heme a3 in E286Q is not simply ferric or ferrous high-spin but, rather, is ferrous low-spin bound to a sixth axial ligand, as confirmed by our recent room temperature magnetic circular dichroism (MCD) studies in collaboration with the group of A. Arutyunyan (unpublished work). The chemical identity of the sixth ligand cannot be determined by means of absorption or room temperature MCD spectroscopy. Resonance Raman and/or low-temperature infrared-MCD (41) will be required for investigation into this issue.
The line shape of the steady-state spectrum of D132N is of limited usefulness because this mutant is partially active (Table 1). Nevertheless, it is consistent with partial conversion of heme a3 to the ligand-bound state found in the E286Q mutant, which shows an increased absorbance around 590 nm after appropriate normalization and correction for partial reduction of heme a.
Electrogenic Activity.
Figs. 2 and 3 show the kinetics of ΔΨ generation by the wild-type (WT) and mutant forms of COX linked to the single-electron photochemical reduction of the enzymes by RuBpy. The oxidase was converted to the F state prior to the photoreduction by preincubation of the samples with 1 mM H2O2. Control spectrophotometric measurements have verified that it is the ferryl-oxo state that is generated under these conditions in WT, D132N, T359A, and K362M with no evidence of any contribution from the peroxy complex. The peak-to-trough magnitude of the H2O2-induced difference spectra in the Soret band was typically 30–45 mM−1 cm−1 in the WT and in the K channel mutants as compared with ca. 50 mM−1 cm−1 observed for beef heart enzyme (29, 42–44). The size of the peroxide-induced spectroscopic changes in the D132N mutant varied for five different preparations examined (from 30 mM−1 cm−1 to 8 mM−1 cm−1 in the Soret band) without much difference in the photoelectric responses of the samples. The situation is not clear with the E286Q mutant, because the absolute absorption spectrum of E286Q in the “oxidized” (i.e., ferricyanide-treated) state is already rather similar to that of the F state. Accordingly, the addition of H2O2 is not accompanied by the typical spectroscopic changes (45). Similarly, variability of the initial absolute spectrum of the “oxidized” D132N may be a reason for the observed scatter in the apparent yield of the amount of F state induced by H2O2 in this mutant.
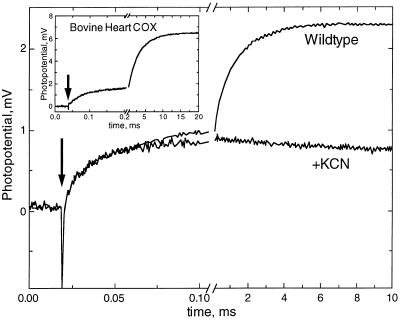
Figure 2.
Rapid kinetics of Δψ generation by the wild-type R. sphaeroides COX. The reaction mixture contains 5 mM Tris-acetate buffer (pH 8), 40 μM RuBpy as photoreductant of COX, and 10 mM aniline as sacrificial electron donor to the photooxidized RuBpy. Before the flash, the sample was incubated for several minutes with 1 mM H2O2 to convert heme a3 to the ferryl-oxo state. After recording a trace (Wildtype), 1 mM potassium cyanide was added and the second trace (+KCN) was recorded. Inset shows a typical photoelectric trace as observed with bovine heart COX under the same conditions except that the H2O2 concentration was 4 mM.
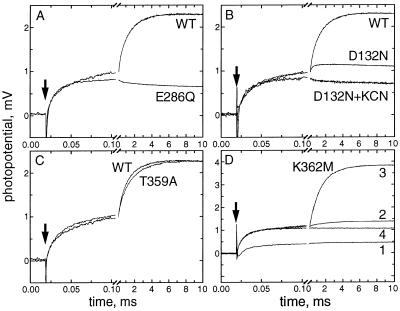
Figure 3.
Effect of mutations in the K channel and the D channel on the membrane potential generation by R. sphaeroides COX following single-electron photoreduction. Where shown on the same panel, the mutant and wild-type traces have been normalized for clarity to the amplitude of the microsecond phase. (A) E286Q. Basic conditions, same as in Fig. 2, except that 0.2 mM ferricyanide has been added in case of E286Q to keep heme a oxidized. (B) D132N. Basic conditions, as in case of E286Q. Where indicated, 1 mM KCN has been added. (C) T359A. Basic conditions, as in Fig. 2. (D) K362M. Basic conditions, as in Fig. 2. The traces (1–4) have been recorded in sequence as follows. Traces: 1, no additions; 2, 5-min incubation with 0.2 mM ferricyanide; 3, addition of 1 mM H2O2; 4, addition of 0.5 mM KCN.
Wild-type oxidase.
Kinetics of ΔΨ generation by the wild-type bacterial COX (Fig. 2) is similar to that observed previously with the beef heart enzyme (31, 32) (Fig. 2 Inset). A rapid phase linked to the reduction of heme a by CuA (τ ≈ 15 μs) (characteristic time, τ = 1/k rather than t½ is used throughout the paper) is followed by a millisecond portion of the response that is inhibited by potassium cyanide and is associated with intraprotein proton transfer (31). As with the beef heart oxidase, the latter is resolved into two components—an intermediate phase (≈0.4 ms) and a slow phase (≈1.5 ms at pH 8). The rates of all three electrogenic phases observed with the R. sphaeroides oxidase are 2- to 3-fold higher than those found for the beef heart COX (31), which is in agreement with higher turnover rates of the bacterial enzyme. The relative amplitudes of the rapid, intermediate, and slow phases (ca. 30, 20, and 50%, respectively) are similar but not identical to those in bovine COX (20, 20, and 60%) (31), so that the relative contribution of the rapid phase is reproducibly higher and that of the slow phase is lower in the bacterial wild-type COX. This can be explained, in part, by a somewhat lower yield of the F state in R. sphaeroides enzyme as compared with the beef heart COX.
D channel mutants.
The photoelectric responses of the E286Q and D132N mutants are shown in Fig. 3 (A and B, respectively), with the responses of the wild-type oxidase included for comparison. Pretreatment with ferricyanide was required to elicit the full size of the response in the mutants due to partial reduction of heme a even after prolonged incubation under aerobic conditions. In both mutants, there is a normal CuA-to-heme a microsecond phase. In contrast, the millisecond part of the electric response is suppressed completely (E286Q) or nearly completely (D132N). The data suggest that in both of these mutants, vectorial proton transfer at this stage of the reaction is fully or largely impaired.
In E286Q, the millisecond phase of the electric response is completely absent, and the response is unaffected either by the addition of excess peroxide or by KCN. As previously mentioned, it is not clear whether E286Q forms the ferryl-oxo state upon addition of H2O2. Therefore, the lack of the millisecond part of the response in this mutant could be a trivial consequence of E286Q simply not being able to generate the ferryl-oxo form of the oxidase under these conditions.
As mentioned above, the ferryl-oxo state of the D132N mutant is generated to quite a significant extent upon the addition of H2O2. Nevertheless, the millisecond portion of the electrogenic response is severely reduced in magnitude (<10% of the wild-type control) (Fig. 3B). Like the millisecond electrogenic phases observed with the wild-type oxidase, the residual millisecond part of the response of D132N is suppressed by KCN so that the response exhibited by D132N (plus KCN) becomes indistinguishable from that of E286Q (with or without KCN), with a characteristic small negative transient following the microsecond phase.
The millisecond part of the electric response in D132N is essentially monoexponential, with τ of 0.5–0.6 ms. This is definitely faster than the slow phase and is similar to the rate typically observed for the intermediate phase with the wild-type oxidase. Hence, the small millisecond transient observed with D132N is not likely to be due to a small portion of the enzyme with wild-type characteristics. The comparison of the experimental data with simulations suggests that in D132N, the slow phase is not simply decelerated ≈30-fold, as could be expected from the turnover rate of the mutant (Table 1), but, rather, is completely eliminated. In addition there is a 2- to 3-fold decrease in the amplitude of the intermediate phase.
K channel mutants.
In contrast to the behavior of the D channel mutants, the photoelectric response associated with the F-to-Ox transition of the R. sphaeroides COX with mutations in the K channel (T359A and K362M) appears to be similar to the response exhibited by the wild-type enzyme. The photoelectric trace of T359A (Fig. 3C) is almost superimposable with the trace from the wild-type oxidase. It is particularly striking that the “dead” K362M mutant (i.e., virtually no steady-state turnover) reveals a fully developed proton translocation phase in the presence of excess peroxide (Fig. 3D). However, the K362M enzyme, as isolated and even after reconstitution in the liposomes, is in a form where heme a (and perhaps CuA as well) remains reduced even under aerobic conditions. This renders electron photoinjection by RuBpy into heme a not possible, and, indeed, in the absence of peroxide or another oxidant, the mutant shows almost no flash-induced electric response (Fig. 3D, trace 1). The addition of ferricyanide to the K362M mutant reoxidizes heme a and, as expected, restores the microsecond but not the millisecond portions of the developing membrane potential (Fig. 3D, trace 2). The millisecond phase of K362M appears upon the addition of H2O2 (Fig. 3D, trace 3) and is fully inhibited by KCN (Fig. 3D, trace 4) or catalase (not shown). It is noteworthy that the normal full electric response is restored with the K362M mutant upon addition of H2O2 even in the absence of ferricyanide (not shown). This is in accordance with the observation that hydrogen peroxide, unlike molecular oxygen, is able to reoxidize heme a readily in the isolated K362M enzyme (45).
Apparently, the mutations in the K channel do not inhibit the electrogenic proton translocation associated with the F-to-Ox transition, at least in a single turnover of the enzyme.
DISCUSSION
The current work demonstrates that mutations in the D and K channels block COX at different steps in the catalytic cycle. First, this is suggested by the absorption spectra indicating different states of heme a3 in the K362M (K channel) and E286Q (D channel) mutants in the aerobic “steady state.” Second, whereas in the mutant within the D channel (D132N) the intraprotein movement of protons associated with the F-to-Ox transition appears to be virtually absent, the mutations in the K channel (K362M and T359A), in contrast, have no significant effect on the electrogenic reactions coupled to this redox step. Because the F-to-Ox transition requires the movement of both chemical and pumped protons (3, 4, 23, 46, 47), mutations that block either a “chemical” or “pumping” channel would impede this step. Hence, the hypothesis (4) that the K channel is required for the delivery of all “chemical” protons destined to be consumed in the formation of H2O (i.e., the “chemical” channel) is probably incorrect. The current work suggests, rather, that the two channels are functionally associated with different steps in the catalytic cycle, and the F-to-Ox transition, at least for a single turnover, does not require the K channel.
To discuss the possible roles of the D and K channels it is worthwhile to highlight the similarity of the COX reaction mechanism to peroxidases and cytochromes P450 and consider the catalytic cycle of COX as two half-reactions (see Scheme II, below). (i) Eu-oxidase half-reaction. This includes the two-electron reduction of the enzyme and the reaction with O2 to yield a bound peroxide product, or P state. The eu-oxidase reaction is not proton-pumping (23), but the initial reduction of the enzyme is accompanied by the uptake of two protons (cf. ref. 48 and references therein). (ii) Peroxidase half-reaction. This includes sequential reduction of the peroxide adduct to the ferric state in two single-electron steps, P-to-F and F-to-O, analogous to the classical heme peroxidase reaction.
It is the peroxidase half-reaction that is coupled to proton pumping (23), as first indicated by the results of the Orii group (49) and demonstrated recently by Vygodina et al. (50). ![]()
The initial product of two-electron oxygen reduction (as well as the initial product of H2O2 binding to ferric COX), with heme iron in the ferric state and an intact O—O bond [heme a3 Fe3+O2 = (2H+)], is analogous to compound 0 in peroxidases (51, 52) and is designated in Scheme II as P0. The “peroxy” state of cytochrome oxidase is poorly defined structurally and can be, in fact, a collective name for more than one state differing in redox state of CuB, protonation of the binuclear center, and even in the oxidation state of the heme a3 iron (3, 42, 53–60).
Traditionally, the transition of P0 to the ferryl-oxo state and formation of the first H2O molecule is believed to require the addition of the third electron to the binuclear center (presumably, via CuB) followed by reductive scission of the O—O bond (not shown in Scheme II; see refs. 3, 53–55, 61, 62 for detailed schemes and discussion).
An alternative mechanism of the peroxidase half-reaction of COX is illustrated by Scheme II. As in the peroxidases or cytochromes P450, the formation of P0 can be followed by proton-assisted heterolytic cleavage of bound peroxide, even in the absence of the third “exogenous” electron. This will yield water plus a ferryl-oxene intermediate, which is two oxidation equivalents above the resting ferric state (formally, Fe5+=O2− or R+ ⋅ Fe4+=O2−), where the second redox equivalent may be donated by an aromatic amino acid residue (denoted R) in the vicinity of heme a3 or CuB (42, 56, 60, 63). This ferryl intermediate will be analogous to compound I of peroxidases and is, thus, denoted in Scheme II as compound FI. It is possible that some of the experimentally observed “peroxy” intermediates(s) of COX discussed in the literature include the FI state. Subsequently, two single-electron reduction steps convert FI first to a compound II-like intermediate ferryl-oxo state (580 nm form, heme a3 Fe4+= O2−), denoted here FII, and then to Ox (heme a3 Fe3+) plus heme-bound hydroxide-anion (or water). It is worth noting that, as shown in Scheme II, the formation of water at the P0-to-FI step can be coupled to the electrogenic uptake of two protons from the negative phase (i.e., bacterial cytoplasm/mitochondrial matrix).
Spontaneous conversion of the ferric peroxy adduct, P0, to the ferryl FI is likely to take place in experiments with the addition of hydrogen peroxide to the oxidized enzyme, especially at pH below 7 (42, 56, 58, 64, 65). Also, recent resonance Raman spectroscopy studies have revealed a ferryl intermediate analogous to compound I and preceding the formation of the conventional F state (580 nm form, FII in Scheme II) during COX reoxidation by molecular oxygen (60).
It is possible that under physiological conditions the P0-to-FI step transition in Scheme II is promoted kinetically or thermodynamically by the addition of the third electron to CuB. In this connection, one can contrast the stability of the “peroxy state” (607 nm species with oxidized CuB) generated from the mixed-valence enzyme (66–68), whereas rapid conversion of P-to-F takes place during oxidation of the fully-reduced COX, in which it is a P state with CuB reduced that is formed (55, 62, 69). It is, however, of critical importance to establish whether the 607-nm form of COX is indeed “peroxy” (P0) with intact O—O bond and ferric heme a3 or ferryl-oxo (FI).
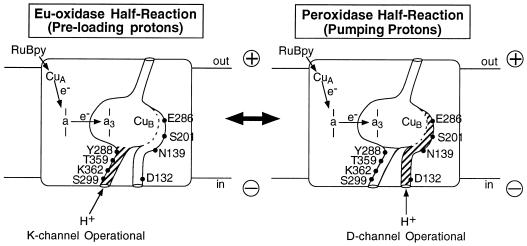
It is tempting to propose that the nonpumping eu-oxidase (Ox-to- P0 or perhaps FI) and proton-translocating peroxidase (P0 or FI-to-Ox) half-reactions may be catalyzed by two conformational states of COX, differing in the status of the proton channels (Fig. 4). In the nonpumping eu-oxidase conformation, the K channel is operational, whereas, during the proton-pumping peroxidase half-reaction it is D channel that is primary. Doubtless, this is an oversimplification, but it represents a reasonable starting point and can be experimentally tested.
Figure 4.
Possible roles of the two input proton channels in cytochrome oxidase. Two proposed states of cytochrome oxidase are illustrated, differing with respect to which of the proton-conducting channels is operational. On the left is the preloading or eu-oxidase conformation, when the K channel appears to be dominant. (Right) The D channel dominates during the second (peroxidase) portion of the catalytic cycle, during which proton pumping occurs.
Although the experiments presented in the current work address mainly the F-to-Ox transition, it is reasonable to suggest that the K channel, if it is a proton channel at all, is required for the uptake of protons during the initial reduction of the oxidized heme-copper center (Ox-to-R) (48), as most other possibilities are ruled out by the existing data. Indeed, oxidation of the fully reduced oxidase by O2 proceeds normally to the ferryl-oxo state, as evidenced by previous studies with the K362M and T359A mutants of the cytochrome bo3 quinol oxidase from E. coli (70). The current work indicates that the F-to-Ox transition is also not impeded by the K channel mutations. So, the step inhibited must involve the initial reduction of the heme-copper center, which has been shown to be rate-limited by the coupled proton uptake (71). This conclusion is consistent with the steady-state spectra (Fig. 1) showing that in the presence of reductant and O2, heme a is fully reduced and heme a3 is in the ferric high-spin state in the K362M mutant.
The D channel is obviously necessary for proton pumping coupled to the F-to-Ox transition and, perhaps, for the P-to-F step(s) as well. Absorption spectra suggest that heme a3 is isolated in E286Q as some adduct that fails to readily undergo reduction to the ferric state by the third or/and fourth electrons in the peroxidase part of the catalytic cycle. The D channel must be effectively closed by the mutation at position D132, which is observed in the x-ray structures to be near the entrance of this channel (4, 6). The small, residual slow phase of the electric response in D132N is of interest (Fig. 3B). The sensitivity to KCN implies that this process is linked to the reduction of heme a3, and its monoexponential kinetics is rather close to that of the intermediate electrogenic phase assigned provisionally to the uptake of “chemical” protons (31). However, the amplitude of this residual “intermediate” phase is less than 1/2 that observed in the wild-type oxidase (normalized to the magnitude of the fast phase). Possibly, this charge movement in the D132N mutant is due to the displacement of protons toward heme a3 within the proton “wire” of the closed D channel upon reduction and the accompanying vicinal protonation of the heme-bound oxene.
It is interesting that although the current work points to the functional importance of residue E286, the significance of this residue in facilitating proton conduction has been questioned (6). In the structure of the bovine heart oxidase, the residue equivalent to E286 does not appear to be within the hydrogen bond network defining the D channel (6). Also, the connections leading from E286 are unclear in the structure of the bacterial oxidase (4). Although such uncertainties might be expected at the level of resolution of the current structural information, it should be noted that both of the x-ray structures are of the fully oxidized form of the enzyme and, hence, may represent a conformational state where the D channel is not fully connected. Perhaps a transition to the “peroxidase conformation” (Fig. 4) brings about subtle structural rearrangements that complete the connection of E286 to the binuclear center and/or the proton exit pathway.
The signal to switch from the K channel to the D channel can only be speculated. Oxygen binding is an attractive possibility for invoking an allosteric change in the enzyme. The analogy with hemoglobin allostery is evident. Another possible trigger signal for channel switching could be the formation of heme-bound oxene in the P0-to-FI transition. Being a strong ligand, oxene could dictate the allosteric state of the enzyme with the D channel open. In this case, the K channel could also be involved in the uptake of two protons required for water formation at the P0-to-FI step of the reaction in Scheme II.
Acknowledgments
We thank Dr. Dmitry Zaslavsky for helpful discussions and advice. We would like to thank Dr. Tatiana Vygodina for her kind help at some stages of this work and donation of beef heart COX, and Mr. Andrey Zaspa for his expert care over the instruments in Moscow. This work has been supported by grants from the National Institutes of Health (Fogarty International Research Collaboration Award PHS 1 R03 TW 00349 and HL16101 to R.B.G.) and Russian Fund for Basic Research (94-04-12726a and 97-04-49765 to A.A.K.).
ABBREVIATIONS
- COX
cytochrome oxidase
- RuBpy
ru(II)-tris-bipyridine
References
- 1.Saraste M, Holm L, Lemieux L, Lubben M, van der Oost J. Biochem Soc Trans. 1991;19:608–612. doi: 10.1042/bst0190608. [DOI] [PubMed] [Google Scholar]
- 2.García-Horsman J A, Barquera B, Rumbley J, Ma J, Gennis R B. J Bacteriol. 1994;176:5587–5600. doi: 10.1128/jb.176.18.5587-5600.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Babcock G T, Wikström M. Nature (London) 1992;356:301–309. doi: 10.1038/356301a0. [DOI] [PubMed] [Google Scholar]
- 4.Iwata S, Ostermeier C, Ludwig B, Michel H. Nature (London) 1995;376:660–669. doi: 10.1038/376660a0. [DOI] [PubMed] [Google Scholar]
- 5.Tsukihara T, Aoyama H, Yamashita E, Tomizaki T, Yamaguchi H, Shinzawa-Itoh K, Nakashima T, Yaono R, Yoshikawa S. Science. 1995;269:1069–1074. doi: 10.1126/science.7652554. [DOI] [PubMed] [Google Scholar]
- 6.Tsukihara T, Aoyama H, Yamashita E, Takashi T, Yamaguichi H, Shinzawa-Itoh K, Nakashima R, Yaono R, Yoshikawa S. Science. 1996;272:1136–1144. doi: 10.1126/science.272.5265.1136. [DOI] [PubMed] [Google Scholar]
- 7.Mitchell P. Chemiosmotic Coupling in Oxidative and Photosynthetic Phosphorylation. Bodmin, U.K.: Glynn Research; 1966. [DOI] [PubMed] [Google Scholar]
- 8.Liberman E A. Biophysics (Moscow) 1977;22:1115–1128. [PubMed] [Google Scholar]
- 9.Rich P R. Biosci Rep. 1991;11:539–571. doi: 10.1007/BF01130217. [DOI] [PubMed] [Google Scholar]
- 10.Konstantinov A A. Dokl Akad Nauk SSSR. 1977;237:713–716. [PubMed] [Google Scholar]
- 11.Artzatbanov V Y, Konstantinov A A, Skulachev V P. FEBS Lett. 1978;87:180–185. doi: 10.1016/0014-5793(78)80327-5. [DOI] [PubMed] [Google Scholar]
- 12.Konstantinov A A, Vygodina T V, Andreev I M. FEBS Lett. 1986;202:229–234. doi: 10.1016/0014-5793(86)80692-5. [DOI] [PubMed] [Google Scholar]
- 13.Wikström M. FEBS Lett. 1988;231:247–252. doi: 10.1016/0014-5793(88)80741-5. [DOI] [PubMed] [Google Scholar]
- 14.Hallén S, Brzezinski P, Malmström B G. Biochemistry. 1994;33:1467–1472. doi: 10.1021/bi00172a024. [DOI] [PubMed] [Google Scholar]
- 15.Ådelroth P, Brzezinski P, Malmström B G. Biochemistry. 1995;34:2844–2849. doi: 10.1021/bi00009a014. [DOI] [PubMed] [Google Scholar]
- 16.Hosler J P, Ferguson-Miller S, Calhoun M W, Thomas J W, Hill J, Lemieux L, Ma J, Georgiou C, Fetter J, Shapleigh J, Tecklenburg M M J, Babcock G T, Gennis R B. J Bioenerg Biomembr. 1993;25:121–136. doi: 10.1007/BF00762854. [DOI] [PubMed] [Google Scholar]
- 17.Brown S, Moody A J, Mitchell R, Rich P R. FEBS Lett. 1993;316:216–223. doi: 10.1016/0014-5793(93)81296-c. [DOI] [PubMed] [Google Scholar]
- 18.Fetter J R, Qian J, Shapleigh J, Thomas J W, Garcia-Horsman A, Schmidt E, Hosler J, Babcock G T, Gennis R B, Ferguson-Miller S. Proc Natl Acad Sci USA. 1995;92:1604–1608. doi: 10.1073/pnas.92.5.1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thomas J W, Puustinen A, Alben J O, Gennis R B, Wikström M. Biochemistry. 1993;32:10923–10928. doi: 10.1021/bi00091a048. [DOI] [PubMed] [Google Scholar]
- 20.Garcia-Horsman J A, Puustinen A, Gennis R B, Wikström M. Biochemistry. 1995;34:4428–4433. doi: 10.1021/bi00013a035. [DOI] [PubMed] [Google Scholar]
- 21.Hosler J P, Shapleigh J P, Mitchell D M, Kim Y, Pressler M, Georgiou C, Babcock G T, Alben J O, Ferguson-Miller S, Gennis R B. Biochemistry. 1996;35:10776–10783. doi: 10.1021/bi9606511. [DOI] [PubMed] [Google Scholar]
- 22.Mitchell R, Mitchell P, Rich P R. Biochim Biophys Acta. 1992;1101:188–191. [PubMed] [Google Scholar]
- 23.Wikström M. Nature (London) 1989;338:776–778. doi: 10.1038/338776a0. [DOI] [PubMed] [Google Scholar]
- 24.Drachev L A, Kaulen A D, Skulachev V P. FEBS Lett. 1978;87:161–167. doi: 10.1016/0014-5793(78)80157-4. [DOI] [PubMed] [Google Scholar]
- 25.Drachev L A, Kaulen A D, Seminov A Y, Severina I I, Skulachev V P. Anal Biochem. 1979;96:250–262. doi: 10.1016/0003-2697(79)90580-3. [DOI] [PubMed] [Google Scholar]
- 26.Drachev S M, Drachev L A, Konstantinov A A, Semenov A Y, Skulachev V P, Arutjunjan M, Shulalov V A, Zaberezhnaya S M. Eur J Biochem. 1988;171:253–264. doi: 10.1111/j.1432-1033.1988.tb13784.x. [DOI] [PubMed] [Google Scholar]
- 27.Wrigglesworth J. Biochem J. 1984;217:715–719. doi: 10.1042/bj2170715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vygodina T V, Konstantinov A A. Ann NY Acad Sci. 1988;550:124–138. doi: 10.1111/j.1749-6632.1988.tb35329.x. [DOI] [PubMed] [Google Scholar]
- 29.Vygodina T, Konstantinov A. Biochim Biophys Acta. 1989;973:390–398. doi: 10.1016/s0005-2728(89)80380-9. [DOI] [PubMed] [Google Scholar]
- 30.Nilsson T. Proc Natl Acad Sci USA. 1992;89:6497–6501. doi: 10.1073/pnas.89.14.6497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zaslavsky D, Kaulen A, Smirnova I A. FEBS Lett. 1993;336:389–393. doi: 10.1016/0014-5793(93)80843-j. [DOI] [PubMed] [Google Scholar]
- 32.Zaslavsky D L, Smirnova I A, Siletsky S A, Kaulen A D, Millett F, Konstantinov A A. FEBS Lett. 1995;359:27–30. doi: 10.1016/0014-5793(94)01443-5. [DOI] [PubMed] [Google Scholar]
- 33.Shapleigh J P, Gennis R B. Mol Microbiol. 1992;6:635–642. doi: 10.1111/j.1365-2958.1992.tb01511.x. [DOI] [PubMed] [Google Scholar]
- 34.Vandeyar M A, Weiner M P, Hutton C J, Batt C A. Gene. 1988;65:129–133. doi: 10.1016/0378-1119(88)90425-8. [DOI] [PubMed] [Google Scholar]
- 35.Mitchell D M, Aasa R, Ådelroth P, Brzezinski P, Gennis R B, Malmström B G. FEBS Lett. 1995;374:371–374. doi: 10.1016/0014-5793(95)01149-9. [DOI] [PubMed] [Google Scholar]
- 36.Mitchell D M, Gennis R B. FEBS Lett. 1995;368:148–150. doi: 10.1016/0014-5793(95)00626-k. [DOI] [PubMed] [Google Scholar]
- 37.Hinkle P C. Methods Enzymol. 1979;55:751–776. doi: 10.1016/0076-6879(79)55083-6. [DOI] [PubMed] [Google Scholar]
- 38.Provencher S W. Biophys J. 1976;16:27–50. doi: 10.1016/S0006-3495(76)85660-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nicholls P, Hildebrandt V A. Biochem J. 1978;173:65–72. doi: 10.1042/bj1730065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cooper C E, Jünemann S, Ioannidis N, Wrigglesworth J M. Biochim Biophys Acta. 1993;1144:149–160. doi: 10.1016/0005-2728(93)90167-e. [DOI] [PubMed] [Google Scholar]
- 41.Cheesman M R, Greenwood C, Thomson A J. Adv Inorg Chem. 1991;36:201–255. [Google Scholar]
- 42.Fabian M, Palmer G. Biochemistry. 1995;34:13802–13810. doi: 10.1021/bi00042a011. [DOI] [PubMed] [Google Scholar]
- 43.Bickar D, Bonaventura J, Bonaventura C. Biochemistry. 1982;21:2661–2666. doi: 10.1021/bi00540a013. [DOI] [PubMed] [Google Scholar]
- 44.Rich P, Moody A J. In: Bioelectrochemistry: Principles and Practice. Graber P, Milazzo G, editors. Basel: Birkhäuser; 1997. pp. 419–456. [Google Scholar]
- 45.Vygodina T, Mitchell D, Pecoraro C, Gennis R, Konstantinov A. EBEC Reports. 1996;9:93. [Google Scholar]
- 46.Morgan J E, Verkhovsky M I, Wikström M. J Bioenerg Biomembr. 1994;26:599–608. doi: 10.1007/BF00831534. [DOI] [PubMed] [Google Scholar]
- 47.Rich P R. Aust J Plant Physiol. 1995;22:479–486. [Google Scholar]
- 48.Mitchell R, Rich P R. Biochim Biophys Acta. 1994;1186:19–26. doi: 10.1016/0005-2728(94)90130-9. [DOI] [PubMed] [Google Scholar]
- 49.Miki T, Orii Y. J Biochem. 1986;100:735–745. doi: 10.1093/oxfordjournals.jbchem.a121766. [DOI] [PubMed] [Google Scholar]
- 50.Vygodina T V, Capitanio N, Papa S, Konstantinov A A. FEBS Lett. 1997;412:405–409. doi: 10.1016/s0014-5793(97)00649-2. [DOI] [PubMed] [Google Scholar]
- 51.Baek H K, Van Wart H E. Biochemistry. 1989;28:5714–5719. doi: 10.1021/bi00440a003. [DOI] [PubMed] [Google Scholar]
- 52.Baek H K, Van Wart H E. J Am Chem Soc. 1992;114:718–725. [Google Scholar]
- 53.Wikström M, Krab K, Saraste M. Cytochrome Oxidase—A Synthesis. New York: Academic; 1981. [Google Scholar]
- 54.Blair D F, Witt S N, Chan S I. J Am Chem Soc. 1985;107:7389–7399. [Google Scholar]
- 55.Morgan J E, Verkhovsky M I, Wikström M. Biochemistry. 1996;35:12235–12240. doi: 10.1021/bi961634e. [DOI] [PubMed] [Google Scholar]
- 56.Weng L, Baker G M. Biochemistry. 1991;30:5727–5733. doi: 10.1021/bi00237a014. [DOI] [PubMed] [Google Scholar]
- 57.Wrigglesworth J M, Ioannidis N, Nicholls P. Ann NY Acad Sci. 1988;550:150–160. doi: 10.1111/j.1749-6632.1988.tb35331.x. [DOI] [PubMed] [Google Scholar]
- 58.Vygodina T V, Schmidmaier K, Konstantinov A A. Biol Membr (Russia) 1993;6(7):883–906. [Google Scholar]
- 59.Proshlyakov D A, Ogura T, Shinzawa-Itoh K, Yoshikawa S, Kitagawa T. Biochemistry. 1996;35:8580–8586. doi: 10.1021/bi952096t. [DOI] [PubMed] [Google Scholar]
- 60.Ogura T, Hirota S, Proshlyakov D A, Shinzawa-Itoh K, Yoshikawa S, Kitagawa T. J Am Chem Soc. 1996;118:5443–5449. [Google Scholar]
- 61.Varotsis C, Zhang Y, Appelman E H, Babcock G T. Proc Natl Acad Sci USA. 1993;90:237–241. doi: 10.1073/pnas.90.1.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Einarsd-ttir O. Biochim Biophys Acta. 1995;1229:129–147. doi: 10.1016/0005-2728(94)00196-c. [DOI] [PubMed] [Google Scholar]
- 63.Cheesman M R, Watmough N J, Gennis R B, Greenwood C, Thomson A J. Eur J Biochem. 1994;219:595–602. doi: 10.1111/j.1432-1033.1994.tb19975.x. [DOI] [PubMed] [Google Scholar]
- 64.Vygodina T V, Konstantinov A A. FEBS Lett. 1987;219:387–392. doi: 10.1016/0014-5793(87)80258-2. [DOI] [PubMed] [Google Scholar]
- 65.Proshlyakov D A, Ogura T, Shinzawa-Itoh K, Yoshikawa S, Kitagawa T. Biochemistry. 1996;35:76–82. doi: 10.1021/bi9511705. [DOI] [PubMed] [Google Scholar]
- 66.Han S, Ching Y-c, Rousseau D L. J Am Chem Soc. 1990;112:9445–9551. [Google Scholar]
- 67.Larsen R W, Li W, Copeland R A, Witt S N, Lou B-S, Chan S I, Ondrias M R. Biochemistry. 1990;29:10135–10140. doi: 10.1021/bi00495a018. [DOI] [PubMed] [Google Scholar]
- 68.Lauraeus M, Morgan J E, Wikström M. Biochemistry. 1993;32:2664–2670. doi: 10.1021/bi00061a026. [DOI] [PubMed] [Google Scholar]
- 69.Han S, Ching Y-C, Rousseau D L. Proc Natl Acad Sci USA. 1990;87:2491–2495. doi: 10.1073/pnas.87.7.2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Svensson M, Hallen S, Thomas J W, Lemieux L, Gennis R B, Nilsson T. Biochemistry. 1995;34:5252–5258. doi: 10.1021/bi00015a040. [DOI] [PubMed] [Google Scholar]
- 71.Verkhovsky M I, Morgan J E, Wikström M. Biochemistry. 1995;34:7483–7491. doi: 10.1021/bi00022a023. [DOI] [PubMed] [Google Scholar]