Abstract
The phosphorelay signal transduction system activates developmental transcription in sporulation of Bacillus subtilis by phosphorylation of aspartyl residues of the Spo0F and Spo0A response regulators. The phosphorylation level of these response regulators is determined by the opposing activities of protein kinases and protein aspartate phosphatases that interpret positive and negative signals for development in a signal integration circuit. The RapA protein aspartate phosphatase of the phosphorelay is regulated by a peptide that directly inhibits its activity. This peptide is proteolytically processed from an inactive pre-inhibitor protein encoded in the phrA gene. The pre-inhibitor is cleaved by the protein export apparatus to a putative pro-inhibitor that is further processed to the active inhibitor peptide and internalized by the oligopeptide permease. This export–import circuit is postulated to be a mechanism for timing phosphatase activity where the processing enzymes regulate the rate of formation of the active inhibitor. The processing events may, in turn, be controlled by a regulatory hierarchy. Chromosome sequencing has revealed several other phosphatase–prepeptide gene pairs in B. subtilis, suggesting that the use of this mechanism may be widespread in signal transduction.
The initiation of sporulation of Bacillus subtilis is a complex process responsive to the state of the metabolism, the environment, and the cell cycle of an organism that has to make the decision whether to cease vegetative growth and direct its energies toward development. The earliest indication of the activation of development is the transcription of genes under the control of the Spo0A transcription factor (1). This protein requires phosphorylation to be active and the level of Spo0A phosphorylation is regulated by the phosphorelay signal transduction system (2). The phosphorelay is a more complex version of the typical two-component signal transduction system used to interpret a variety of metabolic, environmental and cell cycle signals in bacteria, fungi and plants (3–8). At least two kinases, KinA and KinB, respond to some, as yet unknown, sporulation-inducing signals and promote the phosphorylation of the Spo0F response regulator. Spo0F∼P is the substrate for a phosphotransferase, Spo0B, that transfers the phosphate to Spo0A. The level of phosphorylated Spo0A would reflect the activation of the kinases were it not for a series of phosphatases that specifically dephosphorylate either Spo0A∼P or Spo0F∼P. Spo0E is a phosphatase specifically active on Spo0A∼P, but the signals regulating its activity are unknown (9). Spo0F∼P is the target for RapA and RapB, belonging to the Rap family of phosphatases. RapA and RapB are known to be differentially activated by physiological processes alternative to sporulation (10). These phosphatases function to drain the phosphorelay, lower the Spo0A∼P level in the cell, and prevent sporulation. Therefore the phosphatases allow signals contrary to sporulation to impact on the phosphorelay and its output product Spo0A∼P. Thus, the phosphorelay acts as a signal integration circuit permitting a variety of signals both positive and negative to influence the decision to initiate cellular development via competition between kinases and phosphatases (9–12).
The chemical nature of the signals acting on the kinases and phosphatases of the phosphorelay is still obscure. It has been proposed that extracellular factors act as positive signals of sporulation (13). The extracellular competence factor of B. subtilis has been shown to be a peptide (14). Extracellular peptides are signals for competence of Streptococcus pneumoniae, for virulence in Staphyloccus aureus, and for plasmid conjugation in Enterococcus faecalis and have been implicated in control of many processes in eukaryotes (15–18). In a previous study, it was shown that the Rap family of phosphatases for the phosphorelay are cotranscribed with a gene whose protein is ultimately cleaved to produce an exported peptide that regulates phosphatase function (19).
The phosphatase activity of RapA is modulated by a small protein, PhrA, encoded by a gene on the same transcript as RapA (19, 20). PhrA is a 44-amino acid protein whose C-terminal half is exported from the cell and then presumably reimported by the oligopeptide transport system (Opp) (19, 21, 22). Deletion of the phrA gene causes uncontrolled RapA phosphatase activity with the consequence that sporulation is not initiated. Using synthetic peptides in an earlier study, it was shown that the sporulation deficiency caused by the deregulated RapA activity of a phrA mutant can be complemented in vivo using peptides comprising the last six or more C-terminal residues of PhrA. However, none of the synthetic peptides that were active in vivo showed inhibition of RapA phosphatase in vitro, leaving open the question of the mechanism by which the peptides regulated phosphatase activity (19). This report shows that the PhrA C-terminal pentapeptide is the active agent that inhibits RapA phosphatase activity in vitro and that the amino acid sequence of the peptide determines target specificity.
MATERIALS AND METHODS
Bacterial Strains and Growth Conditions.
The B. subtilis strains used in this study are the wild-type JH642 (trpC2, phe-1) and the phrA deletion strain JH12954 (trpC2, phe-1, phrA:: pJM9233, cat) (13). Sporulation assays were carried out by the Sterlini and Mandelstam resuspension method as described (23). After resuspension at OD600 0.7, peptides were added at the concentrations indicated in the figures. Cells were grown for 24 h and then treated with CHCl3 before plating on Schaeffer’s sporulation medium (24).
Protein Expression and Purification.
PCR-generated fragments containing the RapA, RapA892, and RapB coding sequences were cloned in the pET16b expression vector (Novagen), thereby adding 10 histidine codons to the 5′ end of the genes. The modified proteins were expressed in Escherichia coli BL21 (DE3) (Novagen) by induction at OD600 0.7 with 2 mM isopropyl β-d-thiogalactopyranoside for 3 h at 37°C. Proteins were purified in 50 mM Tris⋅HCl (pH 8), 50 mM KCl, 5 mM 2-mercaptoethanol, 1 mM phenylmethylsulfonyl fluoride, and 5% glycerol. Elution from the Ni-NTA agarose column (Qiagen, Chatsworth, CA) was obtained with a gradient of imidazole ranging from 0 to 200 mM. Proteins were then dialyzed in 25 mM Tris⋅HCl (pH 8), 1 mM DTT, and 5% glycerol. Purification of KinA and Spo0F was as described by Grimsley, et al. (25).
In Vitro Assay Conditions.
Spo0F (10 μM) was phosphorylated by KinA (0.1 μM) in reaction mixtures containing 50 mM N-(2-hydroxyethyl) piperazine-N′-(3-propanesulfonic acid) (EPPS) (pH 8.5), 20 mM MgCl2, 100 μM EDTA, 5% glycerol, 1.8 mM ATP, and 1.8 μCi/ml of [γ-32P]ATP (>6,000 Ci/mmol; 1 Ci = 37 GBq). Reactions were incubated at room temperature for 1 h. Rap proteins (5 μM) or premixed (5 min at 0°C) Rap proteins and Phr peptides were then added to 20 μl aliquots of phosphorylation mix and further incubated for 30 min at room temperature. Reactions were stopped by the addition of loading buffer and 10 μl were run on 15% SDS/glycine/polyacrylamide gels at constant current (25 mAmp) for 1.5 h. Gels were immediately exposed to Kodak X-OMAT RP films for 1.5 h at −80°C with an intensifying screen and then exposed to PhosphorImager (Molecular Dynamics) for quantitative analysis.
Synthetic Phr peptides were resuspended in 50 mM K-phosphate buffer (pH 7.8). Peptide concentrations were determined by amino acid analysis.
RESULTS
The PhrA Peptide Specifically Inhibits RapA Phosphatase Activity.
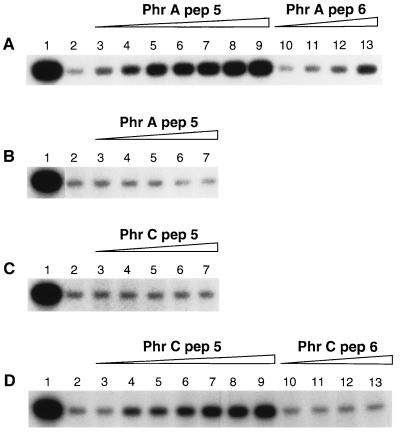
To assess the effect of residue length as a possible determinant in peptide inhibition, a chemically synthesized PhrA peptide comprising the last five amino acids at the C-terminal end of the PhrA protein (PhrApep5: ARNQT) was tested in vitro on RapA activity. Spo0F was phosphorylated by KinA and [γ-32P]ATP and then exposed to dephosphorylation by RapA. In the presence of increasing concentrations of PhrApep5, dephosphorylation of Spo0F∼P by RapA was inhibited (Fig. 1A, lanes 3–9). PhrApep5 at 200 μM versus RapA at 5 μM resulted in about 50% inhibition of phosphatase activity. The PhrA hexapeptide (PhrApep6: AARNQT) was also tested and some activity was observed, but no more than 10% inhibition of RapA dephosphorylation activity was detected when used at 200 μM concentration (Fig. 1A, lanes 10–13). To determine if the inhibitory activity was specific for RapA, the PhrA pentapeptide was tested in vitro on another member of the Rap family, RapB. RapB-specific dephosphorylation of Spo0F∼P was not inhibited by PhrApep5 at any of the concentrations inhibiting RapA (Fig. 1B). To test whether RapA could be inhibited by other related pentapeptides, we synthesized a peptide whose sequence corresponds to the last five amino acids of the PhrC protein. PhrC belongs to the Phr family of secreted peptides associated with the phosphatases of the Rap family and has been isolated from culture supernatants as a pentapeptide (26). The PhrC pentapeptide (PhrCpep5: ERGMT) was tested in vitro against the RapA protein, but no inhibition of phosphatase activity was observed (Fig. 1C). PhrCpep5, along with a PhrC hexapeptide as a control (PhrCpep6: TERGMT) was also tested on RapB. RapB is an exception among the Rap family of phosphatases since its structural gene is not followed by an active peptide-encoding gene (12). RapB was slightly sensitive to PhrCpep5, and no more than 25% inhibition of phosphatase activity was obtained with a 40-fold excess of peptide (Fig. 1D, lanes 3–9). RapB was not inhibited by PhrCpep6 (Fig. 1D, lanes 10–13). To test whether the inhibition of RapA by PhrA was affected by the presence of KinA, fast protein liquid chromatography-purified Spo0F∼P was incubated with the phosphatase in the presence or absence of increasing concentrations of PhrA pentapeptide. The results were comparable to the ones obtained in unpurified conditions, indicating that PhrA activity is independent of KinA (data not shown). Thus, the PhrA pentapeptide is the active molecule specifically inhibiting RapA phosphatase activity and there is no cross-reactivity with the PhrC peptide.
Figure 1.
Specificity of Rap phosphatase inhibition by Phr pentapeptides. Inhibition of Rap phosphatase activity by the Phr peptides results in decreased dephosphorylation of Spo0F∼P. Reactions in lane 1 are the control containing KinA (0.1 μM) and Spo0F (10 μM) only, whereas lane 2 shows the level of dephosphorylation obtained by RapA (5 μM) (A and C) or RapB (5 μM) (B and D). Reactions were carried out as described. (A) Increasing concentrations of PhrApep5 result in decreased dephosphorylation of Spo0F∼P by RapA. PhrApep6 also shows some inhibition but at extremely reduced efficacy. PhrApep5 was added at 10, 20, 30, 40, 50, 100, and 200 μM, in lanes 3–9, respectively; PhrApep6 was added at 10, 50, 100, and 200 μM in lanes 10–13, respectively. (B) PhrApep5 does not inhibit RapB activity. PhrApep5 was added at 10, 30, 50, 100, and 200 μM in lanes 3–7, respectively. (C) PhrCpep5 does not inhibit RapA activity. PhrCpep5 was added at increasing concentrations as in B. (D) PhrCpep5 moderately inhibits Spo0F∼P dephosphorylation by RapB. Increasing concentrations of PhrCpep5 (lanes 3–9) and PhrCpep6 (lanes 10–13) were added as in A.
PhrA pep5 Activity in Vivo.
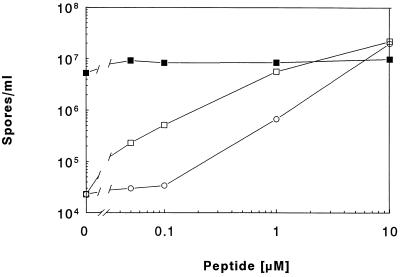
The potential for inhibition of RapA activity by the synthetic peptides was also tested in vivo by means of a sporulation assay. It was previously shown that the sporulation-deficient phenotype of a phrA deletion mutant (strain JH12954) could be complemented by exogenously provided synthetic peptides corresponding in sequence to the C terminus of PhrA (19). Increasing concentrations of PhrApep5 or PhrApep6 were added to JH12954 cells and the sporulation efficiency was tested. As shown in Fig. 2, PhrApep5 and PhrApep6 were equally efficient in complementing the phrA mutation, and sporulation proficiencies comparable to the wild-type strain were achieved by the addition of peptides at 10 μM concentration. Perhaps an aminopeptidase activity converts PhrApep6 to the pentapeptide in vivo accounting for their equal sporulation-inducing efficiencies. The PhrC peptides were also tested on strain JH12954, but no significant increase in sporulation was observed (data not shown). No effect was also observed when PhrApep5 was provided to a culture of the wild-type strain JH642 (Fig. 2).
Figure 2.
In vivo complementation of a phrA mutant (JH12954) (open symbols) by exogenously provided Phr peptides. Sporulation assays were performed by the Sterlini and Mandelstam resuspension method (23). Peptides were added at the concentration indicated. □, PhrApep5; ○, PhrApep6; ▪, JH642.
The RapA892 Phosphatase Is Insensitive to PhrA Inhibition.
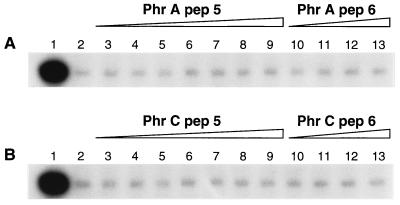
The spo0L892 allele of the rapA gene was originally identified as missense mutation P259L (10). Because this protein retains the same enzymatic activity as the wild-type form in in vitro dephosphorylation assays of Spo0F∼P, we predicted that the sporulation-deficient phenotype in vivo was due to the inability to respond to modulation carried out by an effector molecule. In the same assay conditions used to test the sensitivity of RapA wild-type protein to peptide inhibition, the phosphatase activity of RapA892 was unaffected by the PhrA penta- and hexapeptides (Fig. 3A). Insensitivity was shown toward the PhrC penta and hexapeptides as well (Fig. 3B). The insensitivity of RapA892 to inhibition by PhrA in vitro is thus consistent with the phenotype caused by the spo0L892 mutation in vivo.
Figure 3.
The spo0L892 mutant form of RapA (P259L) is insensitive to PhrA peptide inhibition. Phosphorylation of Spo0F (10 μM) was carried out as in Fig. 1 (lane 1) and as described in Materials and Methods. Purified RapA892 was used at 5 μM final concentration and added to the reaction in the absence (lane 2) or presence of pentapeptides at 10, 20, 30, 40, 50, 100, and 200 μM final concentration (lanes 3–9, respectively) or exapeptide at 10, 50, 100, and 200 μM (lanes 10–13, respectively). (A) PhrApep5 (lanes 3–9) or PhrApep6 (lanes 10–13). (B) PhrCpep5 (lanes 3–9) or PhrCpep6 (lanes 10–13).
Target Specificity Is Encoded in the Pentapeptide Amino Acid Sequence.
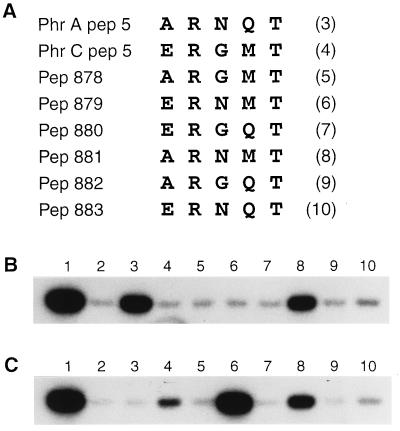
The observation that PhrApep5 is active on RapA but inactive on RapB, and that PhrCpep5 inhibits RapB but not RapA, raises the question of what mechanism rules the specificity of target recognition. A peptide structure-based mechanism seems unlikely because pentapeptides are unlikely to have a stable quaternary structure. Therefore, it seems more probable that target specificity is amino acid sequence-dependent. In an attempt to decipher the five amino acid code embedded in the sequence of the pentapeptides, a series of modified peptides was synthesized and tested for specificity versus RapA or RapB. PhrApep5 and PhrCpep5 have in common an arginine residue at position 2, from the N-terminal end, and a threonine residue at position 5. These residues are present in the same location in the majority of phr genes defined as small ORFs located downstream of genes encoding other members of the Rap family of phosphatases (12). Hence, the amino acid residues at the remaining three positions are candidates for being the determinants of peptide specificity. Modified PhrA or PhrC pentapeptides were designed to contain a single amino acid substitution at position 1, 3, or 4, replacing the residue on one peptide with the corresponding one on the other peptide (Fig. 4A). The ability of these modified peptides to inhibit phosphatase activity was tested on RapA and RapB and the results are shown in Fig. 4 B and C. The modified forms of PhrCpep5 (pep 878, 879, and 880) did not acquire any ability to inhibit RapA (Fig. 4B, lanes 5–7). However, the G to N modification in position 3 resulted in a peptide (pep 879) more highly active in inhibiting RapB than the PhrCpep5 itself (Fig. 4C, lane 6 versus lane 4; 85% and 10% inhibition, respectively). The amino acid replacements in pep 878 and 880, on the contrary, resulted in loss of activity toward RapB (Fig. 4C, lanes 5 and 7). The Q-to-M modification at position 4 of PhrApep5 generated a peptide (pep 881) retaining partial activity toward RapA (60% compared with PhrApep5) (Fig. 4B, lane 8) but also 3-fold more active than PhrCpep5 in inhibiting RapB (Fig. 4C, lane 8). Pep 882 (N to G modification in position 3 of PhrApep5) was inactive while pep 883 (A to E change at position 1) slightly inhibited both RapA and RapB (lane 10, Fig. 4 B and C, respectively). The modified peptides were also tested on RapA892, but no inhibitory activity was detected (data not shown).
Figure 4.
Phr peptide sequence-dependent specificity for target recognition. (A) Amino acid sequence of PhrA and PhrC pentapeptides and their modified forms obtained by single amino acid replacement at position 1, 3, and 4 of PhrApep5 with the corresponding residue of PhrCpep5 and vice versa. The ability of the mutant peptides to inhibit RapA (B) or RapB (C) phosphatase activity (5 μM) on Spo0F∼P (10 μM) was tested in the standard reaction conditions described in Materials and Methods. Peptides were all used at 200 μM final concentration. The control level of Spo0F phosphorylation is shown in lane 1 whereas the dephosphorylation of Spo0F∼P by RapA or RapB is in lane 2. Lanes 3–10 contain the peptides in the order indicated by the numbers in parentheses.
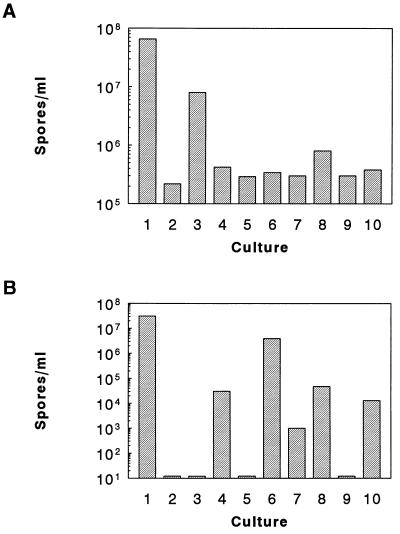
Peptide activities in vivo were tested by means of complementation assays of the sporulation-deficient phenotype of the phrA mutant strain JH12954. When the modified peptides (pep 878–883) were individually added to a culture of JH12954 at a concentration of 10 μM, sporulation was restored by pep 881 at ≈50% of efficiency compared with PhrApep5. The remaining peptides, however, showed some low level of activity ranging between 1% and 6% compared with the activity of PhrApep5 (Fig. 5A). These in vivo results could be accounted for by incidental activity of the peptides on Rap phosphatases acting on signal transduction systems that indirectly affect sporulation. To test in vivo peptide activity on RapB, we took advantage of the observation that a multicopy vector carrying the rapB coding gene produces a stage 0 sporulation defect in otherwise wild-type cells. When the wild-type strain JH642 carrying the multicopy rapB plasmid pIPB213A (10) was grown in the presence of 10 μM pentapeptides, sporulation was partially restored by peptide 879 (13% of spores compared with the control strain carrying only the vector without the rapB gene). Less than 1% of spores were obtained with pep 880, 881, and 883 as well as PhrCpep5, whereas PhrApep5, pep 878, and pep 882 were entirely inactive (Fig. 5B). This analysis shows that single amino acid changes at the variable residues within PhrApep5 and PhrCpep5 remarkably affect peptide activity and specificity toward the target phosphatase in vitro and in vivo.
Figure 5.
In vivo assay of modified pentapeptides. Sporulation assays were carried out with the resuspension method of Sterlini and Mandelstam (23). (A) Strain JH12954 carrying a deletion of the PhrA coding gene was resuspended in the sporulation medium in the presence of peptides at 10 μM final concentration. The wild-type level of sporulation is given by strain JH642 (lane 1), whereas strain JH12954 grown in the absence of peptides is shown in lane 2. PhrApep5, PhrCpep5, pep 878, pep 879, pep 880, pep 881, pep 882, and pep 883 were added to the cultures represented in lanes 3–10, respectively, and the number of spores per ml is shown by the bars. (B) Strain JH642 carrying the rapB multicopy plasmid pIPB213A was grown in the presence of 5 μg/ml chloramphenicol (Cm). The wild-type control strain was JH642 carrying the multicopy vector pBS19 (lane 1). Lane 2 shows the level of sporulation of JH642/pIPB213A in the absence of peptides, whereas lanes 3–10 show the number of spores per ml obtained by the addition of peptides (10 μM) as in A.
DISCUSSION
The experiments described in this communication were designed to shed light on the regulation of RapA phosphatase activity by the product of the phrA gene. Fig. 6 schematically depicts what we know and what we believe about the regulation of this system. The rapA operon is induced by the ComA regulator whose role is little understood but necessary for the induction of the competent state. We believe that competence and sporulation are processes that cannot occur simultaneously in the same cell and in order to ensure this, ComA induces the rapA operon to prevent sporulation. The RapA phosphatase is clearly an inhibitor of sporulation in vivo and functions by dephosphorylating the Spo0F∼P response regulator (10). The phrA gene is known to regulate the RapA phosphatase but not by regulating transcription of the rapA operon (19). Through complementation studies it was shown that the last six residues of the phrA gene product could function as a regulator of RapA phosphatase in vivo (19). The studies presented here show that the regulation is accomplished by direct inhibition of the phosphatase activity by the last five residues of the phrA gene product.
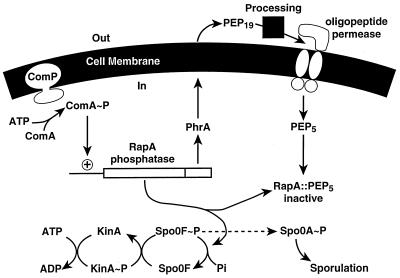
Figure 6.
Modulation of the phosphorelay by the PhrA peptide export–import circuit that regulates the RapA phosphatase activity. RapA is induced by ComA∼P and prevents sporulation during competence development by dephosphorylating Spo0F∼P. PhrA is first exported, processed, and then imported by the oligopeptide transport system as a pentapeptide. The PhrA pentapeptide directly inhibits RapA activity, thereby allowing sporulation to initiate.
There are several reasons to believe that the journey of the phrA gene product to a final pentapeptide inhibitor involves export from the cytoplasm and subsequent importation back to the cytoplasm. The PhrA sequence consists of an amino-terminal signal sequence and signal peptidase consensus site followed by 19–20 C-terminal residues suggesting that the C-terminal residues are exported (19, 27). The oligopeptide permease (Opp) is required for the system to function (19, 21, 22). An opp mutant is sporulation deficient and may be restored to sporulation proficiency by deletion of the rapA gene suggesting that the opp mutation disturbs the regulation of RapA (19). Suppression of the sporulation deficiency of the opp mutant may also be accomplished by the Y13S mutation of Spo0F which prevents the RapA phosphatase from dephosphorylating Spo0F∼P (10, 19). Thus, the oligopeptide permease is deeply implicated in the PhrA regulatory circuit. Unless this permease has some function other than import of peptides, the data are best interpreted by an obligation to import the final inhibitor from the outside.
The formation of the active peptide inhibitor appears to require a sequence of proteolytic events. The primary protein product of the phrA gene may be viewed as a pre-inhibitor peptide translated from the same transcript as the RapA phosphatase. The pre-inhibitor is inactive allowing the RapA phosphatase to function. The pre-inhibitor peptide is exported to the outside of the cell membrane, presumably in pro-inhibitor form consisting of the C-terminal 19–20 amino acids of PhrA. To produce the 5-amino acid active inhibitor, the pro-inhibitor must be subject to at least one more proteolytic cleavage. This cleavage is likely to occur on the outside surface of the membrane because the uptake of the active peptide inhibitor depends on the oligopeptide permease, OppA, the structure of which is designed for peptides up to 5 amino acids long and is very unlikely to bind and transport a peptide the size of the pro-inhibitor (28). Internalization of the inhibitor peptide results in inhibition of the phosphatase and suppression of the inhibition of sporulation. Because the stoichiometry of inhibition of RapA by chemically synthesized PhrA peptide in vitro significantly differs from unity, it remains possible that the natural peptide is subject to further posttranslational modification before it is fully active in vivo.
The rationale for the cell developing an export–import control circuit for regulating the RapA phosphatase is open to speculation. Does the peptide or one of its precursor forms carry out an extra cytoplasmic function? We originally thought that the peptide could act as a quorum sensor (19), but there are other, equally likely possibilities that fit all the available data. The peptide may affect some periplasmic enzymes or surface proteins and function as an extra cytoplasmic regulator by acting as a communication pathway from the inside to the outside. Alternatively, the processing of the pre-inhibitor to the final inhibitor pentapeptide could represent a means for extra cytoplasmic events to regulate the length of time the phosphatase is active. If ComA-induced events required both internal and external processes to be completed, the induction of sporulation would be delayed until the production and importation of the peptide that inhibits the phosphatase and allows the phosphorelay to induce sporulation. The processing of the pre-inhibitor would therefore be a control circuit with the processing enzymes acting as checkpoints. The removal of signal peptides occurs by signal peptidases that are redundantly encoded by at least three genes in B. subtilis (29, 30). Genetic studies have revealed that the capacity for protein secretion in this organism may be modulated through temporally controlled expression of signal peptidase genes and the specificity for secretion is dependent on the particular signal peptidase being expressed (29). Thus, the processing of the pre-inhibitor to pro-inhibitor may be dependent on the presence of a specific signal peptidase whose control affects the time interval that the phosphatase is active. Interestingly, the sipS gene for one signal peptidase is regulated by the DegS–DegU signal transduction system that is highly associated with later events in the competence pathway (29, 31).
Regardless of the ultimate role of the peptide, its journey through this export–import circuit represents a timing device that controls the length of time the RapA phosphatase is active. Transcription of the rapA operon has the immediate effect of blocking sporulation and this block is not released until the PhrA pentapeptide is reimported. Therefore any delay in importation of the peptide for extracellular or communication purposes will prolong the block in sporulation by increasing the time interval the phosphatase is active. There is no evidence that the peptide is a pheromone or quorum sensor for sporulation (32); in fact, in the rapA–phrA mutant, the cells sporulate perfectly well, even better than a wild-type strain. Furthermore, in this mutant competence development is severely reduced in length and efficiency. In contrast, in a mutant carrying a RapA protein insensitive to the PhrA peptide, sporulation is inhibited and competence is prolonged in time (unpublished results). These observations are consistent with the idea that PhrA is required for timing coordination of the competence and sporulation events.
Although attempts to isolate the PhrA peptide have never been made our previous studies showed that little physiologically detectable peptide is found in culture supernatants of wild-type strains compared with opp deficient strains (19). We believe that the PhrA peptide is only exported to the cell wall and not necessarily secreted into the medium and its activity is a self-regulatory mechanism for timing intracellular developmental events.
Phr specificity for target recognition is dependent upon the amino acid sequence of the peptides. Single amino acid substitutions can severely affect pentapeptide activity and specificity. This suggests that the active site for peptide binding on the phosphatase must be shaped to accommodate only specific peptides on the basis of potentially favorable interactions with the ligand. The highly conserved R and T residues at positions 2 and 5 of the Phr peptides may represent the sites of common interaction which define an orientation for bound peptides to Rap phosphatases. Amino acids at positions 1, 3, and 4 of Phr peptides are the determinants of specificity presumably through the interactions established by their side chains. This is reminiscent of the binding of phosphorylated peptides to the Src SH2 domain of Src-related tyrosine kinases (33).
It is clear that phosphorylation-dependent signal transduction systems are modulated by the competition of kinases and phosphatases to regulate the output of the system. The functions of peptides are well known in high eukaryotes and now they are assuming an increasingly important role in the control of signal transduction in lower eukaryotes and bacteria as well. In the scheme of cell development in B. subtilis, peptides serve as direct regulators of phosphatases in signal transduction.
Acknowledgments
I would like to thank Dr. Xiao Zhen Zhou of the Hoch Laboratory for purifying the wild-type RapA and RapB proteins. This work was supported by Grant GM19416 from the National Institute of General Medical Sciences, National Institutes of Health, awarded to Dr. James A. Hoch.
ABBREVIATION
- Opp
oligopeptide transport system
References
- 1.Hoch J A. Annu Rev Microbiol. 1993;47:441–465. doi: 10.1146/annurev.mi.47.100193.002301. [DOI] [PubMed] [Google Scholar]
- 2.Burbulys D, Trach K A, Hoch J A. Cell. 1991;64:545–552. doi: 10.1016/0092-8674(91)90238-t. [DOI] [PubMed] [Google Scholar]
- 3.Parkinson J S, Kofoid E C. Annu Rev Genet. 1992;26:71–112. doi: 10.1146/annurev.ge.26.120192.000443. [DOI] [PubMed] [Google Scholar]
- 4.Alex L A, Borkovich K A, Simon M I. Proc Natl Acad Sci USA. 1996;93:3416–3421. doi: 10.1073/pnas.93.8.3416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kakimoto T. Science. 1996;274:982–985. doi: 10.1126/science.274.5289.982. [DOI] [PubMed] [Google Scholar]
- 6.Chang C. Trends Biochem Sci. 1996;21:129–133. [PubMed] [Google Scholar]
- 7.Posas F, Wurgler-Murphy S M, Maeda T, Witten E A, Thai T C, Saito J. Cell. 1996;86:865–875. doi: 10.1016/s0092-8674(00)80162-2. [DOI] [PubMed] [Google Scholar]
- 8.Hoch J A, Silhavy T J, editors. Two-Component Signal Transduction. Washington, DC: Am. Soc. for Microbiol.; 1995. [Google Scholar]
- 9.Ohlsen K L, Grimsley J K, Hoch J A. Proc Natl Acad Sci USA. 1994;91:1756–1760. doi: 10.1073/pnas.91.5.1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perego M, Hanstein C G, Welsh K M, Djavakhishvili T, Glaser P, Hoch J A. Cell. 1994;79:1047–1055. doi: 10.1016/0092-8674(94)90035-3. [DOI] [PubMed] [Google Scholar]
- 11.Perego M, Hoch J A. Trends Genet. 1996;12:97–101. doi: 10.1016/0168-9525(96)81420-x. [DOI] [PubMed] [Google Scholar]
- 12.Perego M, Glaser P, Hoch J A. Mol Microbiol. 1996;19:1151–1157. doi: 10.1111/j.1365-2958.1996.tb02460.x. [DOI] [PubMed] [Google Scholar]
- 13.Grossman A D, Losick R. Proc Natl Acad Sci USA. 1988;85:4369–4373. doi: 10.1073/pnas.85.12.4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Magnuson R, Solomon J, Grossman A D. Cell. 1994;77:207–216. doi: 10.1016/0092-8674(94)90313-1. [DOI] [PubMed] [Google Scholar]
- 15.Havarstein L S, Coomaraswamy G, Morrison D. Proc Natl Acad Sci USA. 1995;92:11140–11144. doi: 10.1073/pnas.92.24.11140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Balaban N, Novick R P. Proc Natl Acad Sci USA. 1995;92:1619–1623. doi: 10.1073/pnas.92.5.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leonard B A B, Podbielski A, Hedberg P J, Dunny G M. Proc Natl Acad Sci USA. 1996;93:260–264. doi: 10.1073/pnas.93.1.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van de Sande K, Pawlowski K, Czaja I, Wieneke U, Schell J, Schmidt J, Walden R, Matvienko M, Wellink J, van Kammen A, Franssen H, Bisseling T. Science. 1996;273:370–373. doi: 10.1126/science.273.5273.370. [DOI] [PubMed] [Google Scholar]
- 19.Perego M, Hoch J A. Proc Natl Acad Sci USA. 1996;93:1549–1553. doi: 10.1073/pnas.93.4.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mueller J P, Bukusoglu G, Sonenshein A L. J Bacteriol. 1992;174:4361–4373. doi: 10.1128/jb.174.13.4361-4373.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Perego M, Higgins C F, Pearce S R, Gallagher M P, Hoch J A. Mol Microbiol. 1991;5:173–185. doi: 10.1111/j.1365-2958.1991.tb01838.x. [DOI] [PubMed] [Google Scholar]
- 22.Rudner D Z, Ladeaux J R, Breton K, Grossman A D. J Bacteriol. 1991;173:1388–1398. doi: 10.1128/jb.173.4.1388-1398.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nicholson W L, Setlow P. In: Molecular Biological Methods for Bacillus. Harwood C R, Cutting S M, editors. Chichester, U.K.: Wiley; 1990. pp. 391–450. [Google Scholar]
- 24.Schaeffer P, Millet J, Aubert J. Proc Natl Acad Sci USA. 1965;54:701–711. doi: 10.1073/pnas.54.3.704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grimsley J K, Tjalkens R B, Strauch M A, Bird T H, Spiegelman G B, Hostomsky Z, Whiteley J M, Hoch J A. J Biol Chem. 1994;269:16977–16982. [PubMed] [Google Scholar]
- 26.Solomon J M, Lazazzera B A, Grossman A D. Genes Dev. 1996;10:2014–2024. doi: 10.1101/gad.10.16.2014. [DOI] [PubMed] [Google Scholar]
- 27.von Heijne G. Nucleic Acids Res. 1986;14:4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tame J R H, Murshudov G N, Dodson E J, Neil T K, Dodson G G, Higgins C F, Wilkinson A J. Science. 1994;264:1578–1581. doi: 10.1126/science.8202710. [DOI] [PubMed] [Google Scholar]
- 29.Bolhuis A, Sorokin A, Azevedo V, Ehrlich S D, Braun P G, de Jong A, Venema G, Bron S, Van Dijl J M. Mol Microbiol. 1996;22:605–618. doi: 10.1046/j.1365-2958.1996.d01-4676.x. [DOI] [PubMed] [Google Scholar]
- 30.Akagawa E, Kurita K, Sugawara T, Nakamura K, Kasahara Y, Ogasawara N, Yamane K. Microbiology. 1995;141:3241–3245. doi: 10.1099/13500872-141-12-3241. [DOI] [PubMed] [Google Scholar]
- 31.Msadek T, Kunst F, Rapoport G. In: Two-Component Signal Transduction. Hoch J A, Silhavy T J, editors. Washington, DC: Am. Soc. Microbiol.; 1995. pp. 447–471. [Google Scholar]
- 32.Swift S, Throup J P, Williams P, Salmond G P C, Stewart G S A B. Trends Biochem Sci. 1996;21:214–219. [PubMed] [Google Scholar]
- 33.Waksman G, Shoelson S E, Pant N, Cowburn D, Kuriyan J. Cell. 1993;72:779–790. doi: 10.1016/0092-8674(93)90405-f. [DOI] [PubMed] [Google Scholar]