Abstract
Pairs of transcriptional activators in prokaryotes have been shown to activate transcription synergistically from promoters with two activator binding sites. In some cases, such synergistic effects result from cooperative binding, but in other cases each DNA-bound activator plays a direct role in the activation process by interacting simultaneously with separate surfaces of RNA polymerase. In such cases, each DNA-bound activator must possess a functional activating region, the surface that mediates the interaction with RNA polymerase. When transcriptional activation depends on two or more identical activators, it is not straightforward to test the requirement of each activator for a functional activating region. Here we describe a method for directing a mutationally altered activator to either one or the other binding site, and we demonstrate the use of this method to examine the mechanism of transcriptional activator synergy by the Escherichia coli cyclic AMP receptor protein (CRP) working at an artificial promoter bearing two CRP-binding sites.
Many transcriptional activators in prokaryotes bind to specific sites within or upstream of a target promoter and contact RNA polymerase (RNAP) (1, 2). Although some promoters can be controlled by a single activator, others are apparently regulated by multiple DNA-bound activators working together synergistically (reviewed in ref. 3). A classical mechanism that accounts for some examples of transcriptional activator synergy is cooperative DNA binding (4). In such cases, only one DNA-bound activator interacts directly with RNAP, and the other activator plays an indirect role by stabilizing the binding of the former to its recognition site. Other examples of transcriptional activator synergy do not involve direct interactions between the activators themselves; instead, an alternative mechanism has been proposed, namely that two (or more) activators contact separate surfaces of RNAP (reviewed in refs. 3 and 5; see also ref. 6). Several artificial promoters have been designed that can be activated by pairs of regulators (either identical or heterologous) working synergistically (7–9). In this study, we use a new method to examine the synergistic action of the Escherichia coli cyclic AMP receptor protein (CRP), working at an artificial promoter bearing two CRP recognition sites, and we show that each CRP dimer plays a direct role in the activation process.
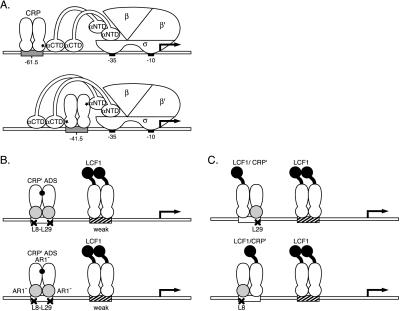
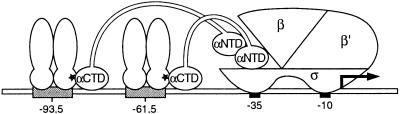
CRP is a two-domain protein that binds as a dimer to its specific recognition sites when complexed with cAMP (reviewed in ref. 10). Many well known promoters in E. coli (for example the lac promoter) are activated by a single DNA-bound CRP dimer. Such CRP-dependent promoters have been divided into two classes: Class I promoters bear a single CRP-recognition site centered approximately at position −61.5 (relative to the start point of transcription) or an integral number of turns of the DNA helix further upstream, and Class II promoters bear a single CRP-recognition site centered approximately at position −41.5 (reviewed in ref. 11). Two separate target sites on RNAP have been identified that can be contacted by CRP, one of which is contacted when CRP is working at a Class I promoter, and both of which are contacted when CRP is working at a Class II promoter (Fig. 1A). RNAP holoenzyme in E. coli consists of an enzymatic core with the subunit composition α2ββ′ and one of several alternative σ factors (the most abundant of which is σ70) that is responsible for promoter recognition (reviewed in ref. 12). The α subunit, which is the target of action of many transcriptional activators including CRP, consists of two independently folded domains, the α amino-terminal domain (α-NTD), which directs the assembly of RNAP, and the α carboxyl-terminal domain (α-CTD), a DNA-binding domain that is connected to the α-NTD by a flexible tether (13). When activating transcription from a Class I promoter, CRP uses an activating region (called AR1) in the promoter-proximal subunit (14) to contact the α-CTD (13). When activating transcription from a Class II promoter, CRP makes two contacts with RNAP by using AR1 in the promoter-distal subunit to contact the α-CTD (15) and a second activating region (called AR2) in the promoter-proximal subunit to contact the α-NTD (16).
Figure 1.
(A) Interactions of CRP with RNAP at Class I and Class II promoters. The specific contacts between AR1 of CRP and the α-CTD and AR2 of CRP and the α-NTD are indicated (*). (B) Two distinct species of CRP homodimers bound to test promoter. The test promoter depicted is that used in the experiment of Table 1. The CRP′ ADS homodimer (with or without the AR1− substitution) can bind to the symmetrically altered L8-L29 CRP-binding site. The LCF1 homodimer (the CRP moiety of which is wild type) binds to the low-affinity promoter-proximal site. (C) Oriented heterodimers experiment. The test promoters (used in the experiment of Table 3) each bear an asymmetrically altered CRP-binding site (L8 or L29) at the promoter-distal position. LCF1/CRP′ AR1− homodimers bind in an oriented fashion to these sites, whereas LCF1 homodimers bind to the low-affinity promoter-proximal site only.
Both artificial and natural promoters have been described that can be activated by two DNA-bound CRP dimers working together (7, 8, 17). In a previous study, we demonstrated that two CRP-binding sites located at Class I positions (−61.5 and −93.5) can function synergistically to mediate transcriptional activation from the lac promoter (plac) (7). We suggested that the observed synergy reflects simultaneous contacts between each DNA-bound CRP dimer and RNAP, and our goal here was to test this hypothesis. The analysis of transcriptional activation under circumstances where two (or more) identical activator binding sites are present is not straightforward. In principle, suitable activation-defective mutants (positive control mutants) can be used to determine whether an individual regulator bound at a particular site plays a direct or an indirect role in the activation process. The difficulty lies in directing the positive control mutant specifically to one or the other binding site. For this purpose, altered DNA-binding specificity mutants are required. Furthermore, with dimeric regulators there is an additional complication, namely that the wild-type and altered DNA-binding specificity mutant forms will assort randomly, forming both homodimeric and heterodimeric species. The formation of such hybrid DNA-binding specificity heterodimers has in fact been exploited in the design of an “oriented heterodimers” approach for analyzing the function of DNA-bound activators (14). Here we describe an alternative method that relies on the use of an altered dimerization specificity (ADS) mutant, which we define as a mutant that can form biologically active homodimers but cannot associate with the wild-type parent to form mutant/wild-type heterodimers (18). By constructing a CRP mutant that is altered in its dimerization specificity as well as in its DNA-binding specificity, we can direct a homodimeric CRP positive-control mutant to the desired DNA site. Both the oriented heterodimers approach and our ADS approach are generalizable, and we discuss considerations that might influence the choice of one or the other method.
Materials and Methods
Bacterial Strains.
E. coli strain JCB43Δcrp39 streptomycin resistant (strR) (F−, λ−, lacZ−, Δcrp39 strR) (9) was used in this study. Antibiotics were used at the following concentrations: tetracycline, 35 μg/ml; carbenicillin, 100 μg/ml; chloramphenicol, 35 μg/ml; streptomycin, 100 μg/ml; and kanamycin, 50 μg/ml.
Plasmids.
Expression vectors.
The pACYC184 derivative pAC LCF1 (18) expressed the λcI-CRP fusion (lcf1) gene, and lacIq. crp alleles were constitutively expressed from derivatives of pHA7E (18). Mutations were introduced into pHA7E with the PCR and confirmed by DNA sequencing. The ADS mutations result in the substitutions T10A and L113R (18). The CRP mutant called CRP′ bears substitution E181V (19, 20). Two positive control mutations were used resulting in substitutions H159L in AR1 of CRP (21) and H19L in AR2 (16). The plasmids pLR1ΔcI and pAD325 served as vector controls (18).
Reporter plasmids.
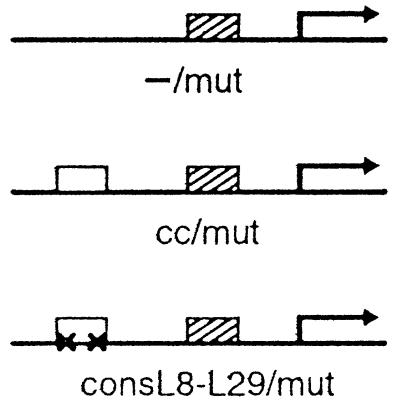
The reporter constructs used in the experiments of Tables 1 and 3 are derivatives of a lac promoter cloned into pRW50 (a low-copy vector also called p5A) (22), driving expression of a trpB∷lacZ fusion. CRP-binding sites were introduced upstream of this promoter. The plasmids p5Amut and p5Alac have a single CRP site centered at position −61.5 (from the start of transcription). The other reporter plasmids have two CRP sites, one at −93.5 and the other at −61.5 (e.g., p5Acc/mut, p5AconsL8-L29/mut). We used CRP-binding sites of varying affinities for CRP, listed from weakest to strongest: mut (7), lac, cc (23), and cons (24). The mutations L8 (G→A at position 7) and L29 (C→T at position 16) were introduced in either the cons, cc, or lac sites; they strongly reduce the binding of wild-type CRP, but have no effect on the binding of CRP′ (20). The upstream promoter sequences for each construct are provided as supplemental Fig. 4 (see www.pnas.org).
Table 1.
Effects of LCF1 and CRP variants on lacZ expression controlled by single- and two-site promoters
The test promoters shown directed expression of a trpB::lacZ fusion gene present on a low copy vector (see Materials and Methods). These plasmid-borne reporter constructs were introduced into strain JCB43Δcrp39 strR together with a pair of compatible plasmids directing the expression of LCF1 and a CRP′ ADS variant (or no CRP). β-Galactosidase assays were done in duplicate on several occasions, and the values shown are the averages from a single representative experiment. Duplicate measurements differed by <10%.
Table 3.
Oriented heterodimer analysis to assess the requirement for AR1 in each subunit of each DNA-bound CRP dimer
(A) Effects of wild-type and mutant LCF1/CRP heterodimers on lacZ expression controlled by test promoters bearing L8 and/or L29 mutations in the promoter-distal CRP-binding site. (B) Effects of wild-type and mutant LCF1/CRP heterodimers on lacZ expression controlled by test promoters bearing L8 and/or L29 mutations in the promoter-proximal CRP-binding site. The test promoters indicated directed expression of a trpB::lacZ fusion gene present on a low copy vector (see Materials and Methods). These plasmid-borne reporter constructs were introduced into strain JCB43Δcrp39 strR together with a pair of compatible plasmids directing the expression of LCF1 and either CRP′ or CRP′ AR1−. β-Galactosidase assays were done in duplicate on several occasions, and the values shown are the averages from a single representative experiment. Duplicate measurements differed by <10%.
Each test promoter was first assembled on plasmid pMH50Δlac (J. K. Joung, personal communication). Downstream of the CRP-binding sites, the promoters differ from the wild-type lac promoter by three mutations: (i) 2-bp substitutions (G→C and A→C at −2 and +2, respectively) that create a SalI site near +1; (ii) an A→G substitution at position −40 that creates a unique SmaI site; and (iii) replacement of the region between +1 and +67 with a synthetic BamHI linker, removing the lac operator and the first 11 codons of lacZ, preventing possible recombination between the test promoters and the lacZ portion of the trpB∷lacZ fusion. After confirmation by DNA sequencing, the promoter regions were excised on an EcoRI-HindIII restriction fragment and cloned into p5A.
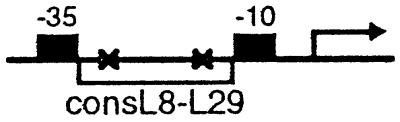
The reporter template used for the in vivo CRP-binding assay of Table 2 was constructed as follows. Two single-stranded oligonucleotides were annealed to form a double-stranded 40-mer encoding a synthetic promoter with a consensus L8-L29 CRP site between the −35 and −10 promoter hexamers. The promoter fragment was inserted upstream of the promoterless lacZ gene on plasmid pFW11-null (25). The resulting plasmid, pFW11 consL8-L29[−22], was transformed into CSH100, and the promoter-lacZ fusion recombined onto an F′ episome and mated into strain JCB43Δcrp39strR in one step (25) to construct the reporter strain.
Table 2.
Occupancy of symmetricaly altered CRP-binding site by CRP variants as measured by in vivo repression assay
| CRP species |
 β-galactosidase units |
|---|---|
| LCF1+ − | 2530 |
| LCF1 + CRPADS | 2420 |
| LCF1 + CRP′ ADS | 640 |
| LCF1 + CRP′ ADS AR1− | 680 |
| LCF1 + CRP′ ADS AR2− | 720 |
The indicated test promoter directed expression of the lacZ gene on an F′ episome. The F′ episome was introduced into strain JCB43Δcrp39 strR together with compatible plasmids directing expression of LCF1 and a CRP variant (or no CRP). The assays were performed in duplicate on three separate occasions, and the values shown are the averages from a single representative experiment. Duplicate measurements differed by <10%.
Electrophoretic Mobility-Shift Analysis.
Labeling of DNA fragment.
A 104-bp DNA fragment containing a consensus CRP-binding site was amplified from plasmid pMH-Cons/RM93 (J. K. Joung, personal communication) and internally labeled with 32P in a reaction containing 20 μCi of [α-32P] dTTP (3,000 Ci/mmol), 20 μM cold dTTP, and 200 μM each dATP, dCTP, and dGTP. The desired fragment was purified after electrophoresis.
Preparation of cell extracts.
Whole-cell extracts were made from strain JCB43Δcrp39 carrying p5Acc/lac, pAC-LCF1 or control plasmid pAD325, and the appropriate pHA7 CRP expression vector or control plasmid pLR1ΔcI. Extracts were prepared as described (18). Total protein concentrations were measured with the Bio-Rad Protein Assay Kit.
Electrophoretic mobility-shift assays.
Assays were performed as described (18).
Results
Use of ADS Mutant to Test Requirement for Functional Activating Region in Upstream CRP Dimer.
To test the hypothesis that each CRP dimer bound to our artificial plac derivative (with CRP-binding sites centered at positions −61.5 and −93.5) interacts directly with RNAP, we first sought to determine whether the stimulatory effect of the upstream CRP dimer depends on either of CRP’s previously characterized activating regions (AR1 or AR2). To answer this question, it was necessary to direct an activation-proficient CRP dimer to the downstream binding site and an activation-defective CRP dimer to the upstream site. We accomplished this through the combined use of two crp mutations, one that confers an altered DNA-binding specificity and the other (actually a pair of mutations) that confers an altered dimerization specificity. Our strategy enabled us to introduce into cells two CRP variants designed to form two functionally distinct homodimeric species with different DNA-binding specificities.
We first constructed a mutant form of CRP altered in both its dimerization and DNA-binding specificities. This CRP mutant bears two amino acid substitutions (T10A and L113R) that confer the ADS phenotype (18) and one amino acid substitution (E181V) that confers a relaxed DNA-binding specificity (20). The CRP (E181V) mutant (called CRP′) was selected for its ability to bind to CRP-recognition sites bearing either the L29 or the symmetrically related L8 mutation (19). Whereas wild-type CRP homodimers do not bind to CRP sites bearing either or both of these mutations, CRP′ homodimers bind equally well to the wild-type and mutant sites (albeit ≈10-fold more weakly than wild-type CRP binds to a wild-type site) (20). Thus, when overexpressed in a cell that also contains wild-type CRP, the CRP mutant bearing all three substitutions (which we call CRP′ ADS) is predicted to form mutant homodimers that are capable of binding to a mutationally altered binding site (L8-L29) that cannot be recognized by wild-type homodimers (Fig. 1B).
We then constructed three test promoters (see Table 1). The first bears a single low-affinity (mut) CRP-binding site at the downstream position, and the second and third bear the low-affinity downstream site and a high-affinity (cc) or a symmetrically altered (consL8-L29) site, respectively, at the upstream position. [We have previously shown that the use of a low-affinity CRP-binding site at the promoter-proximal position, which works relatively poorly on its own, increases the magnitude of the stimulatory effect of a second CRP-binding site provided upstream (7)]. Each of these test promoters was fused to the lacZ gene on a low-copy plasmid vector and introduced into a Δcrp host strain. Our experimental design involved the introduction of two species of CRP into each of the resulting reporter strains: a version with wild-type binding and dimerization specificities and the CRP′ ADS mutant. Because we wished to be able to distinguish between the two CRP species on the basis of size in in vitro assays (see below), we provided wild-type CRP in the form of a λcI-CRP fusion protein (called LCF1) (18) (Fig. 1B). LCF1 consists of the DNA-binding domain and linker region of λcI (residues 1–132) connected to residues 2–209 of CRP. This bifunctional fusion protein (which can bind to both λ operators and CRP-recognition sites) activates transcription efficiently from CRP-dependent promoters. LCF1 and CRP′ ADS were encoded by compatible plasmid vectors and the CRP′ADS mutant either did or did not bear an additional amino acid substitution in either AR1 (H159L) or AR2 (H19L).
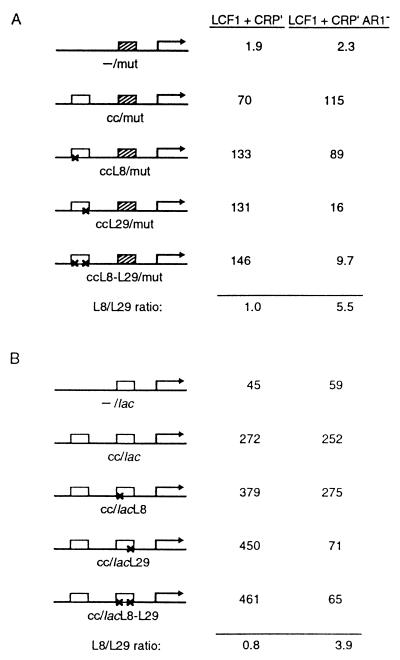
Table 1 shows the results of β-galactosidase assays performed to quantify the effects of the indicated CRP proteins on transcription from each of the three test promoters. In the presence of LCF1 only, transcription from the promoter bearing the single weak CRP-binding site was relatively inefficient, and the addition of a high-affinity (cc) site further upstream resulted in a large (55-fold) increase in transcription. In contrast, addition of the symmetrically altered L8-L29 site at the upstream position resulted in a <4-fold increase in transcription. However, in the presence of both LCF1 and CRP′ ADS, the addition of either the wild-type or the L8-L29 site at the upstream position resulted in a large increase in transcription (58- and 65-fold, respectively), indicating that the CRP′ ADS mutant binds efficiently to the symmetrically altered site. Inactivation of AR1 by the introduction of the H159L substitution into CRP′ ADS abrogated the stimulatory effect of the L8-L29 site, suggesting that a functional AR1 is required either on one or both subunits of the upstream dimer. Finally, inactivation of AR2 by the introduction of the H19L substitution into CRP′ ADS had no inhibitory effect, indicating that this activating region is not involved in the stimulatory effect of CRP bound at the upstream site. We note that introduction of the H159L substitution into CRP′ ADS did not abrogate the stimulatory effect of the wild-type site at the upstream position (compare line 2, columns 2 and 3); this control excludes the possibility that CRP′ ADS is restricting access of LCF1 to the downstream CRP site under the conditions of our experiment. We confirmed that LCF1 and CRP′ ADS are functioning together to stimulate the production of correctly initiated transcripts by performing primer extension analysis of RNA isolated from each of the strains assayed (data not shown).
The conclusion that the CRP dimer bound at the upstream site requires a functional AR1 on at least one subunit to exert its stimulatory effect on transcription depends on the demonstration that CRP′ ADS [H159L] is competent to bind DNA and that CRP′ ADS and CRP′ ADS [H159L] are present in the cells at comparable levels. To address these points, we performed an in vivo repression assay by using a specially designed test promoter bearing the consL8-L29 CRP-recognition site centered at position −22.5 between its −10 and −35 hexamers (called pconsL8-L29[−22]) (see Table 2). The binding of CRP′ derivatives to the mutant CRP-recognition site on this template is predicted to repress transcription from the test promoter; previous experiments with similarly constructed test promoters bearing various protein-binding sites between the −10 and −35 hexamers have established that the magnitude of repression is a sensitive indicator of the intracellular concentration of the cognate DNA-binding protein (25). A Δcrp repression strain was created that harbored an F′ episome bearing pconsL8-L29[-22] fused to the lacZ gene. We introduced into this reporter strain each of the plasmid combinations (encoding LCF1 and the CRP variants) assayed in the experiment of Table 1 as well as two additional control combinations and performed β-galactosidase assays (Table 2). In the presence of plasmid-encoded LCF1 with or without a plasmid-encoded form of CRP with wild-type DNA-binding specificity, the lacZ reporter gene was efficiently transcribed, as expected. Transcription was repressed 5-fold in the presence of LCF1 and each of the CRP′ ADS variants. These results rule out the possibility that the failure of CRP′ ADS [H159L] to exert a strong stimulatory effect on transcription when bound at the consL8-L29 site (Table 1, line 3) reflects a decrease in binding-site occupancy.
Use of Oriented Heterodimers to Test AR1 Requirement in Each Subunit of the Upstream CRP Dimer.
Next, we sought to determine whether the CRP dimer bound at the upstream site on a two-site template requires a functional AR1 on one or both subunits to exert its stimulatory effect on transcription. To do this, we took advantage of the oriented heterodimers approach first described by Zhou et al. (14). This approach is based on the fact that heterodimers composed of a wild-type CRP subunit and a CRP′ subunit bind in an oriented fashion to asymmetric CRP-recognition sites bearing either an L8 or an L29 half-site. It is therefore possible to direct a mutationally altered subunit to either one or the other half-site (Fig. 1C).
A set of five test promoters was used to carry out this experiment (see Table 3A). Each of these bears the low-affinity CRP-binding site at the downstream position. The four two-site templates bear either a high-affinity (cc) site at the upstream position, an asymmetric L8 site (ccL8), an asymmetric L29 site (ccL29), or a symmetrically altered L8-L29 site (ccL8-L29). As before, each of these test promoters was fused to the lacZ gene on a low-copy vector and introduced into a Δcrp host strain. For this experiment, we introduced into the resulting set of reporter strains compatible plasmid vectors encoding LCF1 (which bears a wild-type CRP moiety) and CRP′ (with or without the H159L substitution).
Table 3A shows the results of β-galactosidase assays performed to quantify the effects of the indicated CRP proteins on transcription from each of the five test promoters. In the presence of LCF1 and the activation-proficient form of CRP′, transcription from the promoter bearing the single weak site was relatively inefficient, and addition of a second site at the upstream position resulted in a large (37- to 70-fold) increase irrespective of the presence of either the L8 or the L29 mutation. In particular, the same level of transcription was observed in the ccL8/mut and the ccL29/mut strains. However, a different pattern of results was obtained when LCF1 and the activation-defective form of CRP′ were introduced into these cells. In this case, the addition of the ccL8 site upstream of the weak site resulted in a large (39-fold) increase in transcription, but the addition of the ccL29 site resulted in a much smaller increase, and the addition of the symmetrically altered ccL8-L29 site resulted in a still smaller increase. Primer extension analysis confirmed that each of the test promoters directed the synthesis of correctly initiated transcripts in the presence of each combination of CRP variants (data not shown). A control experiment revealed that the asymmetrically altered upstream sites bound LCF1 homodimers detectably but weakly; in the presence of LCF1 only, addition of the cc site upstream of the mut site resulted in a 55-fold increase in transcription, whereas addition of the ccL8 or the ccL29 site resulted in a 5.0-fold and a 7.6-fold increase, respectively (data not shown).
From these data, we conclude that the CRP dimer bound at the upstream site requires a functional AR1 in its promoter-proximal subunit only. In particular, comparison of the values obtained with the ccL8/mut and the ccL29/mut templates (Table 3A, lines 3 and 4) indicates that transcription is significantly reduced when the activation-defective CRP′[H159L] subunit is bound to the promoter-proximal half-site.
Inspection of Table 3A (line 5) suggests that under the conditions of these assays, the symmetrically altered binding site is occupied primarily by CRP′AR1− homodimers and not by LCF1/CRP′AR1− heterodimers (which would be activation proficient when bound in a nonoriented fashion). This is not surprising, because CRP′ is in significant excess over LCF1 in these cells, as determined by Western blot analysis (data not shown). We note that under these conditions, therefore, it would have been possible to compare the effects of an activation-proficient and an activation-defective CRP′ homodimer at the upstream site without the use of an ADS mutant. Nevertheless, the interpretation of such data is less straightforward because three possible dimeric species must be considered.
Use of Oriented Heterodimers to Test AR1 Requirement in Each Subunit of the Downstream CRP Dimer.
We also used oriented heterodimers to investigate the requirement for AR1 in each subunit of the CRP dimer bound at the downstream binding site. A set of five test promoters was used for this purpose (see Table 3B). In this case, the single-site template bears a higher-affinity binding site (that associated with the wild-type lac promoter) at the downstream position. The two-site templates bear a high-affinity site (cc) at the upstream position and either the lac site or a mutationally altered variant thereof at the downstream position. The salient finding is that in cells containing LCF1 and CRP′, high levels of transcription were observed whether the downstream binding site bears the L8 or the L29 mutation (or both), whereas in cells containing LCF1 and CRP′[H159L], the level of transcription was reduced significantly when the downstream binding site bears the L29 mutation (Table 3B). Again, primer extension analysis confirmed that each of the test promoters directed the synthesis of correctly initiated transcripts in the presence of each combination of CRP variants (data not shown). From these data, we conclude that the CRP dimer bound at the downstream site requires a functional AR1 in its promoter-proximal subunit only.
Thus, when functioning synergistically, both the CRP dimer bound at position −61.5 and that bound at position −93.5 require a functional AR1 in the promoter-proximal subunit only. These findings are in accord with what has been shown previously for CRP dimers working individually at Class I promoters (14) .
Direct Visualization of Predicted Homo- and Heterodimeric CRP Species.
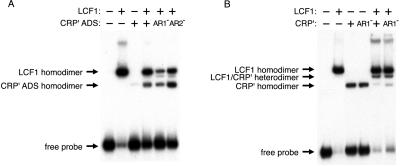
The design of the experiment presented in Table 1 is predicated on our ability to provide in cells two different forms of CRP that preferentially self associate, forming distinct homodimeric species. On the other hand, the design of the experiments presented in Table 3 depends on the formation of heterodimers comprised of two different forms of CRP. To obtain direct confirmation that our test strains contained the predicted homo- and heterodimeric species, we performed electrophoretic mobility shift analysis using cell extracts. The use of the LCF1 protein in our experiments permitted us to distinguish among the three possible species (the two homodimers and the heterodimer) (18). A DNA probe bearing the consensus CRP-binding site was chosen because this site can bind each of the possible homo- and heterodimeric species. Fig. 2A shows the results obtained with extracts prepared from cells containing LCF1 and one or another version of the CRP′ ADS mutant. We incubated extracts containing the indicated proteins with double-stranded radiolabeled DNA probe and subjected the samples to gel electrophoresis. Each extract contained the expected homodimeric species. Fig. 2B shows the results obtained with extracts prepared from cells containing LCF1 and one or another version of CRP′. In this case, we detected both LCF1 homodimers and the predicted heterodimeric species.
Figure 2.
Electrophoretic mobility-shift assays to detect presence of homodimeric and heterodimeric species in cell extracts containing LCF1 and one or another CRP variant. An internally labeled consensus CRP-binding site was incubated with cell extracts from strain JCB43Δcrp39 strR containing the indicated proteins. Cells were grown in the absence of isopropyl-d-thiogalactoside (IPTG) (A) or in the presence of 5 μM IPTG (B). The amount of total cellular protein in each reaction was 5 μg.
We note that these experiments do not permit us to estimate the relative amounts of the homo- and heterodimeric species in each strain. This is because the E181V substitution results in a reduction in overall binding affinity such that wild-type CRP homodimers (and presumably LCF1 homodimers) bind with a higher affinity to the consensus binding site present on the probe than either CRP′ homodimers or heterodimers bearing one CRP′ subunit. Therefore, the results of the electrophoretic mobility shift analysis are expected to provide an overestimate of the amount of LCF1 homodimer relative to the other CRP species.
Discussion
We have investigated the mechanism by which two DNA-bound CRP dimers cooperate to activate transcription from an artificial promoter bearing CRP-binding sites at positions −61.5 and −93.5 (both Class I positions). Using two complementary approaches, we have demonstrated a requirement for a functional activating region (AR1) in the upstream as well as the downstream DNA-bound CRP dimer. These findings are consistent with our previous suggestion that the two dimers function synergistically because they make simultaneous contact with RNAP (7).
As mentioned in the Introduction, the contact site on RNAP for AR1 is located in the α-CTD (for reviews, see refs. 13, 26). When bound at a Class I promoter, CRP uses AR1 of its promoter-proximal subunit to contact the CTD of one α subunit; this interaction stabilizes the binding of the α-CTD to the DNA (reviewed in ref. 13) (see Fig. 1A). At our artificial promoter, we have demonstrated a requirement for a functional AR1 in the promoter-proximal subunit of each DNA-bound CRP dimer. Accordingly, we suggest that under these circumstances both of the α-CTDs in the RNAP holoenzyme serve as activation targets, each one providing a contact site for one DNA-bound CRP dimer. Specifically, we propose that one α-CTD is bound to the DNA downstream of the promoter-proximal CRP dimer, and the other is bound between the two CRP dimers when the activated complex forms at our artificial promoter (see Fig. 3). Additional experiments (e.g., footprint analysis) will be required to determine whether the α-CTDs are bound to the DNA as shown.
Figure 3.
Model depicting proposed interactions between DNA-bound CRP dimers and RNAP. Each CRP dimer uses its promoter-proximal subunit to contact the CTD of one α subunit.
Pairs of CRP dimers have also been shown to function synergistically at artificial promoters bearing CRP-recognition sites at a Class II position (−41.5) and one of a number of different Class I positions (ranging from −74.5 to −102.5) (8, 27). In this case, both AR1 and AR2 are required in the downstream CRP dimer, and AR1 only is required in the upstream dimer. Using the oriented heterodimers approach, Busby and colleagues demonstrated that AR1 is required in the promoter-distal subunit of the downstream CRP dimer and (except in one case) in the promoter-proximal subunit of the upstream CRP dimer; consistent with this proposal, they provided evidence that the α-CTDs bind to the DNA between the DNA-bound CRP dimers (ref. 27; see also ref. 28). There was an interesting exception to this pattern: when the upstream CRP-recognition site was centered at position −74.5, AR1 was required in the promoter-distal subunit of the upstream CRP dimer and, accordingly, the two α-CTDs were separated, with one bound between the CRP dimers and the other bound upstream of the promoter-distal CRP dimer. Our artificial promoter provides a second example in which the two α-CTDs are evidently separated; in both cases, the spacing between the two CRP-recognition sites (32–33 bp) is presumably too close to permit the binding of both α-CTDs between the DNA-bound CRP dimers. The α-CTD is known to be flexibly tethered to the α-NTD, and these studies indicate that together the α-CTDs comprise a particularly accessible pair of activation targets that can be contacted by transcriptional activators positioned in a variety of different configurations. Nevertheless, there is a requirement that the DNA-bound activators be appropriately phased on approximately the same side of the DNA helix (ref. 27 and J. K. Joung and A.H., unpublished data).
Kinetic analysis of the effect of CRP on transcription from the lac promoter has revealed that CRP increases the binding constant (KB) for the initial association of RNAP with double-stranded promoter DNA (29). On the other hand, when bound at a Class II promoter, CRP affects both the initial binding step and the rate constant (kf) for the isomerization of the closed complex to the locally melted open complex by using AR1 and AR2, respectively, to exert these effects (16). On the basis of these observations, we suggest that each CRP dimer bound at our artificial two-site promoter affects KB only. The observed synergy can be explained by postulating that the two DNA-bound CRP dimers bind cooperatively to their targets on RNAP (the α-CTDs).
Synergistic activation has also been demonstrated with heterologous activator pairs that contact different targets on RNAP (9, 17). For example, an artificial promoter was designed bearing binding sites for the bacteriophage λ cI protein (centered at position −42) and CRP (centered at position −93.5) (9). In this case, genetic evidence suggests that the λ cI protein contacts its natural target, the σ subunit, and CRP contacts the α-CTD. A natural example is provided by the E. coli ansB promoter, which depends on both CRP and the related anaerobic regulator FNR. Here, too, genetic evidence suggests that FNR (bound at position −41.5) contacts the σ subunit, and CRP (bound at position −91.5) contacts the α-CTD (17). It is likely that the α-CTD provides a particularly accessible target for an activator bound at an upstream position, and in all of the cases so far examined, the α-CTD provides the contact site for the upstream activator. The demonstration that E. coli RNAP can respond simultaneously to pairs of activators bound in a variety of different configurations provides a mechanism by which multiple regulatory inputs can be integrated. This mechanism has been invoked previously to explain the synergistic action of eukaryotic transcription factors working in both homologous and heterologous combinations (30). Thus, studies in both prokaryotic and eukaryotic systems suggest a common mechanism for generating sensitive transcriptional switches that are triggered by the combined action of multiple regulatory factors.
To dissect the functional requirements of each DNA-bound CRP dimer on our artificial two-site template, we used two genetic methods. The first is new and should be generally applicable in cases requiring the analysis of two or more identical regulators that are dimeric. It relies on the use of a mutant altered in both its DNA-binding and dimerization specificities. We previously described a genetic approach for obtaining ADS mutants of dimeric proteins (18). By constructing a CRP mutant with altered DNA-binding and dimerization specificities, we could direct a CRP homodimer lacking a functional activating region (either AR1 or AR2) to a particular binding site (the upstream site). The use of an ADS mutant prevents the formation of heterodimers composed of one activation-proficient and one activation-deficient subunit. However, we note that under the particular conditions of the experiment presented in Table 3 (CRP′ in excess over LCF1), we would have been able to assess the effect of an activation-defective CRP′ homodimer at the promoter-distal site without specifically restricting heterodimer formation through the use of an ADS mutant (see Results).
The second method we used was designed specifically to exploit the formation of mixed-function heterodimers, which bind in an oriented fashion to appropriate asymmetrically mutated recognition sites (14). The oriented heterodimers approach permits the functional analysis of each subunit of each dimer and therefore can provide more information than is provided by the examination of homodimers only. However, there are circumstances under which the data obtained from an oriented heterodimer experiment alone would be inconclusive. If a functional activating region in either subunit of the dimer could suffice to mediate a productive interaction with RNAP, then the heterodimer (composed of one activation-proficient and one activation-deficient subunit) would be functional no matter what the orientation of the asymmetrically altered recognition site. In such a case, it would be necessary to examine the effect of an activation-deficient homodimer bound at the relevant recognition site to assess the requirement for a functional activating region.
The ADS approach might also provide a technical advantage in some situations. Depending on what sort of altered DNA-binding specificity mutant is available and how well the wild-type protein discriminates between the wild-type and the mutated recognition sites, it may be important to maximize the discrimination by using a symmetrically mutated site that would bind wild-type homodimers with a lower affinity than would recognition sites altered in only one half-site. Finally, we envision the application of these methods to the analysis of transcriptional activation in eukaryotic as well as prokaryotic cells. Such an analysis would generally require the introduction of mutationally altered activators into cells containing an endogenous wild-type activator. The use of an ADS mutant would simplify the analysis when it may be difficult to control the relative concentrations of the two species, a parameter that is critical for the interpretation of an oriented heterodimers experiment.
Supplementary Material
Acknowledgments
We are grateful to J. K. Joung, G. King, and S. Dove for advice and discussions, and to M. Ptashne for comments on the manuscript. This work was supported by National Institutes of Health grant GM44025 and by an established investigatorship from the American Heart Association (A.H.).
Abbreviations
- RNAP
RNA polymerase
- CRP
cyclic AMP receptor protein
- AR
activating region
- NTD
amino-terminal domain
- ADS
altered dimerization specificity
- CTD
carboxyl-terminal domain
- strR
streptomycin resistant
- CPR′
CRP (E181V) mutant
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Busby S, Ebright R H. Cell. 1994;79:743–746. doi: 10.1016/0092-8674(94)90063-9. [DOI] [PubMed] [Google Scholar]
- 2.Hochschild A, Dove S L. Cell. 1998;92:597–600. doi: 10.1016/s0092-8674(00)81126-5. [DOI] [PubMed] [Google Scholar]
- 3.Hochschild A, Joung J K. In: Nucleic Acids and Molecular Biology. Eckstein F, Lilley D M J, editors. Vol. 11. Berlin: Springer; 1997. pp. 101–114. [Google Scholar]
- 4.Ptashne M. A Genetic Switch. Cambridge, MA: Blackwell Scientific; 1992. [Google Scholar]
- 5.Savery N, Belyaeva T, Wing H, Busby S. Biochem Soc Trans. 1996;24:351–353. doi: 10.1042/bst0240351. [DOI] [PubMed] [Google Scholar]
- 6.Carey M, Lin Y-S, Green M R, Ptashne M. Nature (London) 1990;345:361–364. doi: 10.1038/345361a0. [DOI] [PubMed] [Google Scholar]
- 7.Joung J K, Le L U, Hochschild A. Proc Natl Acad Sci USA. 1993;90:3083–3087. doi: 10.1073/pnas.90.7.3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Busby S, West D, Lawes M, Webster C, Ishihama A, Kolb A. J Mol Biol. 1994;241:341–352. doi: 10.1006/jmbi.1994.1511. [DOI] [PubMed] [Google Scholar]
- 9.Joung J K, Koepp D M, Hochschild A. Science. 1994;265:1863–1866. doi: 10.1126/science.8091212. [DOI] [PubMed] [Google Scholar]
- 10.Kolb A, Busby S, Buc H, Garges S, Adhya S. Annu Rev Biochem. 1993;62:749–795. doi: 10.1146/annurev.bi.62.070193.003533. [DOI] [PubMed] [Google Scholar]
- 11.Busby S, Ebright R H. Mol Microbiol. 1997;23:853–859. doi: 10.1046/j.1365-2958.1997.2771641.x. [DOI] [PubMed] [Google Scholar]
- 12.Gross C A, Chan C L, Lonetto M A. Philos Trans R Soc London B. 1996;351:475–482. doi: 10.1098/rstb.1996.0045. [DOI] [PubMed] [Google Scholar]
- 13.Ebright R H, Busby S. Curr Opin Genet Dev. 1995;5:197–203. doi: 10.1016/0959-437x(95)80008-5. [DOI] [PubMed] [Google Scholar]
- 14.Zhou Y, Busby S, Ebright R H. Cell. 1993;73:375–379. doi: 10.1016/0092-8674(93)90236-j. [DOI] [PubMed] [Google Scholar]
- 15.Savery N J, Lloyd G S, Kainz M, Gaal T, Ross W, Ebright R H, Gourse R L, Busby S J. EMBO J. 1998;17:3439–3447. doi: 10.1093/emboj/17.12.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Niu W, Kim Y, Tau G, Heyduk T, Ebright R H. Cell. 1996;87:1123–1134. doi: 10.1016/s0092-8674(00)81806-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scott S, Busby S, Beacham I. Mol Microbiol. 1995;18:521–531. doi: 10.1111/j.1365-2958.1995.mmi_18030521.x. [DOI] [PubMed] [Google Scholar]
- 18.Joung J K, Chung E H, King G, Yu C, Hirsh A S, Hochschild A. Genes Dev. 1995;9:2986–2996. doi: 10.1101/gad.9.23.2986. [DOI] [PubMed] [Google Scholar]
- 19.Ebright R H, Cossart P, Gicquel-Sanzey B, Beckwith J. Nature (London) 1984;311:232–235. doi: 10.1038/311232a0. [DOI] [PubMed] [Google Scholar]
- 20.Ebright R H, Kolb A, Buc H, Kunkel T A, Krakow J S, Beckwith J. Proc Natl Acad Sci USA. 1987;84:6083–6087. doi: 10.1073/pnas.84.17.6083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou Y, Zhang X, Ebright R H. Proc Natl Acad Sci USA. 1993;90:6081–6085. doi: 10.1073/pnas.90.13.6081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lodge J, Fear J, Busby S, Gunasekaran P, Kamini N R. FEMS Microbiol Lett. 1992;74:271–276. doi: 10.1016/0378-1097(92)90441-p. [DOI] [PubMed] [Google Scholar]
- 23.Gaston K, Bell A, Kolb A, Buc H, Busby S. Cell. 1990;62:733–743. doi: 10.1016/0092-8674(90)90118-x. [DOI] [PubMed] [Google Scholar]
- 24.Ebright R H, Ebright Y W, Gunasekera A. Nucleic Acids Res. 1989;17:10295–10305. doi: 10.1093/nar/17.24.10295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Whipple F W. Nucleic Acids Res. 1998;26:3700–3706. doi: 10.1093/nar/26.16.3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ishihama A. J Bacteriol. 1993;175:2483–2489. doi: 10.1128/jb.175.9.2483-2489.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Belyaeva T A, Rhodius V A, Webster C L, Busby S J. J Mol Biol. 1998;277:789–804. doi: 10.1006/jmbi.1998.1666. [DOI] [PubMed] [Google Scholar]
- 28.Murakami K, Owens J T, Belyaeva T A, Meares C F, Busby S J, Ishihama A. Proc Natl Acad Sci USA. 1997;94:11274–11278. doi: 10.1073/pnas.94.21.11274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Malan T P, Kolb A, Buc H, McClure W R. J Mol Biol. 1984;180:881–909. doi: 10.1016/0022-2836(84)90262-6. [DOI] [PubMed] [Google Scholar]
- 30.Ptashne M. Nature (London) 1988;335:683–689. doi: 10.1038/335683a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.