Abstract
The “parallel-up” packing in cellulose Iα and Iβ unit cells was experimentally demonstrated by a combination of direct-staining the reducing ends of cellulose chains and microdiffraction-tilting electron crystallographic analysis. Microdiffraction investigation of nascent bacterial cellulose microfibrils showed that the reducing end of the growing cellulose chains points away from the bacterium, and this provides direct evidence that polymerization by the cellulose synthase takes place at the nonreducing end of the growing cellulose chains. This mechanism is likely to be valid also for a number of processive glycosyltransferases such as chitin synthases, hyaluronan synthases, and proteins involved in the synthesis of nodulation factor backbones.
Keywords: reducing-end staining, microdiffraction/cellulose Iα, cellulose Iβ/chain polarity
The polarity of cellulose chains in a microfibril was debated for decades before two groups independently proved the parallel packing by electron microscopic methods. One involved the silver-labeling of the reducing ends of microfibrils (1), while the other was based on the unidirectional degradation of cellulose microfibrils by a cellobiohydrolase (2). In both studies, the cellulose from Valonia was used because of the high crystallinity and large lateral dimension of the microfibrils. Later, the silver-labeling technique was applied to bacterial cellulose and showed the same parallel packing (3). These microscopic analyses confirmed earlier crystallographic proposals that the most probable mode of packing in the unit cell was parallel (4–6).
The current knowledge on the crystal structure of cellulose is that the native cellulose is a composite of two distinct crystalline phases called Iα and Iβ (7, 8) corresponding to triclinic and monoclinic unit cells, respectively (9). The existence of the Iα and Iβ structures within a microfibril is another confirmation of the parallel packing, because the triclinic unit accepts only one single chain in a unit cell. Furthermore, the fact that Iα and Iβ coexist in a microfibril suggests that the chains in Iα-rich as well as in Iβ-rich cellulose are parallel. Although the parallel packing of the chains in a cellulose microfibril or a unit cell is now firmly established, the molecular directionality of the chains with respect to the unit cells is not known.
Directionality of cellulose chains in a unit cell is frequently defined according to Gardner and Blackwell (4). There are two types of parallel packing, namely, parallel up and parallel down. The parallel-up structure implies that the z coordinate of the O5 atom is greater than that of C5. Two parallel models with opposite molecular directionality thus have been proposed (4) and critically evaluated (10). Molecular dynamics studies recently have suggested that the parallel-up structure was most probable for both cellulose Iα and Iβ (11).
The first aim of the current research was to determine experimentally the chain directionality in a unit cell by using electron crystallography in conjunction with the reducing end staining technique. Once established, the determination of the directionality of cellulose chains can be determined simply by recording a series of microdiffraction diagrams with precise tilts. This technique was applied to ascertain the molecular directionality in nascent bacterial cellulose microfibrils and used to determine at which end of the growing cellulose chains polymerization takes place.
MATERIALS AND METHODS
Preparation of Microcrystalline Suspensions of Cellulose.
The cellulosic cell wall from Cladophora sp., harvested from a sea bed at Chikura, Chiba, Japan, was purified by two cycles of alkali and bleaching treatments. The sample was immersed in 1% KOH at room temperature for 12 hr, then soaked in 0.3% NaClO2 at 70°C for 2 hr buffered at pH 4.9 in acetate buffer. A portion of the purified cell wall material was subjected to hydrothermal annealing, which transforms Iα-rich cellulose into Iβ-dominant type (12).The sample was sealed in a glass vial with aqueous 0.1 N NaOH and heated at 260°C for 30 min. The annealed cell walls were neutralized and washed by centrifugation with distilled water and dialyzed overnight. Both initial and annealed cell wall samples then were hydrolyzed in 40% H2SO4 at room temperature for 48 hr with continuous stirring. The resultant microcrystalline suspension was neutralized by successive centrifugations with distilled water and finally dialyzed overnight. Finally, a microcrystalline suspension from purified Cladophora cell walls was prepared by hydrolysis with 3 N HCl at 95°C for 3 days with continuous stirring. The suspension was neutralized and dialyzed as above.
Selective Enzymatic Degradation by Cellobiohydrolase II.
Recombinant cellobiohydrolase II from Humicola insolens (Novo–Nordisk, Bagsvaerd, Denmark) was used to degrade cellulose microcrystals from their nonreducing ends. Enzymatic hydrolyses were conducted with 1 mg/ml cellulose and 200 μg/ml enzyme in 50 mM of phosphate buffer (pH 7.0) at 40°C for 24 hr. The samples were centrifuged and washed first with 1% NaOH to remove enzymes and then thoroughly washed by three successive centrifugations with distilled water.
Silver Labeling of Reducing Ends.
Initial-, hydrothermally annealed-, and enzymatically degraded Cladophora cellulose microcrystals were stained by a modified protocol of Kuga and Brown (3). The suspension of cellulose microcrystals (1 ml) was mixed with 5 ml of 1% thiosemicarbazide in aqueous 5% acetic acid. After incubation at 65°C for 90 min, the suspension was washed by centrifugation. The sample was resuspended in 1 ml of water and mixed with 5 ml of 1.6% sodium borate with 0.8% silver proteinate (Merck). This mixture was kept in the dark at room temperature for 1 h and washed by centrifugation. The final step was the enhancement of the electron density of the thiosemicarbazide-silver proteinate-treated reducing ends. This was done by mixing 1 ml of sample with 5 ml of silver ammonia solution in a nitric acid-cleaned glass vial for 6–9 min at 95°C until the sample turned brownish.
Production of Bacterial Cellulose on Grids.
Acetobacter aceti strain AJ12368 was used. The culture medium was prepared by dissolving 50 g of sucrose, 5 g of yeast extract, 5 g of (NH4)SO4, 3 g of KH2PO4, and 0.05 g of MgSO4 7H2O in 1 liter of water, and the pH was adjusted to 5.0 by adding a drop of HCl. After 5–7 days of incubation in a flask at 28°C, the newly formed cellulosic pellicle was squeezed in a test tube to release the cells, and a drop of the cell suspension was deposited on a carbon-coated grid. The grid was maintained in a moistened Petri dish for at least 1 hr, briefly washed with distilled water and air-dried.
Electron Microscopy.
All the micrographs and diffraction diagrams were taken with a JEOL-2000EXII operated at 100 kV and recorded on Mitsubishi MEM film. The diffraction contrast imaging in the bright field mode was used to visualize the sample without further contrast enhancement such as negative staining or shadowing. The images were taken at 5,000×–10,000× under low-dose exposure with the use of Minimum Dose System (JEOL).
The diffraction diagrams were obtained by microdiffraction mode. A small condenser aperture of 20 μm was inserted, and the first condenser lens was fully overfocused to achieve an electron probe of approximately 100 nm. The samples were observed at 2,500× under extremely low-dose conditions with a help of image intensifier (Fiber Optics Coupled TV, Gatan). When a microcrystal was found, it was rotated to align the fiber axis to the tilt axis by using the rotation-tilt holder (SRH holder, JEOL). Diffraction patterns then were recorded from the nearest portion of the microcrystal with tilt angles of 0, 40, and −40 degrees.
RESULTS AND DISCUSSION
Cellulose Is Parallel Up.
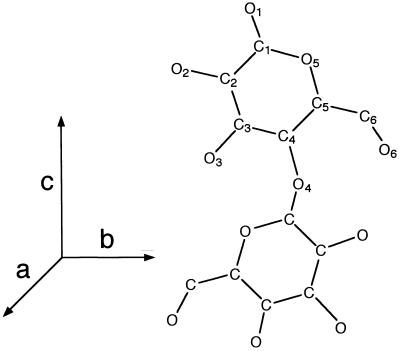
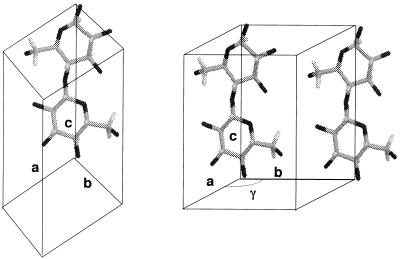
The directionality of a cellulose chain with respect to the crystallographic c axis used throughout this study is given in Fig. 1. The a, b, and c axes were chosen to satisfy the right-hand coordinate system (upper direction is positive for z coordinates), with setting the unique axis to c (for example, ref. 13). The chain sense is defined as parallel up, because the z coordinate of O5 is greater than that of C5. In this case, the reducing end of the chain is oriented in the same direction with respect to the c axis. Conversely, the reverse situation is defined as parallel down.
Figure 1.
The definition of the directionality of the cellobiose repeat unit with respect to the crystallographic axes.
Although recent molecular dynamics investigations favor the parallel-up arrangement (11), no direct experimental evidence has been obtained so far for either arrangement. If the directionality of the chains with respect to the unit cell of cellulose was firmly established, then diffraction experiments alone could directly provide the directionality of chains in a given microfibril.
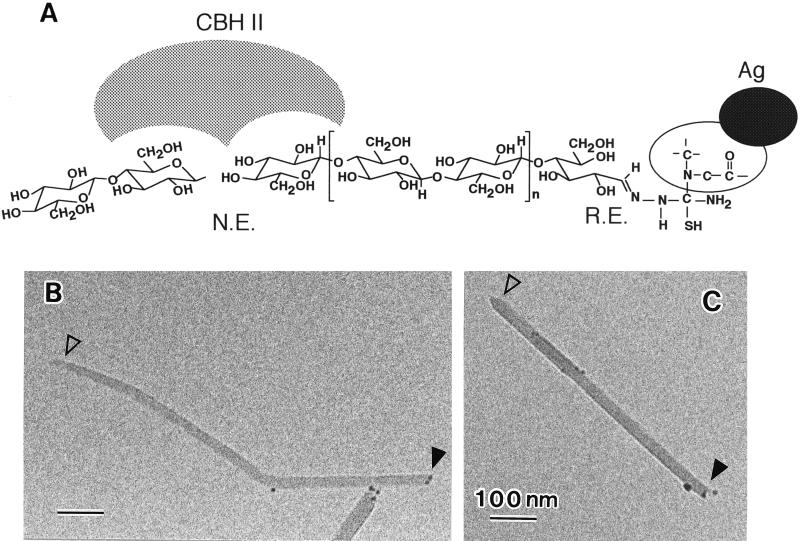
The reducing end of a cellulose microcrystal can be visualized by the silver-staining method. Also the nonreducing end can be visualized by selective degradation with cellobiohydrolase II, because this cellulase hydrolyzes cellulose chains from the nonreducing ends (14) and produces a unidirectional sharpening of the crystal tips (2). By combining these two methods, one can expect to find chemical labeling at one end and the pointed tip at the other end. The microcrystals were treated first with cellobiohydrolase II and then silver-labeled. Microcrystalline cellulose prepared by sulfuric acid treatment was less susceptible to the enzymatic digestion than that obtained by HCl treatment, presumably because of surface charges introduced by the former treatment. The results were, however, the same in both cases. Fig. 2 shows typical electron micrographs of the resulting microcrystals with one end labeled with silver (solid arrowheads) and the other end eroded to a pointed tip (open arrowheads) with the point roughly centered along the long axis of the crystal. No staining was observed when the crystals were first reduced by NaBH4 or when the thiosemicarbazide treatment was skipped.
Figure 2.
Evaluation of the chain directionality by microscopic visualization. (A) Schematic diagram of the mode of reaction used in this study. N.E., nonreducing end; R.E., reducing end. (B and C) Electron micrographs of typical microcrystals of cellulose from Cladophora. Arrowheads, see text.
Like those from Cladophora, microcrystals from Closterium sp. and Halocynthia sp. also were evaluated as a Iβ-type cellulose. Although staining was observed at only one end of the specimen in both cases, the staining efficiency was not as good as with Cladophora cellulose. For Closterium sp., lower labeling was attributed to the smaller lateral size of the microcrystals. The Halocynthia sample did not display the preferential orientation on the supporting carbon necessary to determine the directionality of the c axis. Thus we used annealed Cladophora microcrystals as a Iβ-type cellulose.
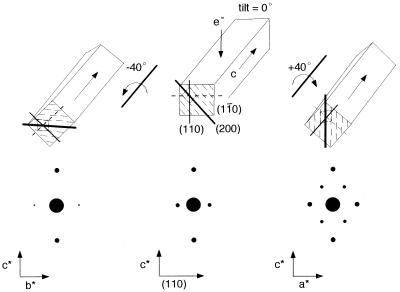
The directionality of the c axis can be defined by electron diffraction together with tilting experiments (15). Because one single microfibril is virtually a single crystal (16, 17), one can assign the c direction from elementary crystallographic considerations as schematically represented in Fig. 3. The directionality of the c axis of the triclinic and monoclinic unit cells is identical if the chains are laterally packed in the same manner. For simplicity, the indexing based on the monoclinic unit cell is used hereafter.‖ The highly crystalline microcrystals from Cladophora have a tendency to lie their 0.61 nm lattice plane (1i0) parallel to the film support for microscopy as in Fig. 3 (middle top). When the c axis points upward, the lattice organization in a microfibril is such that the clockwise rotation around the c axis would bring 0.39 nm (200) lattice plane in Bragg’s position and give the pattern in the bottom right. This situation is totally reversed when the c axis points downward, so as to bring the b*c* projection in Bragg’s position.
Figure 3.
Theoretical background to identify the directionality of c axis by diffraction. (Upper middle) The initial situation of the microcrystal without any tilt. (Lower middle) The corresponding diffraction pattern. The anti-clockwise/clockwise rotation around c axis gives the lower left/lower right diffraction diagrams, respectively.
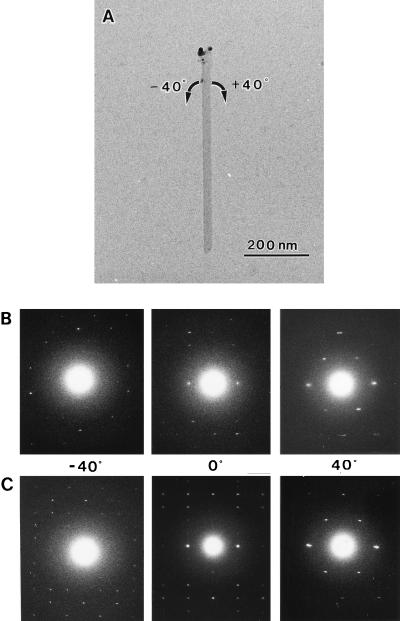
The directionality of reducing end and the c axis of the unit cell can be readily identified by combining the reducing end staining and tilting diffraction experiment. During this experiment, H2SO4-treated crystals were extensively used, because their high dispersion easily allowed us to observed isolated crystals. Typical examples are shown in Fig. 4. The microcrystal with its upper tip labeled by silver grains was rotated in both directions. The diffraction diagrams are in good accord with the situation discussed in Fig. 3, indicating that the c axis of this microcrystal points upward. Analysis of Iα-rich (initial) and Iβ-dominant (annealed) crystals (n > 10 for each type) showed that the reducing end of the chain has the same directionality as the c axis. The packing of the molecule in the unit cells is represented in Fig. 5. Such arrangement is in good accord with the previous parallel-up models for ramie Iβ cellulose (6).
Figure 4.
Two sets of tilt-diffraction patterns from the labeled microcrystal. (A) A microcrystal with its top end labeled with silver grains. (B) Patterns obtained from initial Cladophora. (C) Patterns obtained from annealed Cladophora.
Figure 5.
Schematic representation of mode of chain packing in the unit cell of cellulose. (A) Triclinic unit cell. (B) Monoclinic unit cell. Monoclinic angle γ is obtuse.
Directionality of Cellulose Synthesis in A. aceti.
Having established the parallel-up model of cellulose, the directionality of the chains in a given microfibril can be identified by electron microdiffraction with tilting experiments. Because the technique does not require the visible ends of the microfibrils nor any chemical labeling, the method allows the investigation of long microfibrils or, for instance, the nascent cellulose biosynthesized by an organism such as A. aceti. To avoid problems associated with the twist of the flat bacterial cellulose ribbons around their long axis, which may interfere with the tilting experiments, the diffraction diagrams were recorded specifically in the large flat regions of the ribbons laying parallel to the supporting film. The lattice orientation in these areas is always identical, because the ab plane of the unit cell is a centro-symmetric parallelogram, which therefore superimposes after a 180-degree rotation about the ribbon axis. All of successful tilting analyses (n > 10) gave identical results.
Fig. 6 shows a typical example of tilt-microdiffraction experiment of a nascent bacterial cellulose ribbon. The microdiffractogram taken without any tilt gave (110) equatorial spots, indicating that the bacterial cellulose ribbon behaves like the Cladophora cellulose microcrystals, with the 0.61-nm lattice planes parallel to the supporting film. A clockwise rotation around the molecular axis gave b*c* projection, opposite of that described in Fig. 4. Undoubtedly the result evidences that the cellulose is crystallized with its c axis outward the organisms, more importantly, Acetobacter synthesizes and polymerizes cellulose chains with their reducing ends pointing away from the synthesizing enzymes.
Figure 6.
A sets of tilt-diffraction patterns from the nascent microfibril of Acetobacter. (A) A microfibril ribbon spun out from the cell. Circled areas indicate where a set of diffraction patterns were taken. (B) The corresponding diffraction patterns.
Implications for the Catalytic Mechanism of Cellulose Synthase.
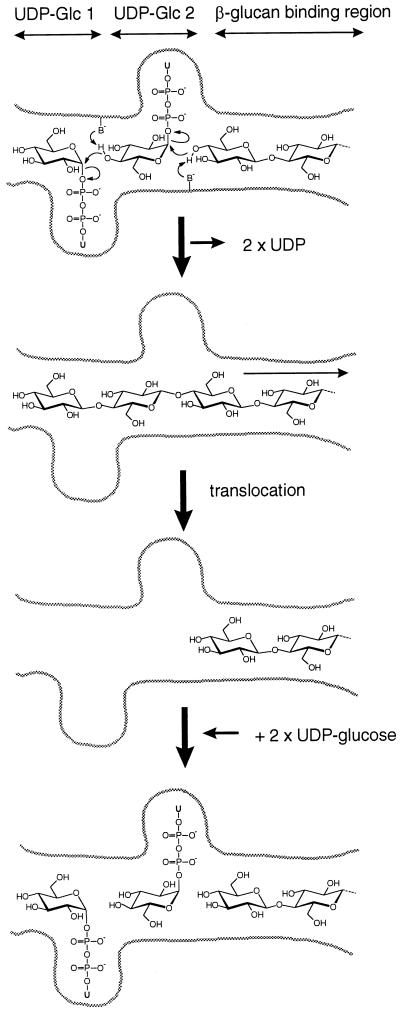
On the basis of sequence analysis of cellulose synthases and other β-glycosyltransferases, Saxena et al. (18) have suggested that cellulose biosynthesis takes place via a dual addition of two UDP-glucose molecules to the growing polymer chain. This mechanism provided a possible explanation for the synthesis of a polymer chain with a regular two-fold screw axis. However, this model is still speculative, and in particular the question of whether the putative dual addition takes place at the nonreducing or at the reducing end of the growing chain could not be solved without experimental evidence for the location of the polymerization site. With the present demonstration that the synthesizing enzymes operate at the nonreducing end of the growing chain, we propose a refined— albeit still speculative—model for the mechanism of cellulose synthase (Fig. 7).
Figure 7.
Model for the polymerization of cellulose chain by addition of sugar residues at the nonreducing end.
The active site of cellulose synthase would contain two UDP-glucose binding sites and a β-1,4-glucan binding region (Fig. 7). The dual addition of two monomer residues on a growing chain enables the synthesis of a chain with a 2-fold screw axis without the enzyme having to rotate by 180°C between consecutive monomer additions as proposed by Saxena et al. (18). Our model, however, is different in that the addition of the sugar residues occurs at the nonreducing end of the growing chain, whereas the model of Saxena et al. (18) displayed a possible addition of the sugar residues on the reducing end of the growing chain. In this respect, our model is similar to that proposed by Albersheim et al. (19), but does not involve transfer of the UDP-glucose to Ser or Thr residues of the glycosyltransferase. Instead, like Saxena et al. (18) we propose that the C-5 hydroxyl group of the acceptor glucose residues would be activated by general base catalysis provided by conserved Asp residues of the glycosyltransferase by a mechanism analogous to that observed during transglycosylation reactions catalyzed by glycosidases. An important aspect of the polymerization mechanism is the translocation of the growing cellulose chain away from the two UDP-glucose binding sites. UDP-glucose has a large aglycon in an α-configuration, and the two UDP-glucose binding sites are likely to base their recognition on the UDP part of the molecule. This is consistent with the fact that UDP is a known inhibitor of cellulose biosynthesis (20). On the other hand, once the glycosyl transfer has taken place, the glycosidic bonds are in the β configuration. The resulting β-1,4-glucan probably has little or no affinity for the UDP-glucose binding sites and would move into a region capable of more favorable interactions (labeled β-glucan binding region in Fig. 7). Two new UDP-glucose molecules then could fill the empty UDP-glucose binding sites, and the dual addition would proceed.
Because plant cellulose synthases are homologous to their Acetobacter counterpart (21), it is likely that plant cellulose biosynthesis also occurs by addition of sugar residues onto the nonreducing ends of growing cellulose chains. Further, Saxena et al. (18) have shown that Acetobacter cellulose synthase displays significant sequence similarities with several other processive β-glycosyltransferases, including chitin synthases, hyaluronan synthases, and N-acetylglucosamine-transferases, involved in the synthesis of the sugar backbone of lipo-oligosaccharides. All these glycosyltransferases therefore are likely to also share the same polymerization mechanism by addition of sugar residues on the nonreducing end of the growing chain.
Acknowledgments
We thank Dr. S. Yamanaka, Ajinomoto, Central Research Center, Kawasaki, Japan, for a gift of the strain of Acetobacter aceti and M. Schülein, Novo-Nordisk, Bagsvaerd, Denmark, for a gift of Humicola insolens CBH II. The help of Dr. James Campbell with the language is gratefully acknowledged. The study was supported by the Joint Research Program between the Japan Society for the Promotion of Science and Centre National de la Recherche Scientifique (J.S. and B.H.), and in part by the Grant-in-Aid for Scientific Research (No. 07456156 to J.S.). W.H. was a recipient of Japan Society for the Promotion of Science Postdoctoral Fellowship.
Footnotes
Throughout this paper, the indexing of the crystallographic planes refers to the monoclinic model of Sugiyama et al. (9), where the three major equatorial planes, assigned with d spacings of 0.60–61, 0.53–54, and 0.39–40 nm, respectively, are indexed as (11̄0), (110), and (200) with the monoclinic cell. The corresponding planes also can be indexed as (100)t, (010)t, and (110)t with the triclinic cell.
References
- 1.Hieta K, Kuga S, Usuda M. Biopolymers. 1984;23:1807–1810. [Google Scholar]
- 2.Chanzy H, Henrissat B. FEBS Lett. 1985;184:285–288. [Google Scholar]
- 3.Kuga S, Brown R M., Jr Carbohydr Res. 1988;180:345–350. [Google Scholar]
- 4.Gardner K H, Blackwell J. Biopolymers. 1974;13:1975–2001. doi: 10.1002/bip.1975.360140804. [DOI] [PubMed] [Google Scholar]
- 5.Sarko A, Muggli R. Macromolecules. 1974;7:486–494. [Google Scholar]
- 6.Woodcock C, Sarko A. Macromolecules. 1980;13:1183–1187. [Google Scholar]
- 7.Attala R H, VandelHart D L. Science. 1984;223:283–285. doi: 10.1126/science.223.4633.283. [DOI] [PubMed] [Google Scholar]
- 8.VandelHart D L, Attala R H. Macromolecules. 1984;17:1465–1472. [Google Scholar]
- 9.Sugiyama J, Vuong R, Chanzy H. Macromolecules. 1991;24:4168–4175. [Google Scholar]
- 10.French A D. Carbohydr Res. 1978;61:67–80. [Google Scholar]
- 11.Kroon-Batenburg L M J, Bouma B, Kroon J. Macromolecules. 1996;29:5695–5699. [Google Scholar]
- 12.Yamamoto H, Horii F, Odani H. Macromolecules. 1989;22:4130–4132. [Google Scholar]
- 13.Tadokoro H. Structure of Crystalline Polymers. New York: Wiley; 1979. pp. 25–35. [Google Scholar]
- 14.Vrsankà M, Biely P. Carbohydr Res. 1992;227:19–27. [Google Scholar]
- 15.Revol J-F, Goring D A I. Polymer. 1983;24:1547–1550. [Google Scholar]
- 16.Revol J-F. Carbohydr Polym. 1982;2:123–134. [Google Scholar]
- 17.Sugiyama J, Harada H, Fujiyoshi Y, Uyeda N. Planta. 1985;166:161–167. doi: 10.1007/BF00397343. [DOI] [PubMed] [Google Scholar]
- 18.Saxena I M, Brown R M, Jr, Fevre M, Geremia R A, Henrissat B. J Bacteriol. 1995;177:1419–1424. doi: 10.1128/jb.177.6.1419-1424.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Albersheim P, Darvill A, Roberts K, Staelin L A, Varner J E. Plant Physiol. 1997;113:1–3. doi: 10.1104/pp.113.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ross P, Mayer R, Benziman M. Microbiol Rev. 1991;55:35–58. doi: 10.1128/mr.55.1.35-58.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pear J R, Kawagoe Y, Schreckengost W E, Delmer D P, Stalker D M. Proc Natl Acad Sci USA. 1996;93:12637–12642. doi: 10.1073/pnas.93.22.12637. [DOI] [PMC free article] [PubMed] [Google Scholar]